Epigenetic regulation of the nuclear-coded GCAT and SHMT2 genes confers
|
|
|
- Basil Goodwin
- 5 years ago
- Views:
Transcription
1 Supplementary Information for Epigenetic regulation of the nuclear-coded GCAT and SHMT2 genes confers human age-associated mitochondrial respiration defects Osamu Hashizume, Sakiko Ohnishi, Takayuki Mito, Akinori Shimizu, Kaori Ishikawa, Kazuto Nakada, Manabu Soda, Hiroyuki Mano, Sumie Togayachi, Hiroyuki Miyoshi, Keisuke Okita & Jun-Ichi Hayashi
2 Supplementary figure legends Supplementary Figure 1. Effects of aging on mtdna copy number and morphology of mitochondria. (a) Comparison of mtdna copy number between young and elderly groups. The young group (black bars) consisted of the fibroblast lines TIG3S (fetus), TIG121 (age 8 months), TIG120 (6 years), and TIG118 (12 years). The elderly group (open bars) consisted of TIG106 (80 years), TIG107 (81 years), TIG101 (86 years), and TIG102 (97 years). Average indicates the average mtdna copy number of four fibroblast lines. (b) Comparison of mitochondrial morphologies. Fibroblasts were treated with Mitotracker Red to stain mitochondria and 4',6-diamidino-2-phenylindole (DAPI) to stain nuclei. The higher intensity of mitochondrial staining with Mitotracker Red reflects the higher membrane potential and higher respiratory function in TIG3S (fetus) than in TIG107 (81 years). Bars, 20 µm. Supplementary Figure 2. Quantitative estimation of mutation frequency at each nucleotide position of mtdna from eight fibroblast lines by deep sequence analysis. Red and blue dots represent the frequencies of mutations at each nucleotide position of mtdna from the young group and the elderly group, respectively. Graphs show mutation frequency versus mutation frequency rank. A higher rank represents a higher frequency of the mutation at each nucleotide position. The young group consists of TIG3S (fetus), TIG121 (8 months), TIG120 (6 years), and TIG118 (12 years). The elderly group consists of TIG106 (80 years), TIG107 (81 years), TIG101 (86 years), and TIG102 (97 years).
3 Supplementary Figure 3. Characterization of human induced pluripotent stem cells isolated from the fibroblast lines TIG3S (fetus), TIG121 (age 8 months), TIG107 (81 years), and TIG102 (97 years). (a) Small colonies with flat embryonic-stem-cell-like morphology were obtained about 4 weeks after transfection with the gene set OCT3/4, SOX2, KFL4, L-MYC, LIN28, and p53shrna. (b) Examination of pluripotency marker genes by using antibodies to Nanog, TRA-1-60, and SSEA4. Expression of Nanog is shown in red (Alexa Fluor 488), and that of TRA-1-60 or SSEA4 is shown in green (Alexa Fluor 594). Bars, 100 µm. Supplementary Figure 4. Selection of six nuclear-coded genes associated with age-associated mitochondrial respiration defects in human fibroblasts. (a) Schematic representation of the process of selection of nuclear-coded genes. From 27,958 genes on the microarray, 1446 genes were selected by using the gene ontology (GO) term mitochondria. From those 1446 genes, 144, 28, 128, and 71 genes were selected by using the GO terms respiration, aging, translation, and amino acids, respectively. Finally, six genes that were commonly more than 1.5-fold upregulated or downregulated in fibroblast lines from elderly subjects were selected by microarray analysis (solid lines). To examine aging phenotypes other than mitochondrial respiration defects, we used the GO terms aging and autophagy. We then searched for genes that were commonly more than 1.5-fold upregulated or downregulated in fibroblast lines from elderly subjects (broken lines). No genes were selected in this part of the
4 microarray analysis. (b) Heatmaps depicting transcripts of genes associated with respiration, aging, translation, and amino acids in mitochondria. Values represent log2 ratios of the average array signals of TIG107 and TIG102 compared with those of TIG3S and TIG121 (TIG107/TIG3S, TIG102/TIG3S, TIG107/TIG121 and TIG102/TIG121). Genes down- and upregulated in the fibroblast lines from elderly subjects (TIG107 and TIG102) are shown in red and green, respectively. Supplementary Figure 5. Comparison of mrna levels of the candidate six genes in the young and elderly groups. The six gene candidates for regulation of age-associated respiration defects were selected by using gene ontology terms and a microarray heatmap (Supplementary Fig. 4b). The young group consisted of fibroblast lines TIG3S (fetus), TIG121 (age 8 months), TIG120 (6 years), and TIG118 (12 years). The elderly group consisted of lines TIG106 (80 years), TIG107 (81 years), TIG101 (86 years), and TIG102 (97 years). Levels of transcripts were normalized against UBC expression. Of the six genes examined, age-associate regulation was confirmed to be present in MRPL28, EHHADH, and GCAT. Supplementary Figure 6. Effects of glycine treatment on the mitochondrial respiratory function of TIG102. Glycine (140 µm) was included in the medium for 10 days before the estimation of O 2 consumption rates. Experiments were performed in triplicate; error bars indicate ± SD. *P < 0.05.
5 Supplementary Figure 1 a mtdna content / cell Donor age TIG line b 0 Fetus 8mo 6y 12y 80y 81y 86y 97y 3S Young group Elderly group Young (TIG121) Young Elderly group group Average Elderly (TIG107) Mitotracker Red DAPI
6 Supplementary Figure 2
7 Supplementary Figure 3 a 3S-iPSCs 121-iPSCs 107-iPSCs 102-iPSCs b Nanog TRA-1-60 SSEA-4
8 Supplementary Figure 4 a Microarray No. of genes 27,958 GO term Mitochondria Aging No. of genes GO term Respiration Aging Translation Amino acids Autophagy No. of genes Heatmap of microarray 1.5 times up- or 1/1.5 times downregulation No. of genes
9 b Gene ontology terms Repiration Translation Aging Amino acid 102/ / /3S 107/3S 102/ / /3S 107/3S 102/ / /3S 107/3S 102/ / /3S 107/3S Total 28 genes Total 71 genes Total 128 genes -4 Total 144 genes -2 0 value 2 4
10 Supplementary Figure 5 mrna (%) ALDH5A1 0 Donor age Fetus 8mo 6y 12y 80y 81y 86y 97y TIG line 3S Young group Elderly group mrna (%) COX7A Fetus 8mo 6y 12y 80y 81y 86y 97y 3S Young group Elderly group mrna (%) CAPRIN Fetus 8mo 6y 12y 80y 81y 86y 97y 3S Young group Elderly group Donor age Fetus mrna (%) TIG line MRPL28 EHHADH GCAT * * * 8mo 6y 12y 80y 81y 86y 97y 3S Young group Elderly group mrna (%) Fetus 8mo 6y 12y 80y 81y 86y 97y 3S Young group Elderly group Fetus 8mo 6y 12y 80y 81y 86y 97y mrna (%) 3S Young group Elderly group
11 Supplementary Figure * O 2 consumption rate (% / s / 10 6 cells / vol.) Glycine TIG3S TIG102
12 Table S1. Characterization of the frequent mutations existing in more than 90% mtdna revealed by the deep sequence analysis of eight fibroblast lines. Human Position a Gene Ref a Alteration Related Total read Alteration Alteration Indel Amino acid fibroblast disease b number read number ratio (%) type change lines TIG3S 103 D-loop A G polymorphism (fetus) 199 C T polymorphism C T polymorphism A G polymorphism RNR1 A G polymorphism A G polymorphism C CA polymorphism ins A G polymorphism RNR2 A G polymorphism T TA polymorphism ins ND1 T C polymorphism ND2 G A polymorphism A G polymorphism T-A 6570 COX1 A AG polymorphism ins ATP8/ATP6 A G polymorphism T-A (ATP6) 8569 ATP6 A G polymorphism G A polymorphism C T polymorphism H-Y 8814 T C polymorphism COX3 C T polymorphism C T polymorphism ND3 G A polymorphism ND4 T C polymorphism C T polymorphism C T polymorphism A G polymorphism TRNH A G polymorphism ND5 G A polymorphism C T polymorphism T C polymorphism ND6 C T polymorphism CYTB C T polymorphism T-I T C polymorphism T C polymorphism G A polymorphism I-V G A polymorphism I-V
13 15912 TRNT A G polymorphism D-loop C T polymorphism C T polymorphism C T polymorphism C T polymorphism G A polymorphism C T polymorphism T A polymorphism C T polymorphism total numbers of alterations 45 TIG D-loop A G polymorphism (8m) 152 T C polymorphism C T polymorphism C T polymorphism T C polymorphism A AC polymorphism ins RNR1 A G polymorphism C CA polymorphism ins A G polymorphism RNR2 A G polymorphism G A polymorphism ND2 C T polymorphism C A polymorphism L-M 6569 COX1 A AG polymorphism ins A AG polymorphism ins COX2 G A polymorphism ATP8 C T polymorphism L-F 8435 T C polymorphism ATP6 A G polymorphism A G polymorphism T-A 8769 G A polymorphism T C polymorphism COX3 C T polymorphism ND3 C T polymorphism C T polymorphism ND4 T C polymorphism T C polymorphism A G polymorphism TRNH A G polymorphism ND5 G A polymorphism C T polymorphism
14 13812 A G polymorphism A G polymorphism ND6 T C polymorphism Y-C C T polymorphism C T polymorphism CYTB C T polymorphism T-I T C polymorphism A G polymorphism T-A G A polymorphism G A polymorphism T C polymorphism G A polymorphism T C polymorphism T C polymorphism G A polymorphism I-V A G polymorphism I-V G A polymorphism TRNT A G polymorphism A G polymorphism D-loop C T polymorphism C T polymorphism G A polymorphism T A polymorphism T C polymorphism C T polymorphism total numbers of alterations 56 TIG D-loop A G polymorphism (6y) 194 C T polymorphism C T polymorphism G A polymorphism T C polymorphism RNR1 A G polymorphism C CA polymorphism ins A G polymorphism RNR2 C T polymorphism G A polymorphism ND1 G A polymorphism ND2 C A polymorphism L-M 5257 G A polymorphism A-T 6570 COX1 A AG polymorphism ins T C polymorphism
15 8399 ATP8 C T polymorphism ATP6 A G polymorphism A G polymorphism T-A 8769 G A polymorphism T C polymorphism COX3 G A polymorphism C T polymorphism ND3 C T polymorphism ND4 T C polymorphism T C polymorphism A G polymorphism TRNH A G polymorphism ND5 G A polymorphism C T polymorphism A G polymorphism T-A ND6 C T polymorphism C T polymorphism CYTB C T polymorphism T-I T C polymorphism G A polymorphism T C polymorphism G A polymorphism T C polymorphism G A polymorphism I-V G A polymorphism I-V TRNT A G polymorphism D-loop C T polymorphism T C polymorphism C T polymorphism C T polymorphism G A polymorphism T A polymorphism A G polymorphism T C polymorphism C T polymorphism total numbers of alterations 50 TIG D-loop A G polymorphism (12y) 152 T C polymorphism C T polymorphism C T polymorphism T C polymorphism
16 706 RNR1 A G polymorphism C CA polymorphism ins A G polymorphism RNR2 G A polymorphism C T polymorphism ND2 C T polymorphism C A polymorphism L-M 6493 COX1 C A polymorphism L-I 6570 A AG polymorphism ins ATP8 C T polymorphism L-F 8458 T C polymorphism ATP6 A G polymorphism A G polymorphism T-A 8769 G A polymorphism T C polymorphism COX3 C T polymorphism ND3 C T polymorphism TRNR T C polymorphism ND4 T C polymorphism T C polymorphism A G polymorphism TRNH A G polymorphism ND5 G A polymorphism C T polymorphism A G polymorphism ND6 C T polymorphism C T polymorphism CYTB C T polymorphism T-I T C polymorphism T C polymorphism G A polymorphism T C polymorphism G A polymorphism T C polymorphism G A polymorphism I-V G A polymorphism I-V TRNT A G polymorphism D-loop G A polymorphism C T polymorphism C T polymorphism G A polymorphism T A polymorphism
17 16304 A G polymorphism T C polymorphism total numbers of alterations 49 TIG D-loop A G polymorphism (80y) 146 T C polymorphism C T polymorphism C T polymorphism G A polymorphism T C polymorphism RNR1 C CA polymorphism ins A G polymorphism ND2 C T polymorphism A G polymorphism T-A 5103 T C polymorphism TRNA C T polymorphism COX1 A AG polymorphism ins A G polymorphism T C polymorphism COX2 G A polymorphism ATP6 A G polymorphism A G polymorphism T-A 8769 G A polymorphism T C polymorphism T C polymorphism L-P 9150 T C polymorphism COX3 A G polymorphism G A polymorphism C T polymorphism ND3 C T polymorphism ND4 T C polymorphism T C polymorphism A G polymorphism TRNH A G polymorphism TRNL2 T C polymorphism ND5 G A polymorphism C T polymorphism A G polymorphism ND6 T C polymorphism C T polymorphism C T polymorphism G A polymorphism
18 14751 CYTB C T polymorphism T-I T C polymorphism G A polymorphism T C polymorphism G A polymorphism T C polymorphism G A polymorphism I-V G A polymorphism I-V TRNT A G polymorphism D-loop C T polymorphism C T polymorphism G A polymorphism A G polymorphism C T polymorphism T A polymorphism A G polymorphism T C polymorphism C T polymorphism total numbers of alterations 56 TIG D-loop A G polymorphism (81y) 150 C T polymorphism T C polymorphism C T polymorphism C T polymorphism G - polymorphism del G A polymorphism T C polymorphism RNR1 A G polymorphism C CA polymorphism ins A G polymorphism RNR2 C T polymorphism T TA polymorphism ins ND1 T A polymorphism ND2 A G polymorphism COX1 G A polymorphism A AG polymorphism ins C A polymorphism COX2 T C polymorphism ATP6 A G polymorphism T-A 8769 G A polymorphism T C polymorphism
19 9530 COX3 A G polymorphism C T polymorphism ND3 C T polymorphism ND4 T C polymorphism T C polymorphism G A polymorphism A-T TRNH A G polymorphism ND5 G A polymorphism A G polymorphism C T polymorphism A G polymorphism ND6 T C polymorphism N-S C T polymorphism CYTB C T polymorphism T-I T C polymorphism G A polymorphism T C polymorphism I-T T C polymorphism G A polymorphism A T polymorphism T C polymorphism G A polymorphism I-V G A polymorphism I-V TRNT A G polymorphism TRNP T C polymorphism D-loop G A polymorphism C T polymorphism G A polymorphism C T polymorphism G A polymorphism T C polymorphism A G polymorphism C T polymorphism total numbers of alterations 55 TIG D-loop A G polymorphism (86y) 152 T C polymorphism C T polymorphism C T polymorphism T C polymorphism RNR1 A G polymorphism C CA polymorphism ins -
20 1594 A G polymorphism RNR2 G A polymorphism C T polymorphism ND1 G A polymorphism ND2 C A polymorphism L-M 6570 COX1 A AG polymorphism ins TRNK A G polymorphism ATP8 C T polymorphism L-F 8458 T C polymorphism ATP6 A G polymorphism A G polymorphism T-A 8769 G A polymorphism T C polymorphism COX3 C T polymorphism TRNG A G polymorphism ND3 C T polymorphism ND4 T C polymorphism T C polymorphism A G polymorphism TRNH A G polymorphism ND5 G A polymorphism C T polymorphism T C polymorphism ND6 C T polymorphism C T polymorphism CYTB C T polymorphism T-I T C polymorphism T C polymorphism G A polymorphism T C polymorphism G A polymorphism T C polymorphism G A polymorphism I-V G A polymorphism I-V TRNT A G polymorphism D-loop G A polymorphism C T polymorphism C T polymorphism G A polymorphism T A polymorphism A G polymorphism T C polymorphism
21 total numbers of alterations 49 TIG D-loop A G polymorphism (97y) 103 A G polymorphism C T polymorphism C T polymorphism A G polymorphism RNR1 C CA polymorphism ins A G polymorphism ND1 A G polymorphism COX1 A AG polymorphism ins TRND T C polymorphism COX2 A G polymorphism ATP6 G A polymorphism T C polymorphism COX3 C T polymorphism G A polymorphism ND4 T C polymorphism G A polymorphism C T polymorphism A G polymorphism TRNH A G polymorphism TRNS2 A G polymorphism ND5 G A polymorphism ND6 C T polymorphism C T polymorphism C T polymorphism C T polymorphism CYTB C T polymorphism T-I T C polymorphism A G polymorphism T C polymorphism G A polymorphism I-V G A polymorphism I-V TRNT A G polymorphism D-loop G A polymorphism T C polymorphism C T polymorphism C T polymorphism G A polymorphism C A polymorphism T A polymorphism
22 16304 A G polymorphism total numbers of alterations 41 a Registered under GenBank accession no. AB b All of the mutations existing in more than 90% of mtdna were polymorphic ones that have been reported in normal human subjects.
23 Table S2. Primers used for PCR and mtdna sequencing. Primer Primer sequence (5-3 ) Primer binding Fragment size (bp) Primer-1 for AAATGTTTAGACGGGCTCACATCA Primer-1 rev CCATTGCGATTAGAATGGGTACAA 3353 Primer-2 for ACCTAACAAACCCACAGGTCCTAA Primer-2 rev TAAAGTGGCTGATTTGCGTTCAGT 5634 Primer-3 for AAAAACTAGCCCCCATCTCAATCA Primer-3 rev AGGGCATACAGGACTAGGAAGCAG 7704 Primer-4 for GTGGCCTGACTGGCATTGTATTAG Primer-4 rev CGGATGAAGCAGATAGTGAGGAAA 9853 Primer-5 for AGCCATGTGATTTCACTTCCACTC Primer-5 rev GGGCATGAGTTAGCAGTTCTTGTG Primer-6 for TCAGCCACATAGCCCTCGTAGTAA Primer-6 rev GGGTTAGGATGAGTGGGAAGAAGA Primer-7 for CTCCAAAGACCACATCATCGAAAC Primer-7 rev GTAGCACTCTTGTGCGGGATATTG Primer-8 for GCGTCCTTGCCCTATTACTATCCA Primer-8 rev TGCGCTTACTTTGTAGCCTTCATC 1301
24 Table S3. Primers used for Real-time quantitative PCR. Gene Symbol Primer sequence (5-3 ) aldehyde dehydrogenase 5 family, member A1 ALDH5A1 F AGACCATCCTGGCTAACACG R GGAGTCTCGCTCTGTCATCC caprin family member 2 CAPRIN2 F CGAATGTGTTTCCCAGACCT R TTGGGAATAAGGTGCTCCAG cytochrome c oxidase subunit VIIa polypeptide 1 COX7A1 F CGCTTTCAGAACCGAGTGC R CCCAGCCAAGGGAGTACAAG enoyl-coa, hydratase/3-hydroxyacyl CoA dehydrogenase EHHADH F CAGCTTCTCCCCAGACTCAC R TGAATTGGCTTGTTGCAGAG glycine C-acetyltransferase GCAT F TCTCGCATGGCGGATGA R GCAGCGGTCAATGTCTTCCT mitochondrial ribosomal protein L28 MRPL28 F ATGCCAACAACGACAAGCTCT R CTGTAGAACTCTCGCTCAAACAGC serine hydroxymethyltransferase 2 SHMT2 F AGTCTATGCCCTATAAGCTCAACCC R GCCGGAAAAGTCGAGCAGT ubiquitin C UBC F TGGAGCCGAGTGACACCATT R TCAGCGAGGGTACGACCATCTTC
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3000 Supplementary Figure 1 Lgr5 expression in the Lgr5-LacZ reporter knock-in mouse. P1: Lgr5-LacZ expression on the ovary surface (black arrow) and sub-surface (arrowheads), as well as
DOI: 10.1038/ncb3000 Supplementary Figure 1 Lgr5 expression in the Lgr5-LacZ reporter knock-in mouse. P1: Lgr5-LacZ expression on the ovary surface (black arrow) and sub-surface (arrowheads), as well as
Yuan_FigS1. Control shrna. Vector. Eset shrna1. Eset shrna2. Eset overexpression. kda. anti-eset. anti-actin
 Yuan_FigS1 kda Vector Control shrna Eset shrna1 Eset shrna2 Eset overexpression 26 16 anti-eset 11 anti-actin Yuan_FigS2 A Relative expression (%) 12 1 8 6 4 2 Nanog Pou5f1 Cdx2 Hand1 Eset B Control shrna
Yuan_FigS1 kda Vector Control shrna Eset shrna1 Eset shrna2 Eset overexpression 26 16 anti-eset 11 anti-actin Yuan_FigS2 A Relative expression (%) 12 1 8 6 4 2 Nanog Pou5f1 Cdx2 Hand1 Eset B Control shrna
Computational Genomics and Molecular Biology, Fall
 Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
Supplementary Material. Cell type-specific termination of transcription by transposable element sequences
 Supplementary Material Cell type-specific termination of transcription by transposable element sequences Andrew B. Conley and I. King Jordan Controls for TTS identification using PET A series of controls
Supplementary Material Cell type-specific termination of transcription by transposable element sequences Andrew B. Conley and I. King Jordan Controls for TTS identification using PET A series of controls
CAP BIOINFORMATICS Su-Shing Chen CISE. 8/19/2005 Su-Shing Chen, CISE 1
 CAP 5510-2 BIOINFORMATICS Su-Shing Chen CISE 8/19/2005 Su-Shing Chen, CISE 1 Building Local Genomic Databases Genomic research integrates sequence data with gene function knowledge. Gene ontology to represent
CAP 5510-2 BIOINFORMATICS Su-Shing Chen CISE 8/19/2005 Su-Shing Chen, CISE 1 Building Local Genomic Databases Genomic research integrates sequence data with gene function knowledge. Gene ontology to represent
Topics of the talk. Biodatabases. Data types. Some sequence terminology...
 Topics of the talk Biodatabases Jarno Tuimala / Eija Korpelainen CSC What data are stored in biological databases? What constitutes a good database? Nucleic acid sequence databases Amino acid sequence
Topics of the talk Biodatabases Jarno Tuimala / Eija Korpelainen CSC What data are stored in biological databases? What constitutes a good database? Nucleic acid sequence databases Amino acid sequence
Supplementary information: Detection of differentially expressed segments in tiling array data
 Supplementary information: Detection of differentially expressed segments in tiling array data Christian Otto 1,2, Kristin Reiche 3,1,4, Jörg Hackermüller 3,1,4 July 1, 212 1 Bioinformatics Group, Department
Supplementary information: Detection of differentially expressed segments in tiling array data Christian Otto 1,2, Kristin Reiche 3,1,4, Jörg Hackermüller 3,1,4 July 1, 212 1 Bioinformatics Group, Department
Introduction to GE Microarray data analysis Practical Course MolBio 2012
 Introduction to GE Microarray data analysis Practical Course MolBio 2012 Claudia Pommerenke Nov-2012 Transkriptomanalyselabor TAL Microarray and Deep Sequencing Core Facility Göttingen University Medical
Introduction to GE Microarray data analysis Practical Course MolBio 2012 Claudia Pommerenke Nov-2012 Transkriptomanalyselabor TAL Microarray and Deep Sequencing Core Facility Göttingen University Medical
User Manual. Ver. 3.0 March 19, 2012
 User Manual Ver. 3.0 March 19, 2012 Table of Contents 1. Introduction... 2 1.1 Rationale... 2 1.2 Software Work-Flow... 3 1.3 New in GenomeGems 3.0... 4 2. Software Description... 5 2.1 Key Features...
User Manual Ver. 3.0 March 19, 2012 Table of Contents 1. Introduction... 2 1.1 Rationale... 2 1.2 Software Work-Flow... 3 1.3 New in GenomeGems 3.0... 4 2. Software Description... 5 2.1 Key Features...
Report Generator User Guide
 12 Tra Report Generator User Guide Agilent Seahorse XF Cell Mito Stress Test For research use only. 1 Table of Contents Introduction... 3 Parameter Calculations... 3 XF Report Generator Overview... 4 How
12 Tra Report Generator User Guide Agilent Seahorse XF Cell Mito Stress Test For research use only. 1 Table of Contents Introduction... 3 Parameter Calculations... 3 XF Report Generator Overview... 4 How
CLC Server. End User USER MANUAL
 CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
Web Resources. iphemap: An atlas of phenotype to genotype relationships of human ipsc models of neurological diseases
 Web Resources iphemap: An atlas of phenotype to genotype relationships of human ipsc models of neurological diseases Ethan W. Hollingsworth 1, 2, Jacob E. Vaughn 1, 2, Josh C. Orack 1,2, Chelsea Skinner
Web Resources iphemap: An atlas of phenotype to genotype relationships of human ipsc models of neurological diseases Ethan W. Hollingsworth 1, 2, Jacob E. Vaughn 1, 2, Josh C. Orack 1,2, Chelsea Skinner
Supplementary Information
 Supplementary Information Supplementary Fig 1. Megalin endocytosis in subconfluent MDCK cells (a) Schematic representation of mmeg-ha endocytic assay. Subconfluent mmeg-mdck cells were labeled for the
Supplementary Information Supplementary Fig 1. Megalin endocytosis in subconfluent MDCK cells (a) Schematic representation of mmeg-ha endocytic assay. Subconfluent mmeg-mdck cells were labeled for the
Tutorial 4 BLAST Searching the CHO Genome
 Tutorial 4 BLAST Searching the CHO Genome Accessing the CHO Genome BLAST Tool The CHO BLAST server can be accessed by clicking on the BLAST button on the home page or by selecting BLAST from the menu bar
Tutorial 4 BLAST Searching the CHO Genome Accessing the CHO Genome BLAST Tool The CHO BLAST server can be accessed by clicking on the BLAST button on the home page or by selecting BLAST from the menu bar
Data Walkthrough: Background
 Data Walkthrough: Background File Types FASTA Files FASTA files are text-based representations of genetic information. They can contain nucleotide or amino acid sequences. For this activity, students will
Data Walkthrough: Background File Types FASTA Files FASTA files are text-based representations of genetic information. They can contain nucleotide or amino acid sequences. For this activity, students will
NGS NEXT GENERATION SEQUENCING
 NGS NEXT GENERATION SEQUENCING Paestum (Sa) 15-16 -17 maggio 2014 Relatore Dr Cataldo Senatore Dr.ssa Emilia Vaccaro Sanger Sequencing Reactions For given template DNA, it s like PCR except: Uses only
NGS NEXT GENERATION SEQUENCING Paestum (Sa) 15-16 -17 maggio 2014 Relatore Dr Cataldo Senatore Dr.ssa Emilia Vaccaro Sanger Sequencing Reactions For given template DNA, it s like PCR except: Uses only
Browser Exercises - I. Alignments and Comparative genomics
 Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
Genome Environment Browser (GEB) user guide
 Genome Environment Browser (GEB) user guide GEB is a Java application developed to provide a dynamic graphical interface to visualise the distribution of genome features and chromosome-wide experimental
Genome Environment Browser (GEB) user guide GEB is a Java application developed to provide a dynamic graphical interface to visualise the distribution of genome features and chromosome-wide experimental
Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex
 CORRECTION NOTICE Nat. Struct. Mol. Biol. advance online publication, doi:10.1038/nsmb.1618 (7 June 2009) Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex
CORRECTION NOTICE Nat. Struct. Mol. Biol. advance online publication, doi:10.1038/nsmb.1618 (7 June 2009) Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex
ViTraM: VIsualization of TRAnscriptional Modules
 ViTraM: VIsualization of TRAnscriptional Modules Version 2.0 October 1st, 2009 KULeuven, Belgium 1 Contents 1 INTRODUCTION AND INSTALLATION... 4 1.1 Introduction...4 1.2 Software structure...5 1.3 Requirements...5
ViTraM: VIsualization of TRAnscriptional Modules Version 2.0 October 1st, 2009 KULeuven, Belgium 1 Contents 1 INTRODUCTION AND INSTALLATION... 4 1.1 Introduction...4 1.2 Software structure...5 1.3 Requirements...5
EBiSC Catalogue User Guide
 EBiSC Catalogue User Guide European Bank for induced pluripotent Stem Cells https://cells.ebisc.org & http://ebisc.org/ The EBiSC - European Bank for induced pluripotent Stem Cells project has received
EBiSC Catalogue User Guide European Bank for induced pluripotent Stem Cells https://cells.ebisc.org & http://ebisc.org/ The EBiSC - European Bank for induced pluripotent Stem Cells project has received
Characters and Character matrices
 Introduction Why? How to Cite Publication Support Credits Help FAQ Web Site Simplicity Files Menus Windows Charts Scripts/Macros Modules How Characters Taxa Trees Glossary New Features Characters and Character
Introduction Why? How to Cite Publication Support Credits Help FAQ Web Site Simplicity Files Menus Windows Charts Scripts/Macros Modules How Characters Taxa Trees Glossary New Features Characters and Character
Supplementary Material VDJServer: A Cloud-Based Analysis Portal and Data Commons for Immune Repertoire Sequences and Rearrangements
 Supplementary Material VDJServer: A Cloud-Based Analysis Portal and Data Commons for Immune Repertoire Sequences and Rearrangements Scott Christley, Walter Scarborough, Eddie Salinas, William H. Rounds,
Supplementary Material VDJServer: A Cloud-Based Analysis Portal and Data Commons for Immune Repertoire Sequences and Rearrangements Scott Christley, Walter Scarborough, Eddie Salinas, William H. Rounds,
Database Searching Using BLAST
 Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of
Finding sequence similarities with genes of known function is a common approach to infer a newly sequenced gene s function
 Outline DNA Sequence Comparison: First Success Stories Change Problem Manhattan Tourist Problem Longest Paths in Graphs Sequence Alignment Edit Distance Longest Common Subsequence Problem Dot Matrices
Outline DNA Sequence Comparison: First Success Stories Change Problem Manhattan Tourist Problem Longest Paths in Graphs Sequence Alignment Edit Distance Longest Common Subsequence Problem Dot Matrices
Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014
 Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014 Dynamic programming is a group of mathematical methods used to sequentially split a complicated problem into
Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014 Dynamic programming is a group of mathematical methods used to sequentially split a complicated problem into
Quantifying Three-Dimensional Deformations of Migrating Fibroblasts
 45 Chapter 4 Quantifying Three-Dimensional Deformations of Migrating Fibroblasts This chapter presents the full-field displacements and tractions of 3T3 fibroblast cells during migration on polyacrylamide
45 Chapter 4 Quantifying Three-Dimensional Deformations of Migrating Fibroblasts This chapter presents the full-field displacements and tractions of 3T3 fibroblast cells during migration on polyacrylamide
Tutorial for Windows and Macintosh SNP Hunting
 Tutorial for Windows and Macintosh SNP Hunting 2010 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Tutorial for Windows and Macintosh SNP Hunting 2010 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Dr. Gabriela Salinas Dr. Orr Shomroni Kaamini Rhaithata
 Analysis of RNA sequencing data sets using the Galaxy environment Dr. Gabriela Salinas Dr. Orr Shomroni Kaamini Rhaithata Microarray and Deep-sequencing core facility 30.10.2017 RNA-seq workflow I Hypothesis
Analysis of RNA sequencing data sets using the Galaxy environment Dr. Gabriela Salinas Dr. Orr Shomroni Kaamini Rhaithata Microarray and Deep-sequencing core facility 30.10.2017 RNA-seq workflow I Hypothesis
and genomic DNA (Mut) and WT of patient healthy women and
 Supplementary Figure 1 TEKT germline variations in the control group and blood sampless of 84 breast cancerr patients. (a) TEKT44 wild-typee (WT), mutant (Mut) and WT to Mut genotype byy Sanger sequencing
Supplementary Figure 1 TEKT germline variations in the control group and blood sampless of 84 breast cancerr patients. (a) TEKT44 wild-typee (WT), mutant (Mut) and WT to Mut genotype byy Sanger sequencing
ROTS: Reproducibility Optimized Test Statistic
 ROTS: Reproducibility Optimized Test Statistic Fatemeh Seyednasrollah, Tomi Suomi, Laura L. Elo fatsey (at) utu.fi March 3, 2016 Contents 1 Introduction 2 2 Algorithm overview 3 3 Input data 3 4 Preprocessing
ROTS: Reproducibility Optimized Test Statistic Fatemeh Seyednasrollah, Tomi Suomi, Laura L. Elo fatsey (at) utu.fi March 3, 2016 Contents 1 Introduction 2 2 Algorithm overview 3 3 Input data 3 4 Preprocessing
Goal-oriented Schema in Biological Database Design
 Goal-oriented Schema in Biological Database Design Ping Chen Department of Computer Science University of Helsinki Helsinki, Finland 00014 EMAIL: pchen@cs.helsinki.fi Abstract In this paper, I reviewed
Goal-oriented Schema in Biological Database Design Ping Chen Department of Computer Science University of Helsinki Helsinki, Finland 00014 EMAIL: pchen@cs.helsinki.fi Abstract In this paper, I reviewed
Genome Browsers - The UCSC Genome Browser
 Genome Browsers - The UCSC Genome Browser Background The UCSC Genome Browser is a well-curated site that provides users with a view of gene or sequence information in genomic context for a specific species,
Genome Browsers - The UCSC Genome Browser Background The UCSC Genome Browser is a well-curated site that provides users with a view of gene or sequence information in genomic context for a specific species,
How to perform module-based differential correlation analysis and GO enrichment using DGCA Andrew McKenzie, Bin Zhang Oct 24, 2016
 How to perform module-based differential correlation analysis and GO enrichment using DGCA Andrew McKenzie, Bin Zhang Oct 24, 2016 Contents 1 Introduction 1 2 Data loading and module construction 1 3 Module-based
How to perform module-based differential correlation analysis and GO enrichment using DGCA Andrew McKenzie, Bin Zhang Oct 24, 2016 Contents 1 Introduction 1 2 Data loading and module construction 1 3 Module-based
VectorBase Web Apollo April Web Apollo 1
 Web Apollo 1 Contents 1. Access points: Web Apollo, Genome Browser and BLAST 2. How to identify genes that need to be annotated? 3. Gene manual annotations 4. Metadata 1. Access points Web Apollo tool
Web Apollo 1 Contents 1. Access points: Web Apollo, Genome Browser and BLAST 2. How to identify genes that need to be annotated? 3. Gene manual annotations 4. Metadata 1. Access points Web Apollo tool
Supplementary Fig. S1. Woloszynska-Read et al.
 Supplementary Fig. S1. Woloszynska-Read et al. A % BORIS Methylat tion Normal Ovary P
Supplementary Fig. S1. Woloszynska-Read et al. A % BORIS Methylat tion Normal Ovary P
Information Resources in Molecular Biology Marcela Davila-Lopez How many and where
 Information Resources in Molecular Biology Marcela Davila-Lopez (marcela.davila@medkem.gu.se) How many and where Data growth DB: What and Why A Database is a shared collection of logically related data,
Information Resources in Molecular Biology Marcela Davila-Lopez (marcela.davila@medkem.gu.se) How many and where Data growth DB: What and Why A Database is a shared collection of logically related data,
Wilson Leung 05/27/2008 A Simple Introduction to NCBI BLAST
 A Simple Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at http://www.ncbi.nih.gov/blast/
A Simple Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at http://www.ncbi.nih.gov/blast/
Definiens. Tissue Studio 4.4. Tutorial 4: Manual ROI Selection and Marker Area Detection
 Definiens Tissue Studio 4.4 Tutorial 4: Manual ROI Selection and Marker Area Detection Tutorial 4: Manual ROI Selection and Marker Area Detection Imprint and Version Copyright 2017 Definiens AG. All rights
Definiens Tissue Studio 4.4 Tutorial 4: Manual ROI Selection and Marker Area Detection Tutorial 4: Manual ROI Selection and Marker Area Detection Imprint and Version Copyright 2017 Definiens AG. All rights
Biostatistics and Bioinformatics Molecular Sequence Databases
 . 1 Description of Module Subject Name Paper Name Module Name/Title 13 03 Dr. Vijaya Khader Dr. MC Varadaraj 2 1. Objectives: In the present module, the students will learn about 1. Encoding linear sequences
. 1 Description of Module Subject Name Paper Name Module Name/Title 13 03 Dr. Vijaya Khader Dr. MC Varadaraj 2 1. Objectives: In the present module, the students will learn about 1. Encoding linear sequences
Tutorial: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and Expression measures
 : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
all M 2M_gt_15 2M_8_15 2M_1_7 gt_2m TopHat2
 Pairs processed per second 6, 4, 2, 6, 4, 2, 6, 4, 2, 6, 4, 2, 6, 4, 2, 6, 4, 2, 72,318 418 1,666 49,495 21,123 69,984 35,694 1,9 71,538 3,5 17,381 61,223 69,39 55 19,579 44,79 65,126 96 5,115 33,6 61,787
Pairs processed per second 6, 4, 2, 6, 4, 2, 6, 4, 2, 6, 4, 2, 6, 4, 2, 6, 4, 2, 72,318 418 1,666 49,495 21,123 69,984 35,694 1,9 71,538 3,5 17,381 61,223 69,39 55 19,579 44,79 65,126 96 5,115 33,6 61,787
Machine Learning. Computational biology: Sequence alignment and profile HMMs
 10-601 Machine Learning Computational biology: Sequence alignment and profile HMMs Central dogma DNA CCTGAGCCAACTATTGATGAA transcription mrna CCUGAGCCAACUAUUGAUGAA translation Protein PEPTIDE 2 Growth
10-601 Machine Learning Computational biology: Sequence alignment and profile HMMs Central dogma DNA CCTGAGCCAACTATTGATGAA transcription mrna CCUGAGCCAACUAUUGAUGAA translation Protein PEPTIDE 2 Growth
Clustering Mixed Numerical and Low Quality Categorical Data: Significance Metrics on a Yeast Example
 Clustering Mixed Numerical and Low Quality Categorical Data: Significance Metrics on a Yeast Example Bill Andreopoulos, Aijun An, Xiaogang Wang York University IQIS 2005 June 17, 2005 What is clustering?
Clustering Mixed Numerical and Low Quality Categorical Data: Significance Metrics on a Yeast Example Bill Andreopoulos, Aijun An, Xiaogang Wang York University IQIS 2005 June 17, 2005 What is clustering?
A tree-structured index algorithm for Expressed Sequence Tags clustering
 A tree-structured index algorithm for Expressed Sequence Tags clustering Benjamin Kumwenda 0408046X Supervisor: Professor Scott Hazelhurst April 21, 2008 Declaration I declare that this dissertation is
A tree-structured index algorithm for Expressed Sequence Tags clustering Benjamin Kumwenda 0408046X Supervisor: Professor Scott Hazelhurst April 21, 2008 Declaration I declare that this dissertation is
The UCSC Genome Browser
 The UCSC Genome Browser Search, retrieve and display the data that you want Materials prepared by Warren C. Lathe, Ph.D. Mary Mangan, Ph.D. www.openhelix.com Updated: Q3 2006 Version_0906 Copyright OpenHelix.
The UCSC Genome Browser Search, retrieve and display the data that you want Materials prepared by Warren C. Lathe, Ph.D. Mary Mangan, Ph.D. www.openhelix.com Updated: Q3 2006 Version_0906 Copyright OpenHelix.
All mirnas (clustered and individual) were inserted into a multiple cloning site in the 3'-UTR of a fluorescent marker.
 Supplementary Figure 1. Vector schematic for UAS-miRNA generation. All mirnas (clustered and individual) were inserted into a multiple cloning site in the 3'-UTR of a fluorescent marker. Supplementary
Supplementary Figure 1. Vector schematic for UAS-miRNA generation. All mirnas (clustered and individual) were inserted into a multiple cloning site in the 3'-UTR of a fluorescent marker. Supplementary
Supplementary Table S1. Sequences of primers used for cloning of neuropeptide-coding genes. Fw: Forward primer; Rv: Reverse primer.
 Supplementary Table S1. Sequences of primers used for cloning of neuropeptide-coding genes. Fw: Forward primer; Rv: Reverse primer. Gene name DjNp3 DjNp4 DjNp9 DjNp12 DjNp17 DjNp19 DjNp23 DjNp25 DjNp28
Supplementary Table S1. Sequences of primers used for cloning of neuropeptide-coding genes. Fw: Forward primer; Rv: Reverse primer. Gene name DjNp3 DjNp4 DjNp9 DjNp12 DjNp17 DjNp19 DjNp23 DjNp25 DjNp28
Tutorial for Windows and Macintosh SNP Hunting
 Tutorial for Windows and Macintosh SNP Hunting 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Tutorial for Windows and Macintosh SNP Hunting 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
ViTraM: VIsualization of TRAnscriptional Modules
 ViTraM: VIsualization of TRAnscriptional Modules Version 1.0 June 1st, 2009 Hong Sun, Karen Lemmens, Tim Van den Bulcke, Kristof Engelen, Bart De Moor and Kathleen Marchal KULeuven, Belgium 1 Contents
ViTraM: VIsualization of TRAnscriptional Modules Version 1.0 June 1st, 2009 Hong Sun, Karen Lemmens, Tim Van den Bulcke, Kristof Engelen, Bart De Moor and Kathleen Marchal KULeuven, Belgium 1 Contents
Dynamic Programming: Edit Distance
 Dynamic Programming: Edit Distance Outline. DNA Sequence Comparison and CF. Change Problem. Manhattan Tourist Problem. Longest Paths in Graphs. Sequence Alignment 6. Edit Distance Section : DNA Sequence
Dynamic Programming: Edit Distance Outline. DNA Sequence Comparison and CF. Change Problem. Manhattan Tourist Problem. Longest Paths in Graphs. Sequence Alignment 6. Edit Distance Section : DNA Sequence
Tutorial 1: Exploring the UCSC Genome Browser
 Last updated: May 12, 2011 Tutorial 1: Exploring the UCSC Genome Browser Open the homepage of the UCSC Genome Browser at: http://genome.ucsc.edu/ In the blue bar at the top, click on the Genomes link.
Last updated: May 12, 2011 Tutorial 1: Exploring the UCSC Genome Browser Open the homepage of the UCSC Genome Browser at: http://genome.ucsc.edu/ In the blue bar at the top, click on the Genomes link.
PLNT4610 BIOINFORMATICS FINAL EXAMINATION
 PLNT4610 BIOINFORMATICS FINAL EXAMINATION 18:00 to 20:00 Thursday December 13, 2012 Answer any combination of questions totalling to exactly 100 points. The questions on the exam sheet total to 120 points.
PLNT4610 BIOINFORMATICS FINAL EXAMINATION 18:00 to 20:00 Thursday December 13, 2012 Answer any combination of questions totalling to exactly 100 points. The questions on the exam sheet total to 120 points.
GeneSifter.Net User s Guide
 GeneSifter.Net User s Guide 1 2 GeneSifter.Net Overview Login Upload Tools Pairwise Analysis Create Projects For more information about a feature see the corresponding page in the User s Guide noted in
GeneSifter.Net User s Guide 1 2 GeneSifter.Net Overview Login Upload Tools Pairwise Analysis Create Projects For more information about a feature see the corresponding page in the User s Guide noted in
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Detailed schematic representation of SuRE methodology. See Methods for detailed description. a. Size-selected and A-tailed random fragments ( queries ) of the human genome are inserted
Supplementary Figure 1 Detailed schematic representation of SuRE methodology. See Methods for detailed description. a. Size-selected and A-tailed random fragments ( queries ) of the human genome are inserted
Managing big biological sequence data with Biostrings and DECIPHER. Erik Wright University of Wisconsin-Madison
 Managing big biological sequence data with Biostrings and DECIPHER Erik Wright University of Wisconsin-Madison What you should learn How to use the Biostrings and DECIPHER packages Creating a database
Managing big biological sequence data with Biostrings and DECIPHER Erik Wright University of Wisconsin-Madison What you should learn How to use the Biostrings and DECIPHER packages Creating a database
The Brigham and Women's Hospital, Inc. and President and Fellows of Harvard College
 2006-2017 The Brigham and Women's Hospital, Inc. and President and Fellows of Harvard College This documentation file supports software published as part of: Michael F. Berger and Martha L. Bulyk. Universal
2006-2017 The Brigham and Women's Hospital, Inc. and President and Fellows of Harvard College This documentation file supports software published as part of: Michael F. Berger and Martha L. Bulyk. Universal
The Human PAX6 Mutation Database
 1998 Oxford University Press Nucleic Acids Research, 1998, Vol. 26, No. 1 259 264 The Human PAX6 Mutation Database Alastair Brown*, Mark McKie, Veronica van Heyningen and Jane Prosser Medical Research
1998 Oxford University Press Nucleic Acids Research, 1998, Vol. 26, No. 1 259 264 The Human PAX6 Mutation Database Alastair Brown*, Mark McKie, Veronica van Heyningen and Jane Prosser Medical Research
TECH NOTE Improving the Sensitivity of Ultra Low Input mrna Seq
 TECH NOTE Improving the Sensitivity of Ultra Low Input mrna Seq SMART Seq v4 Ultra Low Input RNA Kit for Sequencing Powered by SMART and LNA technologies: Locked nucleic acid technology significantly improves
TECH NOTE Improving the Sensitivity of Ultra Low Input mrna Seq SMART Seq v4 Ultra Low Input RNA Kit for Sequencing Powered by SMART and LNA technologies: Locked nucleic acid technology significantly improves
Sept. 9, An Introduction to Bioinformatics. Special Topics BSC5936:
 Special Topics BSC5936: An Introduction to Bioinformatics. Florida State University The Department of Biological Science www.bio.fsu.edu Sept. 9, 2003 The Dot Matrix Method Steven M. Thompson Florida State
Special Topics BSC5936: An Introduction to Bioinformatics. Florida State University The Department of Biological Science www.bio.fsu.edu Sept. 9, 2003 The Dot Matrix Method Steven M. Thompson Florida State
Intro to NGS Tutorial
 Intro to NGS Tutorial Release 8.6.0 Golden Helix, Inc. October 31, 2016 Contents 1. Overview 2 2. Import Variants and Quality Fields 3 3. Quality Filters 10 Generate Alternate Read Ratio.........................................
Intro to NGS Tutorial Release 8.6.0 Golden Helix, Inc. October 31, 2016 Contents 1. Overview 2 2. Import Variants and Quality Fields 3 3. Quality Filters 10 Generate Alternate Read Ratio.........................................
Special course in Computer Science: Advanced Text Algorithms
 Special course in Computer Science: Advanced Text Algorithms Lecture 6: Alignments Elena Czeizler and Ion Petre Department of IT, Abo Akademi Computational Biomodelling Laboratory http://www.users.abo.fi/ipetre/textalg
Special course in Computer Science: Advanced Text Algorithms Lecture 6: Alignments Elena Czeizler and Ion Petre Department of IT, Abo Akademi Computational Biomodelling Laboratory http://www.users.abo.fi/ipetre/textalg
Easy visualization of the read coverage using the CoverageView package
 Easy visualization of the read coverage using the CoverageView package Ernesto Lowy European Bioinformatics Institute EMBL June 13, 2018 > options(width=40) > library(coverageview) 1 Introduction This
Easy visualization of the read coverage using the CoverageView package Ernesto Lowy European Bioinformatics Institute EMBL June 13, 2018 > options(width=40) > library(coverageview) 1 Introduction This
B. Graphing Representation of Data
 B Graphing Representation of Data The second way of displaying data is by use of graphs Although such visual aids are even easier to read than tables, they often do not give the same detail It is essential
B Graphing Representation of Data The second way of displaying data is by use of graphs Although such visual aids are even easier to read than tables, they often do not give the same detail It is essential
Expression pattern of nub-gal4. The expression pattern of nub-gal4 was. assayed by combining it with the reporter UAS-His2ADsRed as in Figure 1.
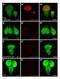 Supplementary Figure 1. Expression pattern of nub-gal4. The expression pattern of nub-gal4 was assayed by combining it with the reporter UAS-His2ADsRed as in Figure 1. His2ADsRed is localized to the nuclei
Supplementary Figure 1. Expression pattern of nub-gal4. The expression pattern of nub-gal4 was assayed by combining it with the reporter UAS-His2ADsRed as in Figure 1. His2ADsRed is localized to the nuclei
Filogeografía BIOL 4211, Universidad de los Andes 25 de enero a 01 de abril 2006
 Laboratory excercise written by Andrew J. Crawford with the support of CIES Fulbright Program and Fulbright Colombia. Enjoy! Filogeografía BIOL 4211, Universidad de los Andes 25 de enero
Laboratory excercise written by Andrew J. Crawford with the support of CIES Fulbright Program and Fulbright Colombia. Enjoy! Filogeografía BIOL 4211, Universidad de los Andes 25 de enero
Documentation for running Normfinder in R
 Documentation for running Normfinder in R December 18, 2013 The Normfinder algorithm NormFinder is an algorithm for identifying the optimal normalization gene among a set of candidates. It ranks the set
Documentation for running Normfinder in R December 18, 2013 The Normfinder algorithm NormFinder is an algorithm for identifying the optimal normalization gene among a set of candidates. It ranks the set
CLUSTERING IN BIOINFORMATICS
 CLUSTERING IN BIOINFORMATICS CSE/BIMM/BENG 8 MAY 4, 0 OVERVIEW Define the clustering problem Motivation: gene expression and microarrays Types of clustering Clustering algorithms Other applications of
CLUSTERING IN BIOINFORMATICS CSE/BIMM/BENG 8 MAY 4, 0 OVERVIEW Define the clustering problem Motivation: gene expression and microarrays Types of clustering Clustering algorithms Other applications of
ChIP-seq Analysis Practical
 ChIP-seq Analysis Practical Vladimir Teif (vteif@essex.ac.uk) An updated version of this document will be available at http://generegulation.info/index.php/teaching In this practical we will learn how
ChIP-seq Analysis Practical Vladimir Teif (vteif@essex.ac.uk) An updated version of this document will be available at http://generegulation.info/index.php/teaching In this practical we will learn how
Supplementary Table S1. mirnas with different expression levels were tested by qrt-pcr.
 Supplementary Table S1. mirnas with different expression levels were tested by qrt-pcr. Average expression level(tpm) >5000 1000-5000 mirna_name HSCs(TPM) imdcs(tpm) madcs(tpm) DCregs(TPM) Average(TPM)
Supplementary Table S1. mirnas with different expression levels were tested by qrt-pcr. Average expression level(tpm) >5000 1000-5000 mirna_name HSCs(TPM) imdcs(tpm) madcs(tpm) DCregs(TPM) Average(TPM)
White, Maximum Symmetry in the Genetic Code.
 White, Maximum Symmetry in the Genetic Code www.codefun.com/genetic_max.htm Title Title, Maximum Symmetry Symmetry refers to a type of pattern that organizes something s shape General shape is invariant
White, Maximum Symmetry in the Genetic Code www.codefun.com/genetic_max.htm Title Title, Maximum Symmetry Symmetry refers to a type of pattern that organizes something s shape General shape is invariant
The Allen Human Brain Atlas offers three types of searches to allow a user to: (1) obtain gene expression data for specific genes (or probes) of
 Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
FunRich Tool Documentation
 FunRich Tool Documentation Version 2.1.2 Shivakumar Keerthikumar, Mohashin Pathan, Johnson Agbinya and Suresh Mathivanan Mathivanan Lab http://www.mathivananlab.org La Trobe University LIMS1 Department
FunRich Tool Documentation Version 2.1.2 Shivakumar Keerthikumar, Mohashin Pathan, Johnson Agbinya and Suresh Mathivanan Mathivanan Lab http://www.mathivananlab.org La Trobe University LIMS1 Department
Wilson Leung 01/03/2018 An Introduction to NCBI BLAST. Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment
 An Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at https://blast.ncbi.nlm.nih.gov/blast.cgi
An Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at https://blast.ncbi.nlm.nih.gov/blast.cgi
Package MiRaGE. October 16, 2018
 Version 1.22.0 Title MiRNA Ranking by Gene Expression Author Y-h. Taguchi Maintainer Y-h. Taguchi Depends R (>= 3.1.0), Biobase(>= 2.23.3) Package MiRaGE October 16,
Version 1.22.0 Title MiRNA Ranking by Gene Expression Author Y-h. Taguchi Maintainer Y-h. Taguchi Depends R (>= 3.1.0), Biobase(>= 2.23.3) Package MiRaGE October 16,
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Courtois et al., http://www.jcb.org/cgi/content/full/jcb.201202135/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Measurement of distribution and size of MTOCs in the
Supplemental material Courtois et al., http://www.jcb.org/cgi/content/full/jcb.201202135/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Measurement of distribution and size of MTOCs in the
1 Introduction ENLIGHT Problem formulation and project aim... 2
 Contents 1 Introduction 2 1.1 ENLIGHT........................................ 2 1.2 Problem formulation and project aim......................... 2 2 Background theory 2 2.1 Biological background.................................
Contents 1 Introduction 2 1.1 ENLIGHT........................................ 2 1.2 Problem formulation and project aim......................... 2 2 Background theory 2 2.1 Biological background.................................
The Dot Matrix Method
 Special Topics BS5936: An Introduction to Bioinformatics. Florida State niversity The Department of Biological Science www.bio.fsu.edu Sept. 9, 2003 The Dot Matrix Method Steven M. Thompson Florida State
Special Topics BS5936: An Introduction to Bioinformatics. Florida State niversity The Department of Biological Science www.bio.fsu.edu Sept. 9, 2003 The Dot Matrix Method Steven M. Thompson Florida State
cbioportal https://www.ncbi.nlm.nih.gov/pubmed/ /5/401
 cbioportal http://www.cbioportal.org/ https://www.ncbi.nlm.nih.gov/pubmed/23550210 http://cancerdiscovery.aacrjournals.org/content/ 2/5/401 Tutorials http://www.cbioportal.org/tutorial.jsp http://www.cbioportal.org/faq.jsp
cbioportal http://www.cbioportal.org/ https://www.ncbi.nlm.nih.gov/pubmed/23550210 http://cancerdiscovery.aacrjournals.org/content/ 2/5/401 Tutorials http://www.cbioportal.org/tutorial.jsp http://www.cbioportal.org/faq.jsp
Colocalization Algorithm. User s Guide
 Colocalization Algorithm User s Guide Copyright 2008 Aperio Technologies, Inc. Part Number/Revision: MAN 0082, Revision A Date: March 7, 2008 This document applies to software versions Release 9.0 and
Colocalization Algorithm User s Guide Copyright 2008 Aperio Technologies, Inc. Part Number/Revision: MAN 0082, Revision A Date: March 7, 2008 This document applies to software versions Release 9.0 and
Incorporating Known Pathways into Gene Clustering Algorithms for Genetic Expression Data
 Incorporating Known Pathways into Gene Clustering Algorithms for Genetic Expression Data Ryan Atallah, John Ryan, David Aeschlimann December 14, 2013 Abstract In this project, we study the problem of classifying
Incorporating Known Pathways into Gene Clustering Algorithms for Genetic Expression Data Ryan Atallah, John Ryan, David Aeschlimann December 14, 2013 Abstract In this project, we study the problem of classifying
/ Computational Genomics. Normalization
 10-810 /02-710 Computational Genomics Normalization Genes and Gene Expression Technology Display of Expression Information Yeast cell cycle expression Experiments (over time) baseline expression program
10-810 /02-710 Computational Genomics Normalization Genes and Gene Expression Technology Display of Expression Information Yeast cell cycle expression Experiments (over time) baseline expression program
SeqScape Software Version 2.7
 Applied Biosystems SeqScape Software SeqScape Software Version 2.7 Quick Reference Card Applied Biosystems SeqScape Software v2.7 is designed for mutation detection and analysis, SNP discovery and validation,
Applied Biosystems SeqScape Software SeqScape Software Version 2.7 Quick Reference Card Applied Biosystems SeqScape Software v2.7 is designed for mutation detection and analysis, SNP discovery and validation,
Report Axon Dynamics during Neocortical Laminar Innervation
 Cell Reports Supplemental Information Report Axon Dynamics during Neocortical Laminar Innervation Randal A. Hand, Syed Khalid, Edric Tam, and Alex L. Kolodkin Figure S1 1 Figure S1. Developmental timing
Cell Reports Supplemental Information Report Axon Dynamics during Neocortical Laminar Innervation Randal A. Hand, Syed Khalid, Edric Tam, and Alex L. Kolodkin Figure S1 1 Figure S1. Developmental timing
Analyzing Variant Call results using EuPathDB Galaxy, Part II
 Analyzing Variant Call results using EuPathDB Galaxy, Part II In this exercise, we will work in groups to examine the results from the SNP analysis workflow that we started yesterday. The first step is
Analyzing Variant Call results using EuPathDB Galaxy, Part II In this exercise, we will work in groups to examine the results from the SNP analysis workflow that we started yesterday. The first step is
Uploading sequences to GenBank
 A primer for practical phylogenetic data gathering. Uconn EEB3899-007. Spring 2015 Session 5 Uploading sequences to GenBank Rafael Medina (rafael.medina.bry@gmail.com) Yang Liu (yang.liu@uconn.edu) confirmation
A primer for practical phylogenetic data gathering. Uconn EEB3899-007. Spring 2015 Session 5 Uploading sequences to GenBank Rafael Medina (rafael.medina.bry@gmail.com) Yang Liu (yang.liu@uconn.edu) confirmation
Computational Molecular Biology
 Computational Molecular Biology Erwin M. Bakker Lecture 2 Materials used from R. Shamir [2] and H.J. Hoogeboom [4]. 1 Molecular Biology Sequences DNA A, T, C, G RNA A, U, C, G Protein A, R, D, N, C E,
Computational Molecular Biology Erwin M. Bakker Lecture 2 Materials used from R. Shamir [2] and H.J. Hoogeboom [4]. 1 Molecular Biology Sequences DNA A, T, C, G RNA A, U, C, G Protein A, R, D, N, C E,
When we search a nucleic acid databases, there is no need for you to carry out your own six frame translation. Mascot always performs a 6 frame
 1 When we search a nucleic acid databases, there is no need for you to carry out your own six frame translation. Mascot always performs a 6 frame translation on the fly. That is, 3 reading frames from
1 When we search a nucleic acid databases, there is no need for you to carry out your own six frame translation. Mascot always performs a 6 frame translation on the fly. That is, 3 reading frames from
Tutorial for Windows and Macintosh. Sequencher Connections
 Tutorial for Windows and Macintosh Sequencher Connections 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Tutorial for Windows and Macintosh Sequencher Connections 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Supplementary Figures
 Supplementary Figures re co rd ed ch annels darkf ield MPM2 PI cell cycle phases G1/S/G2 prophase metaphase anaphase telophase bri ghtfield 55 pixel Supplementary Figure 1 Images of Jurkat cells captured
Supplementary Figures re co rd ed ch annels darkf ield MPM2 PI cell cycle phases G1/S/G2 prophase metaphase anaphase telophase bri ghtfield 55 pixel Supplementary Figure 1 Images of Jurkat cells captured
The GENIA corpus Linguistic and Semantic Annotation of Biomedical Literature. Jin-Dong Kim Tsujii Laboratory, University of Tokyo
 The GENIA corpus Linguistic and Semantic Annotation of Biomedical Literature Jin-Dong Kim Tsujii Laboratory, University of Tokyo Contents Ontology, Corpus and Annotation for IE Annotation and Information
The GENIA corpus Linguistic and Semantic Annotation of Biomedical Literature Jin-Dong Kim Tsujii Laboratory, University of Tokyo Contents Ontology, Corpus and Annotation for IE Annotation and Information
Analyzing ICAT Data. Analyzing ICAT Data
 Analyzing ICAT Data Gary Van Domselaar University of Alberta Analyzing ICAT Data ICAT: Isotope Coded Affinity Tag Introduced in 1999 by Ruedi Aebersold as a method for quantitative analysis of complex
Analyzing ICAT Data Gary Van Domselaar University of Alberta Analyzing ICAT Data ICAT: Isotope Coded Affinity Tag Introduced in 1999 by Ruedi Aebersold as a method for quantitative analysis of complex
ACCEPT MANUAL. Version beta
 ACCEPT MANUAL Version 1.0.0 beta 1 ACCEPT Software 1.1.0 beta Manual The ACCEPT Software developed within the EU Cancer-ID project is an open source image analysis package for the Automated CTC Classification,
ACCEPT MANUAL Version 1.0.0 beta 1 ACCEPT Software 1.1.0 beta Manual The ACCEPT Software developed within the EU Cancer-ID project is an open source image analysis package for the Automated CTC Classification,
SUPPLEMENTARY INFORMATION
 Supplementary Discussion 1: Rationale for Clustering Algorithm Selection Introduction: The process of machine learning is to design and employ computer programs that are capable to deduce patterns, regularities
Supplementary Discussion 1: Rationale for Clustering Algorithm Selection Introduction: The process of machine learning is to design and employ computer programs that are capable to deduce patterns, regularities
- Free fluorophores - Donors without partner acceptors - Acceptors without partner donors
 Determining Distances / Distance Changes Caveats Angular dependence (κ 2 ) Environment dependence (J, ϕd) Distance is a (complicated) average Probe is large, linkages can be long Construct complications
Determining Distances / Distance Changes Caveats Angular dependence (κ 2 ) Environment dependence (J, ϕd) Distance is a (complicated) average Probe is large, linkages can be long Construct complications
Supplementary Information
 Supplementary Information Supplementary Figure S1 The scheme of MtbHadAB/MtbHadBC dehydration reaction. The reaction is reversible. However, in the context of FAS-II elongation cycle, this reaction tends
Supplementary Information Supplementary Figure S1 The scheme of MtbHadAB/MtbHadBC dehydration reaction. The reaction is reversible. However, in the context of FAS-II elongation cycle, this reaction tends
Double Self-Organizing Maps to Cluster Gene Expression Data
 Double Self-Organizing Maps to Cluster Gene Expression Data Dali Wang, Habtom Ressom, Mohamad Musavi, Cristian Domnisoru University of Maine, Department of Electrical & Computer Engineering, Intelligent
Double Self-Organizing Maps to Cluster Gene Expression Data Dali Wang, Habtom Ressom, Mohamad Musavi, Cristian Domnisoru University of Maine, Department of Electrical & Computer Engineering, Intelligent
Lecture 5 Advanced BLAST
 Introduction to Bioinformatics for Medical Research Gideon Greenspan gdg@cs.technion.ac.il Lecture 5 Advanced BLAST BLAST Recap Sequence Alignment Complexity and indexing BLASTN and BLASTP Basic parameters
Introduction to Bioinformatics for Medical Research Gideon Greenspan gdg@cs.technion.ac.il Lecture 5 Advanced BLAST BLAST Recap Sequence Alignment Complexity and indexing BLASTN and BLASTP Basic parameters
(DNA#): Molecular Biology Computation Language Proposal
 (DNA#): Molecular Biology Computation Language Proposal Aalhad Patankar, Min Fan, Nan Yu, Oriana Fuentes, Stan Peceny {ap3536, mf3084, ny2263, oif2102, skp2140} @columbia.edu Motivation Inspired by the
(DNA#): Molecular Biology Computation Language Proposal Aalhad Patankar, Min Fan, Nan Yu, Oriana Fuentes, Stan Peceny {ap3536, mf3084, ny2263, oif2102, skp2140} @columbia.edu Motivation Inspired by the
