An Ultrafast Scalable Many-core Motif Discovery Algorithm for Multiple GPUs
|
|
|
- Owen McCormick
- 6 years ago
- Views:
Transcription
1 2011 IEEE International Parallel & Distributed Processing Symposium An Ultrafast Scalable Many-core Motif Discovery Algorithm for Multiple GPUs Yongchao Liu, Bertil Schmidt, Douglas L. Maskell School of Computer Engineering Nanyang Technological University Singapore {liuy0039, asbschmidt, Abstract The identification of genome-wide transcription factor binding sites is a fundamental and crucial problem to fully understand the transcriptional regulatory processes. However, the high computational cost of many motif discovery algorithms heavily constraints their application for large-scale datasets. The rapid growth of genomic sequences and gene transcription data further deteriorates the situation and establishes a strong requirement for time-efficient scalable motif discovery algorithms. The emergence of many-core architectures, typically CUDA-enabled GPUs, provides an opportunity to reduce the execution time by an order of magnitude without the loss of accuracy. In this paper, we present mcuda-meme, an ultrafast scalable many-core motif discovery algorithm for multiple GPUs based on the MEME algorithm. Our algorithm is implemented using a hybrid combination of the CUDA, OpenMP and MPI parallel programming models in order to harness the powerful compute capability of modern GPU clusters. At present, our algorithm supports OOPS and ZOOPS models, which are sufficient for most motif discovery applications. mcuda- MEME achieves significant speedups for the starting point search stage (and the overall execution) when benchmarked, using real datasets, against parallel MEME running on 32 CPU cores. Speedups of up to 1.4 (1.1) on a single GPU of a Fermi-based Tesla S2050 quad-gpu computing system and up to 10.8 (8.3) on the eight GPUs of a two Tesla S2050 system were observed. Furthermore, our algorithm shows good scalability with respect to dataset size and the number of GPUs (availability: uda-meme). Motif discovery; MEME; CUDA; MPI; OpenMP; GPU I. INTRODUCTION De novo motif discovery is crucial to the complete understanding of transcription regulatory processes by identifying transcription factor binding sites (TFBSs) on a genome-wide scale. Algorithmic approaches for motif discovery can be classified into two categories: iterative and combinatorial. Iterative approaches generally exploit probabilistic matching models, such as Expectation Maximization (EM) [1] and Gibbs sampling [2]. These approaches are often preferred because they use positionspecific scoring matrices to describe the matching between a motif instance and a sequence, instead of a simple Hamming distance. Combinatorial approaches employ deterministic matching models, such as word enumeration [3] and dictionary methods [4]. MEME [5] is a popular and well established motif discovery algorithm, which is primarily comprised of the starting point search (SPS) stage and the EM stage. However, the high computational cost of MEME constrains its application for large-scale datasets, such as motif identification in whole peak regions from ChIP-Seq datasets for transcription factor binding experiments [6]. This encourages the use of high-performance computing solutions to meet the execution time requirement. Several attempts have been made to improve execution speed on conventional computing systems, including distributed-memory workstation clusters [7] and special-purpose hardware [8]. The emerging many-core architectures, typically Compute Unified Device Architecture (CUDA)-enabled GPUs [9], have demonstrated their power for accelerating bioinformatics algorithms [10] [11] [12] [13]. This convinces us to employ CUDA-enabled GPUs to accelerate motif discovery. Previously we have presented CUDA-MEME, based on MEME version 3.5.4, for a single GPU device which accelerates motif discovery using two parallelization approaches, namely: sequence-level parallelization and substring-level parallelization. The detailed implementations of the two approaches have been described in [14]. However, the increasing size and availability of ChIP- Seq datasets established the need for parallel motif discovery with even higher performance. Therefore in this paper, we present mcuda-meme, an ultrafast scalable many-core motif discovery algorithm for multiple GPUs. Our algorithm is designed based on MEME version which incorporates position-specific priors (PSP) to improve accuracy [15]. In order to harness the powerful compute capability of GPU clusters, we employ a hybrid combination of CUDA, Open Multi-Processing (OpenMP) and Message Passing Interface (MPI) parallel programming models to implement this algorithm. Compared to CUDA-MEME, mcuda-meme introduces four new significant features: Supports multiple GPUs in a single host or a GPU cluster through a MPI-based design; Supports starting point search from the reverse complements of DNA sequences; Employs multi-threaded design using OpenMP to accelerate the EM stage; Incorporates PSP prior probabilities to improve accuracy /11 $ IEEE DOI /IPDPS
2 Since the sequence-level parallelization is advantageous to the substring-level parallelization in execution speed [14], our following discussions only refer to the sequence-level parallelization. Real datasets are employed to evaluate the performance of mcuda-meme and parallel MEME (i.e. the MPI version of MEME distributed with the MEME software package). Compared to parallel MEME (version 4.4.0) running on 32 CPU cores, mcuda-meme achieves a speedup of the starting point search stage (and the overall execution) of up to 1.4 (1.1) times on a single GPU of a Fermi-based Tesla S2050 quad-gpu computing system and up to 10.8 (8.3) times on eight GPUs of a two Tesla S2050 system. Furthermore, our algorithm shows good scalability with respect to dataset size and the number of GPUs. The rest of this paper is organized as follows. Section 2 briefly introduces the MEME algorithm and the CUDA, OpenMP and MPI parallel programming models. Section 3 details the new features of mcuda-meme. Section 4 evaluates the performance using real datasets, and Section 5 concludes this paper. II. BACKGROUND A. The MEME Algorithm Given a set of protein or DNA sequences, MEME attempts to search for statistically significant (unknown) motif occurrences, which are believed to be shared in the sequences, by optimizing the parameters of statistical motif models using the EM approach. MEME provides support for three types of search modes: one occurrence per sequence (OOPS), zero or one occurrence per sequence (ZOOPS), and two component mixture (TCM). The OOPS model postulates that there is exactly one motif occurrence per sequence in the dataset, the ZOOPS model postulates zero or one motif occurrence per sequence, and the TCM model postulates that there can be any number of non-overlapping occurrences per sequence [5]. Since the OOPS and ZOOPS models are sufficient for most motif finding applications, our algorithm in this paper concentrates on these two models. MEME begins a motif search with the creation of a set of motif models. Each motif model θ is a position specific probability matrix representing frequency estimates of letters occurring in different positions. Given a motif of width w defined over an alphabet Σ = {A 1, A 2,..., A Σ }, each value θ(i, j) (1 i Σ and 0 j w) of the matrix is defined as: probability of Ai at position j of the motif, 1 j w θ (, i j) = (1) probability of A not in the motif, j = 0 i A starting point is an initial motif model θ (0) from which the EM stage runs for a fixed number of iterations, or until convergence, to find the final motif model θ (q) with maximal posterior probability. To find the set of starting points for a given motif width, MEME converts each substring in a sequence dataset into a potential motif model and then determines its statistical significance by calculating the weighted log likelihood ratio [15] on different numbers of predicted sites, subject to the model used. The potential motif models with the highest weighted log likelihood ratio are selected as starting points for the successive EM algorithm. When performing the starting point search, independent computation from each substring of length w in the dataset S={S 1, S 2,..., S n } of n input sequences is conducted to determine a set of initial motif models. The following notations are used for the convenience of discussion: l i denotes the length of S i, S i denotes the reverse complement of S i, S i,j denotes the substring of length w starting from position j of S i, S i (j) denotes the j-th letter of S i, where 1 i n and 0 j l i -w. The starting point search process is primarily comprised of three steps for the OOPS and ZOOPS models: Calculate the probability score P(S i,j, S k,l ) from the forward strand (or P(S i,j, S k,l) from the reverse complement), which is the probability that a site starts at position l in S k when a site starts at position j in S i. The time complexity is O(l i l k ) for each sequence pair S i and S k. Identify the highest-scoring substring S k,maxk (as well as its strand orientation) for each S k. The time complexity is O(l k ) for each sequence S k. Sort the n highest-scoring substrings {S k,maxk } in decreasing order of scores and determine the potential starting points. The time complexity is O(nlogn) for OOPS and O(n 2 w) for ZOOPS. The probability score P(S i,j, S k,l ) is computed as: w-1 i, j k, l i k p= 0 PS (, S ) = mats [ ( j+ p)][ S( l+ p)] (2) where mat denotes the letter frequency matrix of size Σ Σ. To reduce computation redundancy, (2) can be further simplified to (3), where the computation of the probability scores {P(S i,j, S k,l )} in the j-th iteration depends on the resulting scores {P(S i,j-1, S k,l-1 )} in the (j-1)-th iteration. However, P(S i,j, S k,0 ) needs to be computed individually using (2). PS ( i, j, Sk, l) = PS ( i, j 1, Sk, l 1) + mats [ i( j+ w 1)][ Sk( l+ w 1)] - mat[ S ( j 1)][ S ( l 1)] i B. The CUDA Programming Model More than a software and hardware co-processing architecture, CUDA is also a parallel programming language extending general programming languages, such as C, C++ and Fortran with a minimalist set of abstractions for expressing parallelism. CUDA enables users to write parallel scalable programs for CUDA-enabled processors with familiar languages [16]. A CUDA program is comprised of two parts: a host program running one or more sequential threads on a host CPU, and one or more parallel kernels able to execute on Tesla [17] and Fermi [18] unified graphics and computing architectures. k (3)
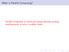
3 A kernel is a sequential program launched on a set of lightweight concurrent threads. The parallel threads are organized into a grid of thread blocks, where all threads in a thread block can synchronize through barriers and communicate via a high-speed, per block shared memory (PBSM). This hierarchical organization of threads enables thread blocks to implement coarse-grained task and data parallelism and lightweight threads, comprising a thread block, to provide fine-grained thread-level parallelism. Threads from different thread blocks in the same grid are able to cooperate through atomic operations on global memory shared by all threads. The CUDA-enabled processors are built around a fully programmable scalable processor array, organized into a number of streaming multiprocessors (SMs). For the Tesla architecture, each SM contains 8 scalar processors (SPs) and shares a fixed 16 KB of PBSM. For the Tesla-based GPU series, the number of SMs per device varies from generation to generation. For the Fermi architecture, it contains 16 SMs with each SM having 32 SPs. Each SM in the Fermi architecture has a configurable PBSM size from the 64 KB on-chip memory. This on-chip memory can be configured as 48 KB of PBSM with 16 KB of L1 cache or as 16 KB of PBSM with 48 KB of L1 cache. When executing a thread block, both architectures split all the threads in the thread block into small groups of 32 parallel threads, called warps, which are scheduled in a single instruction, multiple thread fashion. Divergence of execution paths is allowed for threads in a warp, but SMs realize full efficiency and performance when all threads of a warp take the same execution path. C. The OpenMP Programming Model OpenMP is a compiler-directive-based application program interface for explicitly directing multi-threaded, shared-memory parallelism [19]. This explicit programming model enables programmers to have a full control over parallelism. OpenMP is primarily comprised of compiler directives, runtime library routines and environment variables that are specified for C/C++ and Fortran programming languages. As an open specification and standard for expressing explicit parallelism, OpenMP is portable across a variety of shared memory architectures and platforms. The basic idea behind OpenMP is the existence of multiple threads in the shared memory programming paradigm which enables data-shared parallel execution. In OpenMP, the unit of work is a thread and the fork-join model is used for parallel execution. An OpenMP program begins as the master thread, and executes sequentially until encountering a parallel region construct specified by the programmer. Successively, the master thread creates a team of parallel threads to execute in parallel the statements that are enclosed by the parallel region construct. Having completed the statements in the parallel region construct, the team threads synchronize and terminate, leaving only the master thread. D. The MPI Programming Model MPI is a de facto standard for developing portable parallel applications using the message passing mechanism [20]. MPI works on both shared and distributed memory architectures and platforms, offering a highly portable solution to parallel programming on a variety of hardware topologies. In MPI, it defines each worker as a process and enables the processes to execute different programs. This multiple program, multiple data model offers more flexibility for data-shared or data-distributed parallel program design. Within a computation, processes communicate data by calling runtime library routines, specified for the C/C++ and Fortran programming languages, including point-to-point and collective communication routines. Point-to-point communication is used to send and receive messages between two named processes, suitable for local and unstructured communications. Collective (global) communication is used to perform commonly used global operations (e.g. reduction and broadcast operations). III. METHODS mcuda-meme employs a hybrid CPU+GPU computing framework for each of its MPI processes in order to maximize performance by overlapping the CPU and GPU computation. A MPI process therefore consists of two threads: a CPU thread and a GPU thread. mcuda-meme requires a non-overlapping one-to-one correspondence between a MPI process and a GPU device. Unfortunately, the MPI runtime environment does not provide a mechanism to perform the automatic mapping between MPI processes and GPU devices. In this case, mcuda-meme employs a registration-based management mechanism (as shown in Fig. 1) to dynamically map GPU devices to MPI processes. WORKER 1 WORKER 2 ASSIGN 1 5 GPU ID 5 SAVE HOST DATA 2 HOST DATABASE MASTER VALIDITY 4 1 REGISTER VERIFY 3 VERIFICATION GPU MAPPING MANAGEMENT Figure 1. Registration-based GPU mapping management diagram
4 Firstly, a master process is specified and each worker process (including the master process) registers the name of the host machine in which it resides, as well as the number of qualified GPUs in the host, to the master process. Secondly, the master process iterates each registered host and checks the number of processes distributed to it and the number of qualified GPUs it has. The registration is acceptable if each registered host does not have more processes than qualified GPUs, and is unacceptable, otherwise. If the mapping between processes and GPUs is valid, the master process enumerates each host and assigns a unique GPU identifier to each process running in the host. For the starting point search, most execution time is spent on the computation of probability scores to select the highest-scoring substring S k,maxk for each S k. mcuda- MEME selects the highest-scoring substrings while computing the scores from the forward or reverse strands on GPUs. When using the reverse complements, mcuda- MEME calculates the probability scores {P(S i,j, S k,l )} and {P(S i,j, S k,l)} simultaneously and selects the highest-scoring substring in a single CUDA kernel launch. This is different to MEME which performs two iterations of probability score calculation separately from the two strands of S k. Fig. 2 shows the CUDA kernel pseudocode. Hence, when searching from two strands, MEME will (nearly) double the execution time compared with that from a single strand, whereas mcuda-meme is able to save some execution time by reusing common resources (e.g., the reference substring S i,j is fetched only once from texture memory instead of twice). Our benchmarking shows that when searching from two strands, the execution time of mcuda- MEME only increases by about 60% compared to MEME. The sequence-level parallelization uses multiple iterations to complete the starting point search [14]. Generally, the execution time of all iterations heavily depends on the grid size (dimgrid) and the thread block size (dimblock) used in each iteration. When determining the grid ***************************************************************** Define S i to denote the reverse complement of S i. ***************************************************************** Step 1: Load letter frequency matrix to shared memory from constant memory; Step 2: Get the sequences information; Step 3: All threads calculate the probability scores{p(s i,0, S k,l)} and {P(S i,0, S k,l)} in parallel, and select and save the highest-scoring substring S k,maxk for S i,0, as well as its strand orientation. Step 4: for j from 1 to l i w All threads calculate the probability scores {P(S i,j, S k,l)} and {P(S i,j, S k,l)} in parallel using {P(S i,j-1, S k,l-1)} and {P(S i,j-1, S k,l-1)}, and select and save the highest-scoring substring S k,maxk for S i,j, as well as its strand orientation. end Figure 2. CUDA kernel pseudocode for probability score calculation from two strands. size, we consider the maximum number of resident threads per SM. As in [14], we assume the maximum number of resident threads per SM is 1024, even though Fermi-based GPUs support a maximum number of 1536 resident threads per multiprocessor. Thus, mcuda-meme calculates the grid size per iteration as 1024 dimgrid = 2 #SM (4) dimblock where #SM is the number of SMs in a single GPU device and dimblock is set to 64. For the sequence-level parallelization, the GPU device memory consumption only depends on the grid size and the maximum sequence length. The consumed device memory size can be estimated as memsize = 32 dimgrid max{ l }, 1 i n (5) From (5), we can see that the device memory consumption is directly proportional to the maximum sequence length for a specific GPU device. Thus, mcuda-meme sets a maximum sequence length threshold (by default 64K) to determine the use of the sequence-level parallelization or the substring-level parallelization that is slower but much more memory efficient. When employing multiple GPUs, an appropriate load balancing policy is critical for high overall performance. In this paper, we assume that mcuda-meme is deployed in a homogenous GPU cluster, which is often the case. Since the workload is aware of S, our assumption simplifies the workload distribution between GPUs. In mcuda-meme, both the SPS stage and the EM stages are parallelized using MPI. For the SPS stage, to achieved good workload balance, we calculate the total number of bases in the input sequences, and (nearly) equally distribute these bases to all processes using a sequence as a unit, because a thread block in the CUDA kernel processes a sequence pair every time [14]. This makes each process hold (nearly) equal number of bases in (likely) different number of sequences distributed to it, thus having (nearly) equal number of substrings for a specific motif width. In this case, each MPI process holds a non-overlapping sequence subset of S, where the process converts each substring of this subset into a potential motif model and calculates the highest-scoring substrings of all sequences in S for this substring by invoking the CUDA kernels. After each process obtains the best starting points in its subset, a reduction operation is conducted across all processes to compute the new best starting points. For the EM stage, as we are aware of the number of initial models, we simply divide the set of initial models equally among all processes as in [7]. Unfortunately, because the execution time of the EM algorithm from each initial model is not equal and is dependent on the real data, we might not be able to balance the workloads perfectly. However, considering that the EM stage takes only a very small portion of the total execution time, the imperfect workload balancing does not impact significantly. After obtaining the best motif model for each MPI process from its subset of initial models, all i
5 processes conduct a reduction operation to get the final motif model. Since all processes only need to communicate to collect the final results in the SPS and EM stages, the communication overhead can be neglected. When executing mcuda-meme with only one MPI process, the sequential EM stage limits the overall speedup as per Amdahl's law, after accelerating the SPS stage. As multi-core CPUs have become commonplace, we employ a multi-threaded design using OpenMP to parallelize the EM stage to exploit the compute power of multi-core CPUs. Since the number of threads used can be specified using OpenMP runtime routines, the multi-threaded design is well exploited when running multiple MPI processes. As mentioned above, each MPI process consists of a CPU thread and a GPU thread. We generally recommend that each MPI process runs the two threads on two CPU cores to maximize the performance. In this case, after completing the SPS stage, the one CPU core for the GPU thread of each MPI process will be wasted if the successive EM stage does not use it. Hence, it is viable to employ two threads to parallelize the EM algorithm within each MPI process. By default, mcuda-meme uses as many threads as the available CPU cores in a single machine when running our algorithm with only one MPI process, and two threads when using multiple MPI processes. Note that the number of threads used can also be specified by parameters. As mentioned above, the accuracy of MEME has been improved by incorporating PSP, as described in [15], which defines prior probabilities that a site starts at each position of each input sequence. The PSP approach enables the incorporation of multiple types of additional information into motif discovery and converts the additional information into the measure of the likelihood that a motif starts at each position of each input sequence. When incorporating PSP, for the SPS stage, the highest-scoring substring of each sequence S k is determined using scores calculated from P(S i,j, S k,l ) (or P(S i,j, S k,l)) and PSP prior probabilities, and for the EM algorithm, it substitutes the uniform assumption with the PSP prior probabilities. Details about the PSP approach in MEME can be obtained from [15]. Since mcuda-meme is implemented based on MEME 4.4.0, the PSP approach is also incorporated in our algorithm. IV. PERFORMANCE EVALUATION We use the peaks identified by MICSA [6] in the ChIP- Seq data for neuron-restrictive silencer factor (the peaks are available at to evaluate the performance of mcuda-meme. To evaluate the scalability of our algorithm with respect to dataset size, three subsets of peaks with different numbers of sequences and base pairs (bps) are selected from the whole peaks (as shown in Table 1). TABLE I. PEAK DATASETS FOR PERFORMANCE EVALUATION Datasets No. of Sequences Min (bps) Max (bps) Total (bps) NRSF NRSF NRSF All the following tests are conducted on a computing cluster with eight compute nodes that are connected by a high-speed Infiniband switch. Each compute node consists of an AMD Opteron 2378 quad-core 2.4 GHz processor and 8 GB RAM, running the Linux OS with the MPICH2 library. Furthermore, two Fermi-based Tesla S2050 quad-gpu computing systems are installed and connected to four nodes of the cluster, with each node having access to two GPUs. A single GPU of a Tesla S2050 consists of 14 SMs (a total of 448 SPs) with a core frequency of 1.15GHz and has 2.6 GB user available device memory. The common parameters used for all tests are "-mod zoops -revcomp", and the other parameters use the default values. We have evaluated the performance of mcuda-meme running on a single GPU and on eight GPUs respectively, compared to parallel MEME (version 4.4.0) running on the eight compute nodes with a total of 32 CPU cores (as shown in Table 2). From Table 2, the mcuda-meme speedups, either on a single GPU or eight GPUs, increase as the dataset size grows, suggesting a good scalability for dealing with large-scale datasets. On eight GPUs, mcuda-meme significantly outperforms parallel MEME on 32 CPU cores for all datasets, where it achieves an average speedup of 8.8 (6.5) with a highest of 10.8 (8.3) for the SPS stage alone (and for the overall execution). Even on a single GPU, our algorithm still holds its own compared to parallel MEME on 32 CPU cores, with a highest speedup of 1.4 (1.1) for the SPS stage (and for the overall execution) for the large dataset, even though it only gives a speedup of 0.9 (0.7) for NRSF500 dataset. In addition to the scalability with respect to dataset scale, we further evaluated the scalability of our algorithm with respect to the number of GPUs. Figures 3 and 4 show the speedups of mcuda-meme on different numbers of GPUs compared to parallel MEME on 32 CPU cores for all datasets. From the figures, we can see that for each dataset, the speedup increases (nearly) linearly as the number of GPUs increases both for the SPS stage and for the overall execution. The above evaluation and discussions demonstrate the power of mcuda-meme to accelerate motif discovery for large-scale datasets with relatively inexpensive GPU TABLE II. EXECUTION TIME (IN SECONDS) AND SPEEDUP COMPARISON BETWEEN MCUDA-MEME AND PARALLEL MEME mcuda-meme parallel MEME Speedups Datasets 8 GPUs 1 GPU 32 CPU cores 8 GPUs 1 GPU SPS All SPS All SPS All SPS All SPS All NRSF NRSF NRSF
6 Speedup NRSF500 NRSF1000 NRSF2000 However, our current load balancing policy might not be able to work well for heterogeneous GPU clusters. Hence, designing a robust and appropriate load balancing policies for heterogeneous GPUs clusters is part of our future work. The source code of mcuda-meme and the benchmark datasets are available for download at No. of GPUs Figure 3. The speedups for the SPS stage of mcuda-meme on different numbers of GPUs. Speedup NRSF500 NRSF1000 NRSF No. of GPUs Figure 4. The speedups for the overall execution of mcuda-meme on different numbers of GPUs. clusters. The genome-wide identification of TFBSs has been heavily constrained by the long execution time of motif discovery algorithms. Thus, we believe biologists will greatly benefit from the speedup of our algorithm. V. CONCLUSION In this paper, we have presented mcuda-meme, an ultrafast scalable many-core motif discovery algorithm for multiple GPUs. The use of hybrid CUDA, OpenMP and MPI programming models enables our algorithm to harness the powerful compute capability of GPU clusters. mcuda- MEME achieves significant speedups for both the starting point search stage (and the overall execution) when benchmarked, using real datasets, against parallel MEME running on 32 CPU cores. Speedups of up to 1.4 (1.1) on a single GPU of a Fermi-based Tesla S2050 quad-gpu computing system and up to 10.8 (8.3) on the eight GPUs of a two Tesla S2050 system were observed. Furthermore, our algorithm shows good scalability with respect to dataset size and the number of GPUs. Since the long execution time of motif discovery algorithms has heavily constrained the genome-wide TFBS identification, we believe that biologists will benefit from the high-speed motif discovery of our algorithm on relatively inexpensive GPU clusters. As mentioned above, the load balancing policy of mcuda- MEME is targeted towards homogenous GPU clusters, distributing workload symmetrically between GPUs. REFERENCES [1] C. E. Lawrence, and A. A. Reilly, An expectation maximization (EM) algorithm for the identification and characterization of common sites in unaligned biopolymer sequences, Proteins, vol. 7, no. 1, 1990, pp [2] C. E. Lawrence, S. F. Altschul, M. S. Boguski, J. S. Liu, A. F. Neuwald, and J. C. Wootton, Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment, Science, vol. 262, 1993, pp [3] P. Sumazin, G. Chen, N. Hata, A. D. Smith, T. Zhang, and M. Q. Zhang, DWE: discriminating word enumerator, Bioinformatics, vol. 21, no. 1, 2005, pp [4] C. Sabatti, L. Rohlin, K. Lange, and J. C. Liao, Vocabulon: a dictionary model approach for reconstruction and localization of transcription factor binding sites, Bioinformatics, vol. 21, no. 7, 2005, pp [5] T. L. Bailey, and C. Elkan, Fitting a mixture model by expectation maximization to discover motifs in biopolymers, Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, 1994, pp [6] V. Boeva, D. Surdez, N. Guillon, F. Tirode, A. P. Fejes, O. Delattre, and E. Barillot, De novo motif identification improves the accuracy of predicting transcription factor binding sites in ChIP-Seq data analysis, Nucleic Acids Research, vol. 38, no.11, 2010, pp. e126 [7] W. N. Grundy., T. L. Bailey, and C. P. Elkan, ParaMEME: a parallel implementation and a web interface for a DNA and protein motif discovery tool, Comput. Appl. Biosci., vol. 12, no. 4, 1996, pp [8] G. K. Sandve, M. Nedland, Ø. B. Syrstad, L. A. Eidsheim, O. Abul, and F. Drabløs, Accelerating motif discovery: Motif matching on parallel hardware, LNCS, vol. 4175, 2006, pp [9] NVIDIA CUDA C programming guide version 3.2, /CUDA_C_Programming_Guide.pdf. [10] Y. Liu, D. L. Maskell, and B. Schmidt, CUDASW++: optimizing Smith-Waterman sequence database searches for CUDA-enabled graphics processing units, BMC Research Notes, vol. 2, no. 73, 2009 [11] Y. Liu, B. Schmidt, and D. L. Maskell, CUDASW++2.0: enhanced Smith-Waterman protein database search on CUDA-enabled GPUs based on SIMT and virtualized SIMD abstractions, BMC Research Notes, vol. 3, no. 93, 2010 [12] Y. Liu, B. Schmidt, and D. L. Maskell, MSA-CUDA: Multiple Sequence Alignment on Graphics Processing Units with CUDA, 20th IEEE International Conference on Application-specific Systems, Architectures and Processors, 2009, pp [13] H. Shi, B. Schmidt, W. Liu, and W. Müller-Wittig, A parallel algorithm for error correction in high-throughput short-read data on CUDA-enabled graphics hardware, J Comput Biol., vol. 17, no. 4, 2010, pp [14] Y. Liu, B. Schmidt, W. Liu, and D. L. Maskell, CUDA-MEME: accelerating motif discovery in biological sequences using CUDAenabled graphics processing units, Pattern Recognition Letters, vol. 31, no. 14, 2010, pp [15] T. L. Bailey, M. Bodén, T. Whitington, and P. Machanick, The value of position-specific priors in motif discovery using MEME, BMC Bioinformatics, vol. 11, no. 179,
7 [16] J. Nickolls, I. Buck, M. Garland, and K. Skadron, Scalable parallel programming with CUDA, ACM Queue, vol. 6, no. 2, 2008, pp [17] E. Lindholm, J. Nickolls, S. Oberman, and J. Montrym, NVIDIA Tesla: a unified graphics and computing architecture, IEEE Micro., vol. 28, no. 2, 2008, pp [18] Fermi: NVIDIA s next generation cuda compute architecture, ermi_compute_architecture_whitepaper.pdf [19] OpenMP tutorial, [20] MPI tutorial,
GPU-MEME: Using Graphics Hardware to Accelerate Motif Finding in DNA Sequences
 GPU-MEME: Using Graphics Hardware to Accelerate Motif Finding in DNA Sequences Chen Chen, Bertil Schmidt, Liu Weiguo, and Wolfgang Müller-Wittig School of Computer Engineering, Nanyang Technological University,
GPU-MEME: Using Graphics Hardware to Accelerate Motif Finding in DNA Sequences Chen Chen, Bertil Schmidt, Liu Weiguo, and Wolfgang Müller-Wittig School of Computer Engineering, Nanyang Technological University,
Gene regulation. DNA is merely the blueprint Shared spatially (among all tissues) and temporally But cells manage to differentiate
 Gene regulation DNA is merely the blueprint Shared spatially (among all tissues) and temporally But cells manage to differentiate Especially but not only during developmental stage And cells respond to
Gene regulation DNA is merely the blueprint Shared spatially (among all tissues) and temporally But cells manage to differentiate Especially but not only during developmental stage And cells respond to
PROJECTION Algorithm for Motif Finding on GPUs
 PROJECTION Algorithm for Motif Finding on GPUs Jhoirene B. Clemente, Francis George C. Cabarle, and Henry N. Adorna Algorithms & Complexity Lab Department of Computer Science University of the Philippines
PROJECTION Algorithm for Motif Finding on GPUs Jhoirene B. Clemente, Francis George C. Cabarle, and Henry N. Adorna Algorithms & Complexity Lab Department of Computer Science University of the Philippines
A GPU Algorithm for Comparing Nucleotide Histograms
 A GPU Algorithm for Comparing Nucleotide Histograms Adrienne Breland Harpreet Singh Omid Tutakhil Mike Needham Dickson Luong Grant Hennig Roger Hoang Torborn Loken Sergiu M. Dascalu Frederick C. Harris,
A GPU Algorithm for Comparing Nucleotide Histograms Adrienne Breland Harpreet Singh Omid Tutakhil Mike Needham Dickson Luong Grant Hennig Roger Hoang Torborn Loken Sergiu M. Dascalu Frederick C. Harris,
Portland State University ECE 588/688. Graphics Processors
 Portland State University ECE 588/688 Graphics Processors Copyright by Alaa Alameldeen 2018 Why Graphics Processors? Graphics programs have different characteristics from general purpose programs Highly
Portland State University ECE 588/688 Graphics Processors Copyright by Alaa Alameldeen 2018 Why Graphics Processors? Graphics programs have different characteristics from general purpose programs Highly
Harnessing Associative Computing for Sequence Alignment with Parallel Accelerators
 Harnessing Associative Computing for Sequence Alignment with Parallel Accelerators Shannon I. Steinfadt Doctoral Research Showcase III Room 17 A / B 4:00-4:15 International Conference for High Performance
Harnessing Associative Computing for Sequence Alignment with Parallel Accelerators Shannon I. Steinfadt Doctoral Research Showcase III Room 17 A / B 4:00-4:15 International Conference for High Performance
Fast Sequence Alignment Method Using CUDA-enabled GPU
 Fast Sequence Alignment Method Using CUDA-enabled GPU Yeim-Kuan Chang Department of Computer Science and Information Engineering National Cheng Kung University Tainan, Taiwan ykchang@mail.ncku.edu.tw De-Yu
Fast Sequence Alignment Method Using CUDA-enabled GPU Yeim-Kuan Chang Department of Computer Science and Information Engineering National Cheng Kung University Tainan, Taiwan ykchang@mail.ncku.edu.tw De-Yu
Tesla Architecture, CUDA and Optimization Strategies
 Tesla Architecture, CUDA and Optimization Strategies Lan Shi, Li Yi & Liyuan Zhang Hauptseminar: Multicore Architectures and Programming Page 1 Outline Tesla Architecture & CUDA CUDA Programming Optimization
Tesla Architecture, CUDA and Optimization Strategies Lan Shi, Li Yi & Liyuan Zhang Hauptseminar: Multicore Architectures and Programming Page 1 Outline Tesla Architecture & CUDA CUDA Programming Optimization
GPU 3 Smith-Waterman
 129 211 11 GPU 3 Smith-Waterman Saori SUDO 1 GPU Graphic Processing Unit GPU GPGPUGeneral-purpose computing on GPU 1) CPU GPU GPU GPGPU NVIDIA C CUDACompute Unified Device Architecture 2) OpenCL 3) DNA
129 211 11 GPU 3 Smith-Waterman Saori SUDO 1 GPU Graphic Processing Unit GPU GPGPUGeneral-purpose computing on GPU 1) CPU GPU GPU GPGPU NVIDIA C CUDACompute Unified Device Architecture 2) OpenCL 3) DNA
Profiling-Based L1 Data Cache Bypassing to Improve GPU Performance and Energy Efficiency
 Profiling-Based L1 Data Cache Bypassing to Improve GPU Performance and Energy Efficiency Yijie Huangfu and Wei Zhang Department of Electrical and Computer Engineering Virginia Commonwealth University {huangfuy2,wzhang4}@vcu.edu
Profiling-Based L1 Data Cache Bypassing to Improve GPU Performance and Energy Efficiency Yijie Huangfu and Wei Zhang Department of Electrical and Computer Engineering Virginia Commonwealth University {huangfuy2,wzhang4}@vcu.edu
arxiv: v1 [cs.dc] 24 Feb 2010
![arxiv: v1 [cs.dc] 24 Feb 2010 arxiv: v1 [cs.dc] 24 Feb 2010](/thumbs/95/126041962.jpg) Deterministic Sample Sort For GPUs arxiv:1002.4464v1 [cs.dc] 24 Feb 2010 Frank Dehne School of Computer Science Carleton University Ottawa, Canada K1S 5B6 frank@dehne.net http://www.dehne.net February
Deterministic Sample Sort For GPUs arxiv:1002.4464v1 [cs.dc] 24 Feb 2010 Frank Dehne School of Computer Science Carleton University Ottawa, Canada K1S 5B6 frank@dehne.net http://www.dehne.net February
A TALENTED CPU-TO-GPU MEMORY MAPPING TECHNIQUE
 A TALENTED CPU-TO-GPU MEMORY MAPPING TECHNIQUE Abu Asaduzzaman, Deepthi Gummadi, and Chok M. Yip Department of Electrical Engineering and Computer Science Wichita State University Wichita, Kansas, USA
A TALENTED CPU-TO-GPU MEMORY MAPPING TECHNIQUE Abu Asaduzzaman, Deepthi Gummadi, and Chok M. Yip Department of Electrical Engineering and Computer Science Wichita State University Wichita, Kansas, USA
Parallel Approach for Implementing Data Mining Algorithms
 TITLE OF THE THESIS Parallel Approach for Implementing Data Mining Algorithms A RESEARCH PROPOSAL SUBMITTED TO THE SHRI RAMDEOBABA COLLEGE OF ENGINEERING AND MANAGEMENT, FOR THE DEGREE OF DOCTOR OF PHILOSOPHY
TITLE OF THE THESIS Parallel Approach for Implementing Data Mining Algorithms A RESEARCH PROPOSAL SUBMITTED TO THE SHRI RAMDEOBABA COLLEGE OF ENGINEERING AND MANAGEMENT, FOR THE DEGREE OF DOCTOR OF PHILOSOPHY
OPEN MP-BASED PARALLEL AND SCALABLE GENETIC SEQUENCE ALIGNMENT
 OPEN MP-BASED PARALLEL AND SCALABLE GENETIC SEQUENCE ALIGNMENT Asif Ali Khan*, Laiq Hassan*, Salim Ullah* ABSTRACT: In bioinformatics, sequence alignment is a common and insistent task. Biologists align
OPEN MP-BASED PARALLEL AND SCALABLE GENETIC SEQUENCE ALIGNMENT Asif Ali Khan*, Laiq Hassan*, Salim Ullah* ABSTRACT: In bioinformatics, sequence alignment is a common and insistent task. Biologists align
Exploring the Problem of GPU Programming for Data-Intensive Applications: A Case Study of Multiple Expectation Maximization for Motif Elicitation
 Exploring the Problem of GPU Programming for Data-Intensive Applications: A Case Study of Multiple Expectation Maximization for Motif Elicitation ABSTRACT Yuki Kitsukawa yuki@ertl.jp Shinpei Kato shinpei@is.nagoyau.ac.jp
Exploring the Problem of GPU Programming for Data-Intensive Applications: A Case Study of Multiple Expectation Maximization for Motif Elicitation ABSTRACT Yuki Kitsukawa yuki@ertl.jp Shinpei Kato shinpei@is.nagoyau.ac.jp
Applications of Berkeley s Dwarfs on Nvidia GPUs
 Applications of Berkeley s Dwarfs on Nvidia GPUs Seminar: Topics in High-Performance and Scientific Computing Team N2: Yang Zhang, Haiqing Wang 05.02.2015 Overview CUDA The Dwarfs Dynamic Programming Sparse
Applications of Berkeley s Dwarfs on Nvidia GPUs Seminar: Topics in High-Performance and Scientific Computing Team N2: Yang Zhang, Haiqing Wang 05.02.2015 Overview CUDA The Dwarfs Dynamic Programming Sparse
Scalable GPU Graph Traversal!
 Scalable GPU Graph Traversal Duane Merrill, Michael Garland, and Andrew Grimshaw PPoPP '12 Proceedings of the 17th ACM SIGPLAN symposium on Principles and Practice of Parallel Programming Benwen Zhang
Scalable GPU Graph Traversal Duane Merrill, Michael Garland, and Andrew Grimshaw PPoPP '12 Proceedings of the 17th ACM SIGPLAN symposium on Principles and Practice of Parallel Programming Benwen Zhang
Parallel Programming Principle and Practice. Lecture 9 Introduction to GPGPUs and CUDA Programming Model
 Parallel Programming Principle and Practice Lecture 9 Introduction to GPGPUs and CUDA Programming Model Outline Introduction to GPGPUs and Cuda Programming Model The Cuda Thread Hierarchy / Memory Hierarchy
Parallel Programming Principle and Practice Lecture 9 Introduction to GPGPUs and CUDA Programming Model Outline Introduction to GPGPUs and Cuda Programming Model The Cuda Thread Hierarchy / Memory Hierarchy
THE COMPARISON OF PARALLEL SORTING ALGORITHMS IMPLEMENTED ON DIFFERENT HARDWARE PLATFORMS
 Computer Science 14 (4) 2013 http://dx.doi.org/10.7494/csci.2013.14.4.679 Dominik Żurek Marcin Pietroń Maciej Wielgosz Kazimierz Wiatr THE COMPARISON OF PARALLEL SORTING ALGORITHMS IMPLEMENTED ON DIFFERENT
Computer Science 14 (4) 2013 http://dx.doi.org/10.7494/csci.2013.14.4.679 Dominik Żurek Marcin Pietroń Maciej Wielgosz Kazimierz Wiatr THE COMPARISON OF PARALLEL SORTING ALGORITHMS IMPLEMENTED ON DIFFERENT
Parallel Computing. Hwansoo Han (SKKU)
 Parallel Computing Hwansoo Han (SKKU) Unicore Limitations Performance scaling stopped due to Power consumption Wire delay DRAM latency Limitation in ILP 10000 SPEC CINT2000 2 cores/chip Xeon 3.0GHz Core2duo
Parallel Computing Hwansoo Han (SKKU) Unicore Limitations Performance scaling stopped due to Power consumption Wire delay DRAM latency Limitation in ILP 10000 SPEC CINT2000 2 cores/chip Xeon 3.0GHz Core2duo
Introduction to Parallel Computing
 Portland State University ECE 588/688 Introduction to Parallel Computing Reference: Lawrence Livermore National Lab Tutorial https://computing.llnl.gov/tutorials/parallel_comp/ Copyright by Alaa Alameldeen
Portland State University ECE 588/688 Introduction to Parallel Computing Reference: Lawrence Livermore National Lab Tutorial https://computing.llnl.gov/tutorials/parallel_comp/ Copyright by Alaa Alameldeen
Introduction to CUDA Algoritmi e Calcolo Parallelo. Daniele Loiacono
 Introduction to CUDA Algoritmi e Calcolo Parallelo References q This set of slides is mainly based on: " CUDA Technical Training, Dr. Antonino Tumeo, Pacific Northwest National Laboratory " Slide of Applied
Introduction to CUDA Algoritmi e Calcolo Parallelo References q This set of slides is mainly based on: " CUDA Technical Training, Dr. Antonino Tumeo, Pacific Northwest National Laboratory " Slide of Applied
ENABLING NEW SCIENCE GPU SOLUTIONS
 ENABLING NEW SCIENCE TESLA BIO Workbench The NVIDIA Tesla Bio Workbench enables biophysicists and computational chemists to push the boundaries of life sciences research. It turns a standard PC into a
ENABLING NEW SCIENCE TESLA BIO Workbench The NVIDIA Tesla Bio Workbench enables biophysicists and computational chemists to push the boundaries of life sciences research. It turns a standard PC into a
CUDA Optimizations WS Intelligent Robotics Seminar. Universität Hamburg WS Intelligent Robotics Seminar Praveen Kulkarni
 CUDA Optimizations WS 2014-15 Intelligent Robotics Seminar 1 Table of content 1 Background information 2 Optimizations 3 Summary 2 Table of content 1 Background information 2 Optimizations 3 Summary 3
CUDA Optimizations WS 2014-15 Intelligent Robotics Seminar 1 Table of content 1 Background information 2 Optimizations 3 Summary 2 Table of content 1 Background information 2 Optimizations 3 Summary 3
Towards ParadisEO-MO-GPU: a Framework for GPU-based Local Search Metaheuristics
 Towards ParadisEO-MO-GPU: a Framework for GPU-based Local Search Metaheuristics N. Melab, T-V. Luong, K. Boufaras and E-G. Talbi Dolphin Project INRIA Lille Nord Europe - LIFL/CNRS UMR 8022 - Université
Towards ParadisEO-MO-GPU: a Framework for GPU-based Local Search Metaheuristics N. Melab, T-V. Luong, K. Boufaras and E-G. Talbi Dolphin Project INRIA Lille Nord Europe - LIFL/CNRS UMR 8022 - Université
CUDA PROGRAMMING MODEL Chaithanya Gadiyam Swapnil S Jadhav
 CUDA PROGRAMMING MODEL Chaithanya Gadiyam Swapnil S Jadhav CMPE655 - Multiple Processor Systems Fall 2015 Rochester Institute of Technology Contents What is GPGPU? What s the need? CUDA-Capable GPU Architecture
CUDA PROGRAMMING MODEL Chaithanya Gadiyam Swapnil S Jadhav CMPE655 - Multiple Processor Systems Fall 2015 Rochester Institute of Technology Contents What is GPGPU? What s the need? CUDA-Capable GPU Architecture
A Study of High Performance Computing and the Cray SV1 Supercomputer. Michael Sullivan TJHSST Class of 2004
 A Study of High Performance Computing and the Cray SV1 Supercomputer Michael Sullivan TJHSST Class of 2004 June 2004 0.1 Introduction A supercomputer is a device for turning compute-bound problems into
A Study of High Performance Computing and the Cray SV1 Supercomputer Michael Sullivan TJHSST Class of 2004 June 2004 0.1 Introduction A supercomputer is a device for turning compute-bound problems into
Comparative Analysis of Protein Alignment Algorithms in Parallel environment using CUDA
 Comparative Analysis of Protein Alignment Algorithms in Parallel environment using BLAST versus Smith-Waterman Shadman Fahim shadmanbracu09@gmail.com Shehabul Hossain rudrozzal@gmail.com Gulshan Jubaed
Comparative Analysis of Protein Alignment Algorithms in Parallel environment using BLAST versus Smith-Waterman Shadman Fahim shadmanbracu09@gmail.com Shehabul Hossain rudrozzal@gmail.com Gulshan Jubaed
ACCELERATING THE PRODUCTION OF SYNTHETIC SEISMOGRAMS BY A MULTICORE PROCESSOR CLUSTER WITH MULTIPLE GPUS
 ACCELERATING THE PRODUCTION OF SYNTHETIC SEISMOGRAMS BY A MULTICORE PROCESSOR CLUSTER WITH MULTIPLE GPUS Ferdinando Alessi Annalisa Massini Roberto Basili INGV Introduction The simulation of wave propagation
ACCELERATING THE PRODUCTION OF SYNTHETIC SEISMOGRAMS BY A MULTICORE PROCESSOR CLUSTER WITH MULTIPLE GPUS Ferdinando Alessi Annalisa Massini Roberto Basili INGV Introduction The simulation of wave propagation
high performance medical reconstruction using stream programming paradigms
 high performance medical reconstruction using stream programming paradigms This Paper describes the implementation and results of CT reconstruction using Filtered Back Projection on various stream programming
high performance medical reconstruction using stream programming paradigms This Paper describes the implementation and results of CT reconstruction using Filtered Back Projection on various stream programming
Optimization solutions for the segmented sum algorithmic function
 Optimization solutions for the segmented sum algorithmic function ALEXANDRU PÎRJAN Department of Informatics, Statistics and Mathematics Romanian-American University 1B, Expozitiei Blvd., district 1, code
Optimization solutions for the segmented sum algorithmic function ALEXANDRU PÎRJAN Department of Informatics, Statistics and Mathematics Romanian-American University 1B, Expozitiei Blvd., district 1, code
 By: Tomer Morad Based on: Erik Lindholm, John Nickolls, Stuart Oberman, John Montrym. NVIDIA TESLA: A UNIFIED GRAPHICS AND COMPUTING ARCHITECTURE In IEEE Micro 28(2), 2008 } } Erik Lindholm, John Nickolls,
By: Tomer Morad Based on: Erik Lindholm, John Nickolls, Stuart Oberman, John Montrym. NVIDIA TESLA: A UNIFIED GRAPHICS AND COMPUTING ARCHITECTURE In IEEE Micro 28(2), 2008 } } Erik Lindholm, John Nickolls,
THE Smith-Waterman (SW) algorithm [1] is a wellknown
![THE Smith-Waterman (SW) algorithm [1] is a wellknown THE Smith-Waterman (SW) algorithm [1] is a wellknown](/thumbs/95/125558830.jpg) Design and Implementation of the Smith-Waterman Algorithm on the CUDA-Compatible GPU Yuma Munekawa, Fumihiko Ino, Member, IEEE, and Kenichi Hagihara Abstract This paper describes a design and implementation
Design and Implementation of the Smith-Waterman Algorithm on the CUDA-Compatible GPU Yuma Munekawa, Fumihiko Ino, Member, IEEE, and Kenichi Hagihara Abstract This paper describes a design and implementation
ParalleX. A Cure for Scaling Impaired Parallel Applications. Hartmut Kaiser
 ParalleX A Cure for Scaling Impaired Parallel Applications Hartmut Kaiser (hkaiser@cct.lsu.edu) 2 Tianhe-1A 2.566 Petaflops Rmax Heterogeneous Architecture: 14,336 Intel Xeon CPUs 7,168 Nvidia Tesla M2050
ParalleX A Cure for Scaling Impaired Parallel Applications Hartmut Kaiser (hkaiser@cct.lsu.edu) 2 Tianhe-1A 2.566 Petaflops Rmax Heterogeneous Architecture: 14,336 Intel Xeon CPUs 7,168 Nvidia Tesla M2050
CUDA GPGPU Workshop 2012
 CUDA GPGPU Workshop 2012 Parallel Programming: C thread, Open MP, and Open MPI Presenter: Nasrin Sultana Wichita State University 07/10/2012 Parallel Programming: Open MP, MPI, Open MPI & CUDA Outline
CUDA GPGPU Workshop 2012 Parallel Programming: C thread, Open MP, and Open MPI Presenter: Nasrin Sultana Wichita State University 07/10/2012 Parallel Programming: Open MP, MPI, Open MPI & CUDA Outline
Parallel and Distributed Computing
 Parallel and Distributed Computing NUMA; OpenCL; MapReduce José Monteiro MSc in Information Systems and Computer Engineering DEA in Computational Engineering Department of Computer Science and Engineering
Parallel and Distributed Computing NUMA; OpenCL; MapReduce José Monteiro MSc in Information Systems and Computer Engineering DEA in Computational Engineering Department of Computer Science and Engineering
Sequence Alignment with GPU: Performance and Design Challenges
 Sequence Alignment with GPU: Performance and Design Challenges Gregory M. Striemer and Ali Akoglu Department of Electrical and Computer Engineering University of Arizona, 85721 Tucson, Arizona USA {gmstrie,
Sequence Alignment with GPU: Performance and Design Challenges Gregory M. Striemer and Ali Akoglu Department of Electrical and Computer Engineering University of Arizona, 85721 Tucson, Arizona USA {gmstrie,
CSCI 402: Computer Architectures. Parallel Processors (2) Fengguang Song Department of Computer & Information Science IUPUI.
 CSCI 402: Computer Architectures Parallel Processors (2) Fengguang Song Department of Computer & Information Science IUPUI 6.6 - End Today s Contents GPU Cluster and its network topology The Roofline performance
CSCI 402: Computer Architectures Parallel Processors (2) Fengguang Song Department of Computer & Information Science IUPUI 6.6 - End Today s Contents GPU Cluster and its network topology The Roofline performance
Parallel Computing Using OpenMP/MPI. Presented by - Jyotsna 29/01/2008
 Parallel Computing Using OpenMP/MPI Presented by - Jyotsna 29/01/2008 Serial Computing Serially solving a problem Parallel Computing Parallelly solving a problem Parallel Computer Memory Architecture Shared
Parallel Computing Using OpenMP/MPI Presented by - Jyotsna 29/01/2008 Serial Computing Serially solving a problem Parallel Computing Parallelly solving a problem Parallel Computer Memory Architecture Shared
Accelerating InDel Detection on Modern Multi-Core SIMD CPU Architecture
 Accelerating InDel Detection on Modern Multi-Core SIMD CPU Architecture Da Zhang Collaborators: Hao Wang, Kaixi Hou, Jing Zhang Advisor: Wu-chun Feng Evolution of Genome Sequencing1 In 20032: 1 human genome
Accelerating InDel Detection on Modern Multi-Core SIMD CPU Architecture Da Zhang Collaborators: Hao Wang, Kaixi Hou, Jing Zhang Advisor: Wu-chun Feng Evolution of Genome Sequencing1 In 20032: 1 human genome
COMP 605: Introduction to Parallel Computing Lecture : GPU Architecture
 COMP 605: Introduction to Parallel Computing Lecture : GPU Architecture Mary Thomas Department of Computer Science Computational Science Research Center (CSRC) San Diego State University (SDSU) Posted:
COMP 605: Introduction to Parallel Computing Lecture : GPU Architecture Mary Thomas Department of Computer Science Computational Science Research Center (CSRC) San Diego State University (SDSU) Posted:
Introduction to CUDA Algoritmi e Calcolo Parallelo. Daniele Loiacono
 Introduction to CUDA Algoritmi e Calcolo Parallelo References This set of slides is mainly based on: CUDA Technical Training, Dr. Antonino Tumeo, Pacific Northwest National Laboratory Slide of Applied
Introduction to CUDA Algoritmi e Calcolo Parallelo References This set of slides is mainly based on: CUDA Technical Training, Dr. Antonino Tumeo, Pacific Northwest National Laboratory Slide of Applied
The Art of Parallel Processing
 The Art of Parallel Processing Ahmad Siavashi April 2017 The Software Crisis As long as there were no machines, programming was no problem at all; when we had a few weak computers, programming became a
The Art of Parallel Processing Ahmad Siavashi April 2017 The Software Crisis As long as there were no machines, programming was no problem at all; when we had a few weak computers, programming became a
CMSC 714 Lecture 6 MPI vs. OpenMP and OpenACC. Guest Lecturer: Sukhyun Song (original slides by Alan Sussman)
 CMSC 714 Lecture 6 MPI vs. OpenMP and OpenACC Guest Lecturer: Sukhyun Song (original slides by Alan Sussman) Parallel Programming with Message Passing and Directives 2 MPI + OpenMP Some applications can
CMSC 714 Lecture 6 MPI vs. OpenMP and OpenACC Guest Lecturer: Sukhyun Song (original slides by Alan Sussman) Parallel Programming with Message Passing and Directives 2 MPI + OpenMP Some applications can
Serial. Parallel. CIT 668: System Architecture 2/14/2011. Topics. Serial and Parallel Computation. Parallel Computing
 CIT 668: System Architecture Parallel Computing Topics 1. What is Parallel Computing? 2. Why use Parallel Computing? 3. Types of Parallelism 4. Amdahl s Law 5. Flynn s Taxonomy of Parallel Computers 6.
CIT 668: System Architecture Parallel Computing Topics 1. What is Parallel Computing? 2. Why use Parallel Computing? 3. Types of Parallelism 4. Amdahl s Law 5. Flynn s Taxonomy of Parallel Computers 6.
High Performance Computing on GPUs using NVIDIA CUDA
 High Performance Computing on GPUs using NVIDIA CUDA Slides include some material from GPGPU tutorial at SIGGRAPH2007: http://www.gpgpu.org/s2007 1 Outline Motivation Stream programming Simplified HW and
High Performance Computing on GPUs using NVIDIA CUDA Slides include some material from GPGPU tutorial at SIGGRAPH2007: http://www.gpgpu.org/s2007 1 Outline Motivation Stream programming Simplified HW and
Particle-in-Cell Simulations on Modern Computing Platforms. Viktor K. Decyk and Tajendra V. Singh UCLA
 Particle-in-Cell Simulations on Modern Computing Platforms Viktor K. Decyk and Tajendra V. Singh UCLA Outline of Presentation Abstraction of future computer hardware PIC on GPUs OpenCL and Cuda Fortran
Particle-in-Cell Simulations on Modern Computing Platforms Viktor K. Decyk and Tajendra V. Singh UCLA Outline of Presentation Abstraction of future computer hardware PIC on GPUs OpenCL and Cuda Fortran
COMPUTER ORGANIZATION AND DESIGN The Hardware/Software Interface. 5 th. Edition. Chapter 6. Parallel Processors from Client to Cloud
 COMPUTER ORGANIZATION AND DESIGN The Hardware/Software Interface 5 th Edition Chapter 6 Parallel Processors from Client to Cloud Introduction Goal: connecting multiple computers to get higher performance
COMPUTER ORGANIZATION AND DESIGN The Hardware/Software Interface 5 th Edition Chapter 6 Parallel Processors from Client to Cloud Introduction Goal: connecting multiple computers to get higher performance
Algorithms and Tools for Bioinformatics on GPUs. Bertil SCHMIDT
 Algorithms and Tools for Bioinformatics on GPUs Bertil SCHMIDT Contents Motivation Pairwise Sequence Alignment Multiple Sequence Alignment Short Read Error Correction using CUDA Some other CUDA-enabled
Algorithms and Tools for Bioinformatics on GPUs Bertil SCHMIDT Contents Motivation Pairwise Sequence Alignment Multiple Sequence Alignment Short Read Error Correction using CUDA Some other CUDA-enabled
Modern Processor Architectures. L25: Modern Compiler Design
 Modern Processor Architectures L25: Modern Compiler Design The 1960s - 1970s Instructions took multiple cycles Only one instruction in flight at once Optimisation meant minimising the number of instructions
Modern Processor Architectures L25: Modern Compiler Design The 1960s - 1970s Instructions took multiple cycles Only one instruction in flight at once Optimisation meant minimising the number of instructions
GRAPHICS PROCESSING UNITS
 GRAPHICS PROCESSING UNITS Slides by: Pedro Tomás Additional reading: Computer Architecture: A Quantitative Approach, 5th edition, Chapter 4, John L. Hennessy and David A. Patterson, Morgan Kaufmann, 2011
GRAPHICS PROCESSING UNITS Slides by: Pedro Tomás Additional reading: Computer Architecture: A Quantitative Approach, 5th edition, Chapter 4, John L. Hennessy and David A. Patterson, Morgan Kaufmann, 2011
TUNING CUDA APPLICATIONS FOR MAXWELL
 TUNING CUDA APPLICATIONS FOR MAXWELL DA-07173-001_v6.5 August 2014 Application Note TABLE OF CONTENTS Chapter 1. Maxwell Tuning Guide... 1 1.1. NVIDIA Maxwell Compute Architecture... 1 1.2. CUDA Best Practices...2
TUNING CUDA APPLICATIONS FOR MAXWELL DA-07173-001_v6.5 August 2014 Application Note TABLE OF CONTENTS Chapter 1. Maxwell Tuning Guide... 1 1.1. NVIDIA Maxwell Compute Architecture... 1 1.2. CUDA Best Practices...2
NVIDIA GTX200: TeraFLOPS Visual Computing. August 26, 2008 John Tynefield
 NVIDIA GTX200: TeraFLOPS Visual Computing August 26, 2008 John Tynefield 2 Outline Execution Model Architecture Demo 3 Execution Model 4 Software Architecture Applications DX10 OpenGL OpenCL CUDA C Host
NVIDIA GTX200: TeraFLOPS Visual Computing August 26, 2008 John Tynefield 2 Outline Execution Model Architecture Demo 3 Execution Model 4 Software Architecture Applications DX10 OpenGL OpenCL CUDA C Host
GPU Fundamentals Jeff Larkin November 14, 2016
 GPU Fundamentals Jeff Larkin , November 4, 206 Who Am I? 2002 B.S. Computer Science Furman University 2005 M.S. Computer Science UT Knoxville 2002 Graduate Teaching Assistant 2005 Graduate
GPU Fundamentals Jeff Larkin , November 4, 206 Who Am I? 2002 B.S. Computer Science Furman University 2005 M.S. Computer Science UT Knoxville 2002 Graduate Teaching Assistant 2005 Graduate
Computer Architecture
 Computer Architecture Slide Sets WS 2013/2014 Prof. Dr. Uwe Brinkschulte M.Sc. Benjamin Betting Part 10 Thread and Task Level Parallelism Computer Architecture Part 10 page 1 of 36 Prof. Dr. Uwe Brinkschulte,
Computer Architecture Slide Sets WS 2013/2014 Prof. Dr. Uwe Brinkschulte M.Sc. Benjamin Betting Part 10 Thread and Task Level Parallelism Computer Architecture Part 10 page 1 of 36 Prof. Dr. Uwe Brinkschulte,
HiPANQ Overview of NVIDIA GPU Architecture and Introduction to CUDA/OpenCL Programming, and Parallelization of LDPC codes.
 HiPANQ Overview of NVIDIA GPU Architecture and Introduction to CUDA/OpenCL Programming, and Parallelization of LDPC codes Ian Glendinning Outline NVIDIA GPU cards CUDA & OpenCL Parallel Implementation
HiPANQ Overview of NVIDIA GPU Architecture and Introduction to CUDA/OpenCL Programming, and Parallelization of LDPC codes Ian Glendinning Outline NVIDIA GPU cards CUDA & OpenCL Parallel Implementation
B. Tech. Project Second Stage Report on
 B. Tech. Project Second Stage Report on GPU Based Active Contours Submitted by Sumit Shekhar (05007028) Under the guidance of Prof Subhasis Chaudhuri Table of Contents 1. Introduction... 1 1.1 Graphic
B. Tech. Project Second Stage Report on GPU Based Active Contours Submitted by Sumit Shekhar (05007028) Under the guidance of Prof Subhasis Chaudhuri Table of Contents 1. Introduction... 1 1.1 Graphic
Parallel Computing Why & How?
 Parallel Computing Why & How? Xing Cai Simula Research Laboratory Dept. of Informatics, University of Oslo Winter School on Parallel Computing Geilo January 20 25, 2008 Outline 1 Motivation 2 Parallel
Parallel Computing Why & How? Xing Cai Simula Research Laboratory Dept. of Informatics, University of Oslo Winter School on Parallel Computing Geilo January 20 25, 2008 Outline 1 Motivation 2 Parallel
Parallelising Pipelined Wavefront Computations on the GPU
 Parallelising Pipelined Wavefront Computations on the GPU S.J. Pennycook G.R. Mudalige, S.D. Hammond, and S.A. Jarvis. High Performance Systems Group Department of Computer Science University of Warwick
Parallelising Pipelined Wavefront Computations on the GPU S.J. Pennycook G.R. Mudalige, S.D. Hammond, and S.A. Jarvis. High Performance Systems Group Department of Computer Science University of Warwick
Performance Analysis of Memory Transfers and GEMM Subroutines on NVIDIA TESLA GPU Cluster
 Performance Analysis of Memory Transfers and GEMM Subroutines on NVIDIA TESLA GPU Cluster Veerendra Allada, Troy Benjegerdes Electrical and Computer Engineering, Ames Laboratory Iowa State University &
Performance Analysis of Memory Transfers and GEMM Subroutines on NVIDIA TESLA GPU Cluster Veerendra Allada, Troy Benjegerdes Electrical and Computer Engineering, Ames Laboratory Iowa State University &
Lecture 13: Memory Consistency. + a Course-So-Far Review. Parallel Computer Architecture and Programming CMU , Spring 2013
 Lecture 13: Memory Consistency + a Course-So-Far Review Parallel Computer Architecture and Programming Today: what you should know Understand the motivation for relaxed consistency models Understand the
Lecture 13: Memory Consistency + a Course-So-Far Review Parallel Computer Architecture and Programming Today: what you should know Understand the motivation for relaxed consistency models Understand the
Implementation of GP-GPU with SIMT Architecture in the Embedded Environment
 , pp.221-226 http://dx.doi.org/10.14257/ijmue.2014.9.4.23 Implementation of GP-GPU with SIMT Architecture in the Embedded Environment Kwang-yeob Lee and Jae-chang Kwak 1 * Dept. of Computer Engineering,
, pp.221-226 http://dx.doi.org/10.14257/ijmue.2014.9.4.23 Implementation of GP-GPU with SIMT Architecture in the Embedded Environment Kwang-yeob Lee and Jae-chang Kwak 1 * Dept. of Computer Engineering,
Accelerating the Implicit Integration of Stiff Chemical Systems with Emerging Multi-core Technologies
 Accelerating the Implicit Integration of Stiff Chemical Systems with Emerging Multi-core Technologies John C. Linford John Michalakes Manish Vachharajani Adrian Sandu IMAGe TOY 2009 Workshop 2 Virginia
Accelerating the Implicit Integration of Stiff Chemical Systems with Emerging Multi-core Technologies John C. Linford John Michalakes Manish Vachharajani Adrian Sandu IMAGe TOY 2009 Workshop 2 Virginia
Performance impact of dynamic parallelism on different clustering algorithms
 Performance impact of dynamic parallelism on different clustering algorithms Jeffrey DiMarco and Michela Taufer Computer and Information Sciences, University of Delaware E-mail: jdimarco@udel.edu, taufer@udel.edu
Performance impact of dynamic parallelism on different clustering algorithms Jeffrey DiMarco and Michela Taufer Computer and Information Sciences, University of Delaware E-mail: jdimarco@udel.edu, taufer@udel.edu
How to perform HPL on CPU&GPU clusters. Dr.sc. Draško Tomić
 How to perform HPL on CPU&GPU clusters Dr.sc. Draško Tomić email: drasko.tomic@hp.com Forecasting is not so easy, HPL benchmarking could be even more difficult Agenda TOP500 GPU trends Some basics about
How to perform HPL on CPU&GPU clusters Dr.sc. Draško Tomić email: drasko.tomic@hp.com Forecasting is not so easy, HPL benchmarking could be even more difficult Agenda TOP500 GPU trends Some basics about
Tesla GPU Computing A Revolution in High Performance Computing
 Tesla GPU Computing A Revolution in High Performance Computing Mark Harris, NVIDIA Agenda Tesla GPU Computing CUDA Fermi What is GPU Computing? Introduction to Tesla CUDA Architecture Programming & Memory
Tesla GPU Computing A Revolution in High Performance Computing Mark Harris, NVIDIA Agenda Tesla GPU Computing CUDA Fermi What is GPU Computing? Introduction to Tesla CUDA Architecture Programming & Memory
Accelerating Parameter Sweep Applications Using CUDA
 2 9th International Euromicro Conference on Parallel, Distributed and Network-Based Processing Accelerating Parameter Sweep Applications Using CUDA Masaya Motokubota, Fumihiko Ino and Kenichi Hagihara
2 9th International Euromicro Conference on Parallel, Distributed and Network-Based Processing Accelerating Parameter Sweep Applications Using CUDA Masaya Motokubota, Fumihiko Ino and Kenichi Hagihara
Chapter 4: Threads. Chapter 4: Threads. Overview Multicore Programming Multithreading Models Thread Libraries Implicit Threading Threading Issues
 Chapter 4: Threads Silberschatz, Galvin and Gagne 2013 Chapter 4: Threads Overview Multicore Programming Multithreading Models Thread Libraries Implicit Threading Threading Issues 4.2 Silberschatz, Galvin
Chapter 4: Threads Silberschatz, Galvin and Gagne 2013 Chapter 4: Threads Overview Multicore Programming Multithreading Models Thread Libraries Implicit Threading Threading Issues 4.2 Silberschatz, Galvin
TUNING CUDA APPLICATIONS FOR MAXWELL
 TUNING CUDA APPLICATIONS FOR MAXWELL DA-07173-001_v7.0 March 2015 Application Note TABLE OF CONTENTS Chapter 1. Maxwell Tuning Guide... 1 1.1. NVIDIA Maxwell Compute Architecture... 1 1.2. CUDA Best Practices...2
TUNING CUDA APPLICATIONS FOR MAXWELL DA-07173-001_v7.0 March 2015 Application Note TABLE OF CONTENTS Chapter 1. Maxwell Tuning Guide... 1 1.1. NVIDIA Maxwell Compute Architecture... 1 1.2. CUDA Best Practices...2
Programmable Graphics Hardware (GPU) A Primer
 Programmable Graphics Hardware (GPU) A Primer Klaus Mueller Stony Brook University Computer Science Department Parallel Computing Explained video Parallel Computing Explained Any questions? Parallelism
Programmable Graphics Hardware (GPU) A Primer Klaus Mueller Stony Brook University Computer Science Department Parallel Computing Explained video Parallel Computing Explained Any questions? Parallelism
EE/CSCI 451: Parallel and Distributed Computation
 EE/CSCI 451: Parallel and Distributed Computation Lecture #7 2/5/2017 Xuehai Qian Xuehai.qian@usc.edu http://alchem.usc.edu/portal/xuehaiq.html University of Southern California 1 Outline From last class
EE/CSCI 451: Parallel and Distributed Computation Lecture #7 2/5/2017 Xuehai Qian Xuehai.qian@usc.edu http://alchem.usc.edu/portal/xuehaiq.html University of Southern California 1 Outline From last class
Lecture 2: CUDA Programming
 CS 515 Programming Language and Compilers I Lecture 2: CUDA Programming Zheng (Eddy) Zhang Rutgers University Fall 2017, 9/12/2017 Review: Programming in CUDA Let s look at a sequential program in C first:
CS 515 Programming Language and Compilers I Lecture 2: CUDA Programming Zheng (Eddy) Zhang Rutgers University Fall 2017, 9/12/2017 Review: Programming in CUDA Let s look at a sequential program in C first:
Keywords -Bioinformatics, sequence alignment, Smith- waterman (SW) algorithm, GPU, CUDA
 Volume 5, Issue 5, May 2015 ISSN: 2277 128X International Journal of Advanced Research in Computer Science and Software Engineering Research Paper Available online at: www.ijarcsse.com Accelerating Smith-Waterman
Volume 5, Issue 5, May 2015 ISSN: 2277 128X International Journal of Advanced Research in Computer Science and Software Engineering Research Paper Available online at: www.ijarcsse.com Accelerating Smith-Waterman
Fundamental CUDA Optimization. NVIDIA Corporation
 Fundamental CUDA Optimization NVIDIA Corporation Outline Fermi/Kepler Architecture Kernel optimizations Launch configuration Global memory throughput Shared memory access Instruction throughput / control
Fundamental CUDA Optimization NVIDIA Corporation Outline Fermi/Kepler Architecture Kernel optimizations Launch configuration Global memory throughput Shared memory access Instruction throughput / control
What is Parallel Computing?
 What is Parallel Computing? Parallel Computing is several processing elements working simultaneously to solve a problem faster. 1/33 What is Parallel Computing? Parallel Computing is several processing
What is Parallel Computing? Parallel Computing is several processing elements working simultaneously to solve a problem faster. 1/33 What is Parallel Computing? Parallel Computing is several processing
Computational Genomics and Molecular Biology, Fall
 Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
Execution Strategy and Runtime Support for Regular and Irregular Applications on Emerging Parallel Architectures
 Execution Strategy and Runtime Support for Regular and Irregular Applications on Emerging Parallel Architectures Xin Huo Advisor: Gagan Agrawal Motivation - Architecture Challenges on GPU architecture
Execution Strategy and Runtime Support for Regular and Irregular Applications on Emerging Parallel Architectures Xin Huo Advisor: Gagan Agrawal Motivation - Architecture Challenges on GPU architecture
Introduction to Parallel and Distributed Computing. Linh B. Ngo CPSC 3620
 Introduction to Parallel and Distributed Computing Linh B. Ngo CPSC 3620 Overview: What is Parallel Computing To be run using multiple processors A problem is broken into discrete parts that can be solved
Introduction to Parallel and Distributed Computing Linh B. Ngo CPSC 3620 Overview: What is Parallel Computing To be run using multiple processors A problem is broken into discrete parts that can be solved
Improving CUDASW++, a Parallelization of Smith-Waterman for CUDA Enabled Devices
 2011 IEEE International Parallel & Distributed Processing Symposium Improving CUDASW++, a Parallelization of Smith-Waterman for CUDA Enabled Devices Doug Hains, Zach Cashero, Mark Ottenberg, Wim Bohm and
2011 IEEE International Parallel & Distributed Processing Symposium Improving CUDASW++, a Parallelization of Smith-Waterman for CUDA Enabled Devices Doug Hains, Zach Cashero, Mark Ottenberg, Wim Bohm and
CUDA Architecture & Programming Model
 CUDA Architecture & Programming Model Course on Multi-core Architectures & Programming Oliver Taubmann May 9, 2012 Outline Introduction Architecture Generation Fermi A Brief Look Back At Tesla What s New
CUDA Architecture & Programming Model Course on Multi-core Architectures & Programming Oliver Taubmann May 9, 2012 Outline Introduction Architecture Generation Fermi A Brief Look Back At Tesla What s New
Subset Sum Problem Parallel Solution
 Subset Sum Problem Parallel Solution Project Report Harshit Shah hrs8207@rit.edu Rochester Institute of Technology, NY, USA 1. Overview Subset sum problem is NP-complete problem which can be solved in
Subset Sum Problem Parallel Solution Project Report Harshit Shah hrs8207@rit.edu Rochester Institute of Technology, NY, USA 1. Overview Subset sum problem is NP-complete problem which can be solved in
Parallel Computing: Parallel Architectures Jin, Hai
 Parallel Computing: Parallel Architectures Jin, Hai School of Computer Science and Technology Huazhong University of Science and Technology Peripherals Computer Central Processing Unit Main Memory Computer
Parallel Computing: Parallel Architectures Jin, Hai School of Computer Science and Technology Huazhong University of Science and Technology Peripherals Computer Central Processing Unit Main Memory Computer
OpenMP Optimization and its Translation to OpenGL
 OpenMP Optimization and its Translation to OpenGL Santosh Kumar SITRC-Nashik, India Dr. V.M.Wadhai MAE-Pune, India Prasad S.Halgaonkar MITCOE-Pune, India Kiran P.Gaikwad GHRIEC-Pune, India ABSTRACT For
OpenMP Optimization and its Translation to OpenGL Santosh Kumar SITRC-Nashik, India Dr. V.M.Wadhai MAE-Pune, India Prasad S.Halgaonkar MITCOE-Pune, India Kiran P.Gaikwad GHRIEC-Pune, India ABSTRACT For
The Effect of Inverse Document Frequency Weights on Indexed Sequence Retrieval. Kevin C. O'Kane. Department of Computer Science
 The Effect of Inverse Document Frequency Weights on Indexed Sequence Retrieval Kevin C. O'Kane Department of Computer Science The University of Northern Iowa Cedar Falls, Iowa okane@cs.uni.edu http://www.cs.uni.edu/~okane
The Effect of Inverse Document Frequency Weights on Indexed Sequence Retrieval Kevin C. O'Kane Department of Computer Science The University of Northern Iowa Cedar Falls, Iowa okane@cs.uni.edu http://www.cs.uni.edu/~okane
CS 590: High Performance Computing. Parallel Computer Architectures. Lab 1 Starts Today. Already posted on Canvas (under Assignment) Let s look at it
 Lab 1 Starts Today Already posted on Canvas (under Assignment) Let s look at it CS 590: High Performance Computing Parallel Computer Architectures Fengguang Song Department of Computer Science IUPUI 1
Lab 1 Starts Today Already posted on Canvas (under Assignment) Let s look at it CS 590: High Performance Computing Parallel Computer Architectures Fengguang Song Department of Computer Science IUPUI 1
On the Comparative Performance of Parallel Algorithms on Small GPU/CUDA Clusters
 1 On the Comparative Performance of Parallel Algorithms on Small GPU/CUDA Clusters N. P. Karunadasa & D. N. Ranasinghe University of Colombo School of Computing, Sri Lanka nishantha@opensource.lk, dnr@ucsc.cmb.ac.lk
1 On the Comparative Performance of Parallel Algorithms on Small GPU/CUDA Clusters N. P. Karunadasa & D. N. Ranasinghe University of Colombo School of Computing, Sri Lanka nishantha@opensource.lk, dnr@ucsc.cmb.ac.lk
PROGRAMOVÁNÍ V C++ CVIČENÍ. Michal Brabec
 PROGRAMOVÁNÍ V C++ CVIČENÍ Michal Brabec PARALLELISM CATEGORIES CPU? SSE Multiprocessor SIMT - GPU 2 / 17 PARALLELISM V C++ Weak support in the language itself, powerful libraries Many different parallelization
PROGRAMOVÁNÍ V C++ CVIČENÍ Michal Brabec PARALLELISM CATEGORIES CPU? SSE Multiprocessor SIMT - GPU 2 / 17 PARALLELISM V C++ Weak support in the language itself, powerful libraries Many different parallelization
CUDA. Matthew Joyner, Jeremy Williams
 CUDA Matthew Joyner, Jeremy Williams Agenda What is CUDA? CUDA GPU Architecture CPU/GPU Communication Coding in CUDA Use cases of CUDA Comparison to OpenCL What is CUDA? What is CUDA? CUDA is a parallel
CUDA Matthew Joyner, Jeremy Williams Agenda What is CUDA? CUDA GPU Architecture CPU/GPU Communication Coding in CUDA Use cases of CUDA Comparison to OpenCL What is CUDA? What is CUDA? CUDA is a parallel
OVERHEADS ENHANCEMENT IN MUTIPLE PROCESSING SYSTEMS BY ANURAG REDDY GANKAT KARTHIK REDDY AKKATI
 CMPE 655- MULTIPLE PROCESSOR SYSTEMS OVERHEADS ENHANCEMENT IN MUTIPLE PROCESSING SYSTEMS BY ANURAG REDDY GANKAT KARTHIK REDDY AKKATI What is MULTI PROCESSING?? Multiprocessing is the coordinated processing
CMPE 655- MULTIPLE PROCESSOR SYSTEMS OVERHEADS ENHANCEMENT IN MUTIPLE PROCESSING SYSTEMS BY ANURAG REDDY GANKAT KARTHIK REDDY AKKATI What is MULTI PROCESSING?? Multiprocessing is the coordinated processing
Comparison of Phylogenetic Trees of Multiple Protein Sequence Alignment Methods
 Comparison of Phylogenetic Trees of Multiple Protein Sequence Alignment Methods Khaddouja Boujenfa, Nadia Essoussi, and Mohamed Limam International Science Index, Computer and Information Engineering waset.org/publication/482
Comparison of Phylogenetic Trees of Multiple Protein Sequence Alignment Methods Khaddouja Boujenfa, Nadia Essoussi, and Mohamed Limam International Science Index, Computer and Information Engineering waset.org/publication/482
Lecture 1: Introduction and Computational Thinking
 PASI Summer School Advanced Algorithmic Techniques for GPUs Lecture 1: Introduction and Computational Thinking 1 Course Objective To master the most commonly used algorithm techniques and computational
PASI Summer School Advanced Algorithmic Techniques for GPUs Lecture 1: Introduction and Computational Thinking 1 Course Objective To master the most commonly used algorithm techniques and computational
Compiling for GPUs. Adarsh Yoga Madhav Ramesh
 Compiling for GPUs Adarsh Yoga Madhav Ramesh Agenda Introduction to GPUs Compute Unified Device Architecture (CUDA) Control Structure Optimization Technique for GPGPU Compiler Framework for Automatic Translation
Compiling for GPUs Adarsh Yoga Madhav Ramesh Agenda Introduction to GPUs Compute Unified Device Architecture (CUDA) Control Structure Optimization Technique for GPGPU Compiler Framework for Automatic Translation
What is GPU? CS 590: High Performance Computing. GPU Architectures and CUDA Concepts/Terms
 CS 590: High Performance Computing GPU Architectures and CUDA Concepts/Terms Fengguang Song Department of Computer & Information Science IUPUI What is GPU? Conventional GPUs are used to generate 2D, 3D
CS 590: High Performance Computing GPU Architectures and CUDA Concepts/Terms Fengguang Song Department of Computer & Information Science IUPUI What is GPU? Conventional GPUs are used to generate 2D, 3D
EE/CSCI 451: Parallel and Distributed Computation
 EE/CSCI 451: Parallel and Distributed Computation Lecture #11 2/21/2017 Xuehai Qian Xuehai.qian@usc.edu http://alchem.usc.edu/portal/xuehaiq.html University of Southern California 1 Outline Midterm 1:
EE/CSCI 451: Parallel and Distributed Computation Lecture #11 2/21/2017 Xuehai Qian Xuehai.qian@usc.edu http://alchem.usc.edu/portal/xuehaiq.html University of Southern California 1 Outline Midterm 1:
Double-Precision Matrix Multiply on CUDA
 Double-Precision Matrix Multiply on CUDA Parallel Computation (CSE 60), Assignment Andrew Conegliano (A5055) Matthias Springer (A995007) GID G--665 February, 0 Assumptions All matrices are square matrices
Double-Precision Matrix Multiply on CUDA Parallel Computation (CSE 60), Assignment Andrew Conegliano (A5055) Matthias Springer (A995007) GID G--665 February, 0 Assumptions All matrices are square matrices
GPU Performance Optimisation. Alan Gray EPCC The University of Edinburgh
 GPU Performance Optimisation EPCC The University of Edinburgh Hardware NVIDIA accelerated system: Memory Memory GPU vs CPU: Theoretical Peak capabilities NVIDIA Fermi AMD Magny-Cours (6172) Cores 448 (1.15GHz)
GPU Performance Optimisation EPCC The University of Edinburgh Hardware NVIDIA accelerated system: Memory Memory GPU vs CPU: Theoretical Peak capabilities NVIDIA Fermi AMD Magny-Cours (6172) Cores 448 (1.15GHz)
CUDA Programming Model
 CUDA Xing Zeng, Dongyue Mou Introduction Example Pro & Contra Trend Introduction Example Pro & Contra Trend Introduction What is CUDA? - Compute Unified Device Architecture. - A powerful parallel programming
CUDA Xing Zeng, Dongyue Mou Introduction Example Pro & Contra Trend Introduction Example Pro & Contra Trend Introduction What is CUDA? - Compute Unified Device Architecture. - A powerful parallel programming
Modern Processor Architectures (A compiler writer s perspective) L25: Modern Compiler Design
 Modern Processor Architectures (A compiler writer s perspective) L25: Modern Compiler Design The 1960s - 1970s Instructions took multiple cycles Only one instruction in flight at once Optimisation meant
Modern Processor Architectures (A compiler writer s perspective) L25: Modern Compiler Design The 1960s - 1970s Instructions took multiple cycles Only one instruction in flight at once Optimisation meant
CUDA Performance Optimization. Patrick Legresley
 CUDA Performance Optimization Patrick Legresley Optimizations Kernel optimizations Maximizing global memory throughput Efficient use of shared memory Minimizing divergent warps Intrinsic instructions Optimizations
CUDA Performance Optimization Patrick Legresley Optimizations Kernel optimizations Maximizing global memory throughput Efficient use of shared memory Minimizing divergent warps Intrinsic instructions Optimizations
Introduction to Numerical General Purpose GPU Computing with NVIDIA CUDA. Part 1: Hardware design and programming model
 Introduction to Numerical General Purpose GPU Computing with NVIDIA CUDA Part 1: Hardware design and programming model Dirk Ribbrock Faculty of Mathematics, TU dortmund 2016 Table of Contents Why parallel
Introduction to Numerical General Purpose GPU Computing with NVIDIA CUDA Part 1: Hardware design and programming model Dirk Ribbrock Faculty of Mathematics, TU dortmund 2016 Table of Contents Why parallel
