Dynamic Graph-based Relational Learning of Temporal Patterns in Biological Networks Changing over Time
|
|
|
- Silas Roberts
- 5 years ago
- Views:
Transcription
1 Dynamic Graph-based Relational Learning of Temporal Patterns in Biological Networks Changing over Time Chang Hun You, Lawrence B. Holder and Diane J. Cook School of Electrical Engineering & Computer Science, Washington State University, Pullman, WA, USA {changhun, holder, Abstract We propose a dynamic graph-based relational learning approach using graph-rewriting rules to analyze how biological networks change over time. The analysis of dynamic biological networks is necessary to understand life at the systemlevel, because biological networks continuously change their structures and properties while an organism performs various biological activities to promote reproduction and sustain our lives. Most current graph-based data mining approaches overlook dynamic features of biological networks, because they are focused on only static graphs. First, we generate a dynamic graph, which is a sequence of graphs representing biological networks changing over time. Then, our approach discovers graph rewriting rules, which show how to replace subgraphs, between two sequential graphs. These rewriting rules describe the structural difference between two graphs, and describe how the graphs in the dynamic graph change over time. Temporal patterns discovered in dynamic graphs representing metabolic pathways show that our approach enables the discovery of dynamic patterns in biological networks. keywords: Temporal Graph Mining, Graph Rewriting Rules, Biological Network 1 Introduction Our bodies are well-organized biological networks, which promote reproduction and sustain our lives. Furthermore, biological networks continuously change their structures and properties, while an organism performs various biological activities, such as digestion, respiration and so on. We assume the structures of biological networks change over time as they interact with specific conditions, for instance, a disease. The temporal patterns in the structural changes of biological networks can be significant information about a disease and help researchers develop new drugs. During the development period, the temporal patterns in the structural changes of biological networks after taking the medicine are also used for the development and evaluation of the new drug. Lactose intolerance is the inability to digest lactose because of a lack of the lactase enzyme, breaking down lactose into galactose and glucose [2]. Two major treatments are to minimize the intake of lactose products and take the lactase supplement. Our approach can help us discover the temporal patterns in the structural changes of the galactose metabolism pathway after these treatments, and investigate other treatments (i.e., improving the production of the lactase enzyme in the pathway). We propose a novel approach to analyze structural features along with temporal features in a time series of biological networks to enhance our systems-level understanding of bioorganisms. Our dynamic graph-based data mining approach uses graph-rewriting rules to analyze how biological networks change over time. Graph-rewriting rules define how one graph changes to another in its topology replacing vertices, edges or subgraphs according to the rewriting rules. First, we generate a dynamic graph, which is a sequence of graphs representing biological networks changing over time. Then, our approach discovers rewriting rules, which show how to replace subgraphs, between two sequential graphs. After discovery of whole sets of graph rewriting rules from a dynamic graph, we discover temporal patterns in graph rewriting rules. The temporal patterns show what graph rewriting rule is applied before or after the other is applied. The graph rewriting rules can describe the structural difference between two graphs. The temporal patterns in rewriting rules can describe how the graphs in the dynamic graph changing over time. This approach enables us to investigate dynamic patterns in biological networks. This paper, first, introduce, several preceding approaches related to dynamic analysis of biological networks and temporal data mining. Then, we define the problem of our research. We present our Dynamic Graph Relational Learning (DynGRL) algorithm. Our approach is applied to the glycolysis metabolic pathway in combination with the mathematical modeling. The results section shows our discovered graph rewriting rules and temporal patterns of rewriting rules in two aspects: temporal and structural aspects. 2 Related Works According to the central dogma in molecular biology, the genetic information in DNA is transcribed into RNA (transcription) and protein is synthesized from RNA (translation). These biomolecules (DNA, RNA and proteins) play central roles in the aspects of the function and structure of organisms. However, there are few molecules that can work alone. For an example, a glycolysis (Glucose + 2NAD + + 2ADP +
2 2P i 2P yruvate + 2NADH AT P + 2H 2 O), which is a metabolic pathway converting one molecule of glucose into two molecules of pyruvate with the production of two molecules of ATP (Adenosine TriPhosphate), includes more than 10 biochemical reactions and various enzymes [13]. Biological networks including metabolic pathways, proteinprotein interactions and gene regulatory networks, consist of various molecules and their relationships [10]. In addition to the structural aspect, we also consider the temporal aspect of biological networks, because the biosystems always change their properties and structures while interacting with other conditions. Two approaches have been developed for the analysis of biological networks. One approach is graph-based data mining [11, 18]. This approach represents biological networks as graphs, where vertices represent molecules and edges represent relations between molecules, and discovers frequent patterns in graphs. Many approaches of graph-based data mining discover structural features of biological networks, but they overlook temporal properties. The other approach is mathematical modeling, which is an abstract model to describe a system using mathematical formulae [14]. Most of these approaches, as a type of quantitative analysis, model the kinetics of pathways and analyzes the trends in the amounts of molecules and the flux of biochemical reactions. But most of them disregard relations among multiple molecules. Temporal data mining attempts to learn temporal patterns in sequential data, which is ordered with respect to some index like time stamps, rather than static data [16]. Temporal data mining is focused on discovery of relational aspects in data such as discovery of temporal relations or cause-effect association. In other words, we can understand how or why the object changes rather than merely static properties of the object. There are several approaches to apply temporal data mining in biological data. Ho et al. [8] propose an approach to detect temporal patterns and relations between medical events of Hepatitis data. They represent medical information of patients as sequential events and classify temporal patterns and relations of medical testing results in the sequential events using the Naive Bayes classifier. Farach-Colton et al. [6] introduce an approach of mining temporal relations in protein-protein interactions. They model the assembly pathways of Ribosome using protein-protein interactions. This approach determines the order of molecular connections using the distance measure of each interaction between two proteins. Temporal data mining approaches discover temporal patterns in data, but they disregard relational aspects among entities. For example, they can identify temporal patterns in the appearance of genes such that a gene, YBR218C, appears before another gene, YGL062W, but cannot identify how these two genes interact with each other. There are two main aspects to consider for understanding biological networks. First, we need to focus on relations between molecules as well as a single molecule. Second, we should G1 sce:c PPrel:+p PPrel:--- sce:a PPrel:--- sce:b sce:e R sce:d S G2 sce:c S sce:f PPrel:--- sce:e PPrel:+p sce:d sce:g Figure 1. An example of graph rewriting rules between two graphs G 1 and G 2. S represents the maximal common subgraph between the two graphs. R and A represent the removal and addition substructures. The red edges with labels marked by the boxes represent the connection edges. consider biological networks as dynamic operations rather than static structures because every biological process changes over time and interacts with inner or outer conditions. It is necessary to analyze biological networks not only for structural aspect but also for dynamic aspect for system-level understanding of our organisms. For this reason, we need an approach to analyze graphs which structurally change over time for both aspects: structural and dynamic properties. 3 Problem definition This paper focuses on temporal and structural analysis of biological networks. Our dynamic graph-based relational learning approach discovers graph rewriting rules in a series of graphs changing their structures over time. Each graph rewriting rule represents topological changes between two sequential graphs. Here, we define graph rewriting rules for our approach. Graph rewriting is a method to represent topological changes of graphs using graph rewriting rules [5, 17]. Generally, graph rewriting rules identify subgraphs in a graph and modify them. Each graph rewriting rule defines a transformation between L and R, where L and R are subgraphs in two graphs G and H respectively, such that L is replaced by R, L is deleted, or R is created [15]. There are also several algorithms to discover the node or edge replacement graph grammar using the minimum description length principle [9, 12]. However, their scope is limited to static graphs. Traditional approaches to the identification of graph rewriting rules determine which subgraphs will be replaced by other subgraphs. Our approach is focused on representing changing structures between two graphs rather than just what subgraphs change. We define our graph rewriting rules to represent how substructures change between two graphs rather than just what subgraphs change. First, we discover maximum common subgraphs between two sequential graphs G 1 and G 2. Then, we A
3 derive removal substructures from G 1 and addition substructures from G 2. Figure 1 shows an instance of this process. A maximum common subgraph (denoted by S) is discovered between two graphs, G 1 and G 2. Then the remaining structure in G 1 and G 2 becomes removal (denoted by R) and addition (denoted by A) substructures respectively. These substructures with connection edges (red edges with boxed labels) are elements of graph rewriting rules: removal and addition rules. For this approach, we define several preliminary terms. A directed graph G is defined as G = (V, E), where V is a set of vertices and E is a set of edges. An edge e ( E) is directed from x to y as e = (x, y), where x, y V. Here, we define a dynamic graph DG as a sequence of n graphs as DG = {G 1, G 2,, G n }, where each graph G i is a graph at time i for 1 i n. Then, we define a set of removal substructures RG and a set of addition substructures AG as follows. RG i = G i /S i,i+1, AG i+1 = G i+1 /S i,i+1 RG i denotes a set of removal substructures in a graph G i, AG i+1 denotes a set of addition substructures in the next graph G i+1, and S i,i+1 is a maximum set of common subgraphs between two sequential graphs G i and G i+1 in a dynamic graph DG. A prior graph G i is transformed to a posterior graph G i+1 by application of a set of graph rewriting rules GR i,i+1 as denoted by G i+1 = G i GRi,i+1 A set of graph rewriting rules GR i,i+1 between two sequential graphs G i and G i+1 is defined as follows. GR i,i+1 = {(m, p, CE m, CL m ),, (n, q, CE n, CL n ), } m and n are indices of graph rewriting rules in a set GR i,i+1. p and q are indices of a removal substructure in RG i and an addition substructure in AG i+1 respectively. CE and CL are defined as a set of connection edges and a set of labels of the connection edges. Each element of RG and AG corresponds to a set of CE and CL, unless a removal (addition) substructure does not connect to the G i (G i+1 ). CE k and CL k represent connections between substructures and the original graphs (k = m or n) as follows. CE = {(d, X, Y ), }, CL = {label xy, } d represents whether the edge is directed or undirected using d and u. X and Y denote the starting and ending vertices of the edge. Because the connection edge links the substructure to the original graph, one end of this edge is from the substructure and the other is from the original graph. The end vertex from the substructure starts with s followed by the index of the vertex, and the end vertex from the original graph starts with g followed by the index of the vertex. For example, (d, g1, s3) represents the directed edge from a vertex 1 in the original graph to another vertex 3 in the substructure. label xy represents a label for the corresponding connection edge between two vertices X and Y. The number of elements of CE (CL as well) represents the number of connections between substructures and the original graph. If a substructure is not connected to the original graph, both sets of CE and CL are empty. Using the definitions of graph rewriting rules, we describe more detail about the example in figure 1. As described previously, the graph rewriting rule, GR 1,2, includes one removal rule and one addition rule. The removal rule includes one substructure (denoted by R) and two connection edges (red edges with boxed labels). The addition rule includes one substructure (denoted by A) and one connection edge. G 1 is transformed to G 2 by application of GR 1,2 (G 2 = G 1 GR1,2 ). GR 1,2 is described as follows, GR 1,2 = {(r 1, rsub 1, {(d, s2, g3), (d, s2, g4)}, {P P rel : +p, P P rel : p}), (a 1, asub 2, {(d, g3, s1)}, {P P rel : p})}, where r 1 represents an index into the set of removal rules and a 1 represents an index into the set of addition rules. A removal rule r 1 includes a removal substructure rsub 1 denoted by R in figure 1. rsub 1 was connected to the original graph G 1 by two edges (d, s2, g3) and (d, s2, g4), which are labeled by P P rel : +p and P P rel : p. These connection edges are directed edges (indicated by d ). These two edges are connected from the substructure (denoted by s ) to the original graph (denoted by g ), where each number denotes a vertex number in the substructure or the original graph. For example, (d, s2, g3) denotes a connection from a vertex number 2 in the substructure to a vertex number 3 in the original graph. In a similar way, an addition rule a 1 includes an addition substructure asub 1 (denoted by A in figure 1), which is connected by one connection edge (d, g3, s1) labeled by P P rel : p. The graph rewriting rules show how two sequential graphs are structurally different. After collecting all sets of graph rewriting rules in a dynamic graph, we also discover temporal patterns in graph rewriting rules, which can describe how the graphs change over time as well as what structures change. 4 Discovery of Graph Rewriting Rules The first goal of our research is to discover graph rewriting rules in a dynamic graph representing biological networks changing over time. This section describes our algorithm to discover graph rewriting rules in a dynamic graph. This section describes our graph rewriting rule discovery system, DynGRL, that discovers graph rewriting rules in a dynamic graph. The algorithm starts with a dynamic graph DG consisting of a sequence of n graphs as shown in algorithm 1. First, the algorithm creates a list of n virtual graphs, V GL,
4 corresponding to n time series of graphs at line 1. Our approach uses a virtual graph to specify the application locations of graph rewriting rules. Because a graph may have multiple graph rewriting rules and several same-labeled vertices and edges, the exact locations of connection edges and rewriting rules are important to reduce the discovery error. The next procedure is to create a two-graph set, Graphs, including two sequential graphs G i and G i+1 (line 5) and to specify the limit based on unique labeled vertices and edges of G i and G i+1 (line 6). UV L and UEL denote the number of unique vertex labels and edges in G i and G i+1. The Limit specifies the number of substructures to consider when searching for a common substructure (line 6). The Limit based on the number of labels in the input graph bounds the search space within polynomial time and ensure consideration of most of the possible substructures. The inner loop (lines 7 to 14) represents the procedure to discover common substructures between two sequential graphs. We use the SUBDUE graph-based relational learning approach to discover substructures [3, 4]. SUBDUE evaluates substructures using the Minimum Description Length (MDL) principle to find the best substructure which minimizes the description length of the input graph after being compressed by the substructure. More detail on the evaluation approach is described in [3]. Even though to find the maximum common subgraph is NP-Complete, SUBDUE can be used as a polynomialtime approximation to this problem using Limit and iteration as described later in this section. After discovery of the best substructure, the algorithm checks whether the substructure is a subgraph of both graphs G i and G i+1. In the affirmative case, the best substructure is added into ComSubSet and the two target graphs are compressed by replacing the substructure with a vertex. If the best substructure does not belong to one of the two graphs, the algorithm just compresses the graphs without adding any entry into ComSubSet. After compression, the algorithm discovers another substructure at the next iteration until there is no more compression. Using the complete list of common substructures, ComSubSet, the algorithm acquires removal substructures, remsubs, and addition substructures, addsubs, (lines 15 and 17). First, the algorithm identifies vertices and edges not part of common substructures and finds each disconnected substructure in G i and G i+1 using the modified Breadth First Search (mbfs), which adds each edge as well as each vertex into the queues as visited or to be visited. The marked substructures in G i and G i+1 are removal and addition substructures respectively. While mbfs searches these removal and addition substructures, it also finds connection edges, CE, as described previously. These edges are added into RemCESet and AddCESet, where removal and addition substructures are added into RemSubSet and AddSubSet respectively (in lines 16 and 18). Using these rewriting substructures and connection edges, rewriting rules (RR) are created and stored into RRL (in lines 19 to 20). The main challenge of our algorithm is to discover maximum common subgraphs between two sequential graphs, because this problem is known to be NP-hard [7]. To avoid this problem, first we use the Limit to restrict the number of substructures to consider in each iteration. The Limit is computed using the number of unique labels of vertices and edges in graphs. Second, our algorithm does not try to discover the whole common substructures at once. In each step, the algorithm discovers a portion of common, connected substructure and iterates the discovery process until discovering the whole maximum common subgraphs. Usually, the size of graphs representing biological networks is not too large. Therefore, discovery of graph rewriting rules is still feasible. However, we still have challenges to analyze very large graphs. Algorithm 1: DynGRL discovery algorithm Input: DG = {G 1, G 2,, G n } Output: RRL Create V GL = {V G 1, V G 2,, V G n } RRL = {} for i = 1 to n 1 do RemSubSet = AddSubSet = ComSubSet = {} Graphs = {G i, G i+1 } Limit = UV L + 4(UEL 1) while No more compression do BestSub = DiscoverSub(Limit, Graphs) if BestSub G i & G i+1 then Add BestSub into ComSet end Compress Graphs by BestSub Mark BestSub on V G i and V G i+1 end Get remsubs and CE from V G i Add remsubs into RemSubSet and CE into RemCESet Get addsubs and CE from V G i+1 Add addsubs into AddSubSet and CE into AddCESet Create RR from RemSubSet, AddSubSet, RemCESet, AddCESet Add RR into RRL end Dynamic Graph Generation and Experiment We evaluate our algorithm using a dynamic graph representing the glycolysis metabolic pathway in combination with a mathematical modeling result. The glycolysis pathway is a series of enzyme-catalyzed reactions of degrading a molecule of glucose (6 carbons) to yield two molecules of pyruvate (3 carbons) [13]. In the process of glycolysis, some of the free energy are produced as the forms of ATP and NADH. Glycolysis
5 is the most important pathway in aerobic respiration, which is a process to generate energy in a cell. As described in the previous section, a mathematical modeling approach explores only numerical values, such as the concentration of molecules and the flux of reactions. We propose to combine the result of mathematical modeling and graphs for structural and temporal analysis. We use a result of the simulation of glycolysis pathway of the yeast (Saccharomyces cerevisiae) [14]. This result contains the trends of concentrations of 14 molecules. We normalize these concentrations from 0 to 1, because we are focused on trends of the changes and the concentration of different molecules are various. Figure 2 shows the oscillated curves of the normalized concentration of two molecules: Pyruvate and Acetaldehyde. Because the simulation is performed for 100 seconds, we have 101 time series data from the initial time to the final time. We generate a static graph representing the glycolysis pathway from the KEGG PATHWAY data [1], where vertices represent reactions, compounds and enzymes, and edges represent relations between vertices. Usually, a reaction catalyzed by one or more enzymes contain one or more compounds as substrates and one or more compounds as products. We use a threshold t to activate compounds. At each time, we assume a compound, which has more than t amount, is shown in the graph. The reactions and enzymes are shown in the graph, only when all related substrates and products are activated. In other words, every related compound should be activated to place a reaction in a graph. We try 0.1 and 0.3 as our thresholds. We perform DynGRL with a dynamic graph including 101 graphs representing the glycolysis simulation for 101 seconds. DynGRL discovers 100 sets of graph rewriting rules during 100 time intervals for each threshold: 0.1 and 0.3. Then, we discovers some temporal patterns in the graph rewriting rules to describe temporal and structural aspects of the dynamic graph. 6 Results As described in the previous section, the goal of this research is to discover temporal patterns in graph rewriting rules to describe structural changes of metabolic pathways over time. First, we show temporal patterns in graph rewriting rules. Then, we discuss structural aspects of the graph rewriting rules. Because the modeling result represents the oscillation of glycolysis, we observe several temporal patterns in graph rewriting rules showing the oscillation. In both experiments (threshold 0.1 and 0.3), we discover oscillated temporal patterns. Using 0.1 as the threshold, temporal patterns among three chemical reactants such as C00008 (ADP), C00084 (Acetaldehyde) and C00003 (NAD + ) are discovered as shown in figure 3 (a). The points above the time axis represent the time when the substructures including each compound are removed from the graph representing the glycolysis pathway. The points below the time axis represent the time when the substructures including each compound are added to the pathway graph. For ("!#'"!#&"!#%"!#$"!"!" $!" %!" &!" '!" (!!" )!!!$$" )!!!'%" *+,-" Figure 2. The oscillation curves of changing concentrations of Pyruvate (C00022) and Acetaldehyde (C00084). example, the first red diamond time point close to 0 represent the addition of a substructure including the ADP molecule at time 2. We also observe these time points are ordered by the time of removal and addition. Acetaldehyde is added before ADP is added except the first case, and then NAD + is added if applicable. In case of removals, they are removed following the order of ADP, Acetaldehyde and NAD +. Similarly, figure 3 (b) shows other temporal patterns between Acetaldehyde and Pyruvate (C00022) at the experiment of threshold 0.3. We observe Acetaldehyde is added after Pyruvate is added to the pathway graph. In contrast, Pyruvate is removed after Acetaldehyde is removed from the pathway graph. We can compare these temporal patterns with the mathematical modeling results shown in figure 2. The oscillation curves represent that Pyruvate is increased slightly earlier and decreased slightly later than Acetaldehyde. For this reason, the temporal patterns in the graph rewriting rules of our dynamic graph represent the substructures including Pyruvate are added earlier and removed later. In the temporal aspect, the results discovered by DynGRL show several reactants in the glycolysis pathway are increased and decreased in order and these ordered patterns are repeated periodically as shown in the mathematical modeling. These patterns are observed as addition and removal of substructures of the dynamic graph representing the glycolysis pathway changing over time. The addition substructures include the increasing (over the threshold) reactants, and the removal substructures include the decreasing (under the threshold) reactants. The temporal patterns in the learned graph rewriting rules show the temporal relations that describe how the glycolysis pathway changes over time by showing which elements changes earlier than the other. These temporal patterns and graph rewriting rules help us to understand temporal properties of the glycolysis pathway. In addition to the temporal aspect, our results can depict structural changes of the metabolic pathways. Because an advantage of the graph representation is visualization, we can understand metabolic pathways better using structural analysis as
6 34/."" /0+*""!" #!" $!" %!" &!" '!" (!" )!" *!" +!" #!!"!" #!" $!" %!" &!" '!!",!!!!*"-./0",!!!!*"1220",!!!*&"-./0",!!!*&"1220",!!!!%"-./",!!!!%"122" (!!!&$")*+," (!!!&$"-..," (!!!##")*+," (!!!##"-..," Figure 3. A visualization of time points when the substructure including each compound is removed from or added to graphs representing the glycolysis pathway at the experiment of threshold 0.1 (a) and 0.3 (b). well as temporal analysis. Here we show some instances of substructures in the discovered graph rewriting rules, and how they are related to the original graphs. Figure 4 shows the graphs G i in the dynamic graph from time 24 to 32 at the experiment of threshold 0.3. These graphs show how the glycolysis pathway changes over time. In other words, what substructures are removed and added over time. The graph removes a substructure including two reactants: Acetaldehyde and Pyruvate and three related reactions, and changes its structure to the graph 25. The structure is not changed from time 25 to 31. At time 32, the graph changes more after the addition of a substructure including Pyruvate and a related reaction, R The removal substructure at time 24 is also removed at time 49, 60, 72, 84 and 96. As shown in figure 3 (b), there are six time points (the third point and the last five points above the axis) when Acetaldehyde and Pyruvate are removed at the same time. The addition substructure at time 32 is also added at time 3, 12, 21, 44, 56, 68, 80, and 92 as shown in figure 3 (b). In this substructure, Acetaldehyde (C00084) works as a substrate of a reaction R00754 and as a product of a reaction R Pyruvate (C00022) works as a substrate of the reaction R00224 and as a product of the reaction R GR k are represented as follows, (k = 24, 25 or 31, 32) GR 24,25 = {(r 24, rsub1, CE 24, CL 24 )} GR 31,32 = {(a 32, asub 1, CE 32, CL 32 )} where rsub 1 represents the removal substructure of G 24 with labels marked by -[], and asub 1 represents the addition substructure of G 32 with labels marked by +[]. As described in section 3, CE m(n) and CL m(n) (m = 24 and n = 32) represent sets of connection edges and connection edge labels (denoted by labels marked by () ) as follows, CE 24 = {(d, s2, g5), (d, s2, g9), (d, s2, g11), (d, s7, g8), (d, s7, g10), (d, s7, g12)} CL 24 = {S to Rct, S to Rct, Rct to P, Rct to P, S to Rct, S to Rct} CE 32 = {(d, s2, g5), (d, s2, g9), (d, s2, g8)} CL 32 = {S to Rct, S to Rct, Rct to P } The structural results show how the substructures are related to the original graphs (i.e., which connection edges link the substructures to the original graphs.) as well as what substructures are removed or added. The graph rewriting rules describe the relational aspects between the substructures (some elements) and the original graph (the pathway), but not merely which elements are changed. In summary, our results show temporal and structural aspects of the dynamic graph representing the metabolic pathway. Temporal patterns show some elements are added and removed in order and periodically. Structural patterns show how the original graphs change after applications of removal and addition rules. These temporal patterns and graph rewriting rules help us to understand temporal properties as well as structural properties of biological networks. Some discovered temporal and structural patterns in a specific disease can show us how they are different from normal patterns and help us investigate disease and develop a new treatment. 7 Conclusion In this research, we formalize graph rewriting rules to describe structurally changing biological networks. We represent an algorithm, DynGRL, to discover graph rewriting rules in a dynamic graph. The algorithm is evaluated with a dynamic graph representing the glycolysis pathway in combination with mathematical modeling results. We also discover several temporal patterns in graph rewriting rules of the pathway. Our results are visualized to identify how the metabolic pathway changes structurally over time, and what temporal patterns are discovered repeatedly. Our approaches allow us to identify not only structural changes of pathways but also temporal patterns
7 G 24 EC: EC: EC: cpd:c00236 cpd:c05345 rn:r01518+r00658 cpd:c00074 () () cpd:c [rn:r00200] EC: cpd:c00002 rn:r02848+r00771 () -[] -[] cpd:c00004 () -[rn:r00224] cpd:c00118 rn:r00127 cpd:c00003 EC: [rn:r00754] () -[] -[EC: ] -[] -[] -[] -[] () -[] EC: cpd:c [EC: ] -[cpd:c00022] -[EC: ] -[cpd:c00084] cpd:c [EC: ] G 25 to G 31 G 32 EC: EC: EC: EC: EC: rn:r01518+r00658 rn:r02848+r00771 cpd:c00074 cpd:c00236 cpd:c00221 cpd:c00008 cpd:c00002 cpd:c05345 rn:r00127 cpd:c00004 cpd:c00003 cpd:c00469 cpd:c00118 EC: EC: EC: cpd:c00221 cpd:c05345 cpd:c00469 EC: EC: EC: rn:r02848+r00771 rn:r01518+r [EC: ] cpd:c [] () cpd:c [rn:r00200] () cpd:c00008 () +[] rn:r [cpd:c00022] cpd:c00002 cpd:c00003 cpd:c00118 EC: cpd:c00004 Figure 4. An example of graph transformations from time 24 to 32. Red substructure with labels marked by -[] or +[] represent removal or addition substructures. The blue edges with labels marked by () represent the connection edges between. between multiple structural changes, providing us better understanding of how biological networks change over time. The future works follow several directions. We need more systematic evaluation for the learned graph rewriting rules including regenerating a dynamic graph using the learned graph rewriting rules to compare with the original dynamic graph from real world data. We will also focus on the fully automated approach to learn temporal patterns in the graph rewriting rules. Finally, we will evaluate how this approach can be used to predict future structures of biological networks using the learned patterns. References [1] Kyoto university bioinformatics center, KEGG website. [2] R. Bowen. Lactose intolerance (lactase non-persistence). Pathophysiology of the Digestive System, [3] D. Cook and L. Holder. Substructure discovery using minimum description length and background knowledge. Journal of Artificial Intelligence Research, 1: , [4] D. Cook and L. Holder. Graph-based data mining. IEEE Intelligent Systems, 15(2):32 41, [5] H. Dörr. Efficient Graph Rewriting and Its Implementation. Springer, [6] M. Farach-Colton, Y. Huang, and J. L. L. Woolford. Discovering temporal relations in molecular pathways using protein-protein interactions. In Proceedings of RECOMB, [7] M. R. Garey and D. S. Johnson. Computers and Intractability: A Guide to the Theory of NP-Completeness. W. H. Freeman, [8] T. Ho, C. Nguyen, S. S. Kawasaki, S. Le, and K. Takabayashi. Exploiting temporal relations in mining hepatitis data. Journal of New Generation Computing, 25: , [9] I. Jonyer, L. Holder, and D. Cook. Mdl-based context-free graph grammar induction. In Proceedings of FLAIRS-2003., [10] H. Kitano. Systems biology: A brief overview. Science, 295: , [11] J. Kukluk, C. You, L. Holder, and D. Cook. Learning node replacement graph grammars in metabolic pathways. In Proceedings of BIOCOMP, [12] J. P. Kukluk, L. B. Holder, and D. J. Cook. Inference of node replacement recursive graph grammars. In Proceedings of the Sixth SIAM International Conference on Data Mining, [13] D. L. Nelson and M. M. Cox. Lehniger Principle of Biochemistry. Freeman, New York, 4th edition, [14] K. Nielsen, P. Sørensen, and F. H. H.-G. Busse. Sustained oscillations in glycolysis: an experimental and theoretical study of chaotic and complex periodic behavior and of quenching of simple oscillations. Biophysical Chemistry, 7:49 62, [15] K. Nupponen. The design and implementation of a graph rewrite engine for model transformations. Master s thesis, Helsinki University of Technology, Dept. Com. Sci.and Eng., May [16] J. F. Roddick and M. Spiliopoulou. A survey of temporal knowledge discovery paradigms and methods. IEEE Transactions on Knowledge and Data Engineering, 14(4): , [17] G. Rozenberg. Handbook of Graph Grammars and Computing by Graph Transformation. World Scientific, [18] C. You, L. Holder, and D. Cook. Application of graph-based data mining to metabolic pathways. In Proceedings of IEEE ICDM Workshop on Data Mining in Bioinformatics, 2006.
Temporal and Structural Analysis of Biological Networks in Combination with Microarray Data
 Temporal and Structural Analysis of Biological Networks in Combination with Microarray Data Chang Hun You, Lawrence B. Holder and Diane J. Cook Abstract We introduce a graph-based relational learning approach
Temporal and Structural Analysis of Biological Networks in Combination with Microarray Data Chang Hun You, Lawrence B. Holder and Diane J. Cook Abstract We introduce a graph-based relational learning approach
Dynamic Graph-based Relational Learning of Temporal Patterns in Biological Networks Changing over Time
 Dynamic Graph-based Relational Learning of Temporal Patterns in Biological Networks Changing over Time Chang hun You, Lawrence B. Holder, Diane J. Cook School of Electrical Engineering & Computer Science
Dynamic Graph-based Relational Learning of Temporal Patterns in Biological Networks Changing over Time Chang hun You, Lawrence B. Holder, Diane J. Cook School of Electrical Engineering & Computer Science
V10 Metabolic networks - Graph connectivity
 V10 Metabolic networks - Graph connectivity Graph connectivity is related to analyzing biological networks for - finding cliques - edge betweenness - modular decomposition that have been or will be covered
V10 Metabolic networks - Graph connectivity Graph connectivity is related to analyzing biological networks for - finding cliques - edge betweenness - modular decomposition that have been or will be covered
System Identification Algorithms and Techniques for Systems Biology
 System Identification Algorithms and Techniques for Systems Biology by c Choujun Zhan A Thesis submitted to the School of Graduate Studies in partial fulfillment of the requirements for the degree of Doctor
System Identification Algorithms and Techniques for Systems Biology by c Choujun Zhan A Thesis submitted to the School of Graduate Studies in partial fulfillment of the requirements for the degree of Doctor
Graph-based Learning. Larry Holder Computer Science and Engineering University of Texas at Arlington
 Graph-based Learning Larry Holder Computer Science and Engineering University of Texas at Arlingt 1 Graph-based Learning Multi-relatial data mining and learning SUBDUE graph-based relatial learner Discovery
Graph-based Learning Larry Holder Computer Science and Engineering University of Texas at Arlingt 1 Graph-based Learning Multi-relatial data mining and learning SUBDUE graph-based relatial learner Discovery
bcnql: A Query Language for Biochemical Network Hong Yang, Rajshekhar Sunderraman, Hao Tian Computer Science Department Georgia State University
 bcnql: A Query Language for Biochemical Network Hong Yang, Rajshekhar Sunderraman, Hao Tian Computer Science Department Georgia State University Introduction Outline Graph Data Model Query Language for
bcnql: A Query Language for Biochemical Network Hong Yang, Rajshekhar Sunderraman, Hao Tian Computer Science Department Georgia State University Introduction Outline Graph Data Model Query Language for
Properties of Biological Networks
 Properties of Biological Networks presented by: Ola Hamud June 12, 2013 Supervisor: Prof. Ron Pinter Based on: NETWORK BIOLOGY: UNDERSTANDING THE CELL S FUNCTIONAL ORGANIZATION By Albert-László Barabási
Properties of Biological Networks presented by: Ola Hamud June 12, 2013 Supervisor: Prof. Ron Pinter Based on: NETWORK BIOLOGY: UNDERSTANDING THE CELL S FUNCTIONAL ORGANIZATION By Albert-László Barabási
Complementary Graph Coloring
 International Journal of Computer (IJC) ISSN 2307-4523 (Print & Online) Global Society of Scientific Research and Researchers http://ijcjournal.org/ Complementary Graph Coloring Mohamed Al-Ibrahim a*,
International Journal of Computer (IJC) ISSN 2307-4523 (Print & Online) Global Society of Scientific Research and Researchers http://ijcjournal.org/ Complementary Graph Coloring Mohamed Al-Ibrahim a*,
Pattern Mining in Frequent Dynamic Subgraphs
 Pattern Mining in Frequent Dynamic Subgraphs Karsten M. Borgwardt, Hans-Peter Kriegel, Peter Wackersreuther Institute of Computer Science Ludwig-Maximilians-Universität Munich, Germany kb kriegel wackersr@dbs.ifi.lmu.de
Pattern Mining in Frequent Dynamic Subgraphs Karsten M. Borgwardt, Hans-Peter Kriegel, Peter Wackersreuther Institute of Computer Science Ludwig-Maximilians-Universität Munich, Germany kb kriegel wackersr@dbs.ifi.lmu.de
Modeling with Uncertainty Interval Computations Using Fuzzy Sets
 Modeling with Uncertainty Interval Computations Using Fuzzy Sets J. Honda, R. Tankelevich Department of Mathematical and Computer Sciences, Colorado School of Mines, Golden, CO, U.S.A. Abstract A new method
Modeling with Uncertainty Interval Computations Using Fuzzy Sets J. Honda, R. Tankelevich Department of Mathematical and Computer Sciences, Colorado School of Mines, Golden, CO, U.S.A. Abstract A new method
Numeric Ranges Handling for Graph Based Knowledge Discovery Oscar E. Romero A., Jesús A. González B., Lawrence B. Holder
 Numeric Ranges Handling for Graph Based Knowledge Discovery Oscar E. Romero A., Jesús A. González B., Lawrence B. Holder Reporte Técnico No. CCC-08-003 27 de Febrero de 2008 2008 Coordinación de Ciencias
Numeric Ranges Handling for Graph Based Knowledge Discovery Oscar E. Romero A., Jesús A. González B., Lawrence B. Holder Reporte Técnico No. CCC-08-003 27 de Febrero de 2008 2008 Coordinación de Ciencias
MINING GRAPH DATA EDITED BY. Diane J. Cook School of Electrical Engineering and Computei' Science Washington State University Puliman, Washington
 MINING GRAPH DATA EDITED BY Diane J. Cook School of Electrical Engineering and Computei' Science Washington State University Puliman, Washington Lawrence B. Holder School of Electrical Engineering and
MINING GRAPH DATA EDITED BY Diane J. Cook School of Electrical Engineering and Computei' Science Washington State University Puliman, Washington Lawrence B. Holder School of Electrical Engineering and
Data mining, 4 cu Lecture 8:
 582364 Data mining, 4 cu Lecture 8: Graph mining Spring 2010 Lecturer: Juho Rousu Teaching assistant: Taru Itäpelto Frequent Subgraph Mining Extend association rule mining to finding frequent subgraphs
582364 Data mining, 4 cu Lecture 8: Graph mining Spring 2010 Lecturer: Juho Rousu Teaching assistant: Taru Itäpelto Frequent Subgraph Mining Extend association rule mining to finding frequent subgraphs
Subdue: Compression-Based Frequent Pattern Discovery in Graph Data
 Subdue: Compression-Based Frequent Pattern Discovery in Graph Data Nikhil S. Ketkar University of Texas at Arlington ketkar@cse.uta.edu Lawrence B. Holder University of Texas at Arlington holder@cse.uta.edu
Subdue: Compression-Based Frequent Pattern Discovery in Graph Data Nikhil S. Ketkar University of Texas at Arlington ketkar@cse.uta.edu Lawrence B. Holder University of Texas at Arlington holder@cse.uta.edu
A Roadmap to an Enhanced Graph Based Data mining Approach for Multi-Relational Data mining
 A Roadmap to an Enhanced Graph Based Data mining Approach for Multi-Relational Data mining D.Kavinya 1 Student, Department of CSE, K.S.Rangasamy College of Technology, Tiruchengode, Tamil Nadu, India 1
A Roadmap to an Enhanced Graph Based Data mining Approach for Multi-Relational Data mining D.Kavinya 1 Student, Department of CSE, K.S.Rangasamy College of Technology, Tiruchengode, Tamil Nadu, India 1
SBMLmerge, a system for combining biochemical network models
 SBMLmerge 1 SBMLmerge, a system for combining biochemical network models Marvin Schulz schulzma@molgen.mpg.de Jannis Uhlendorf uhlndorf@molgen.mpg.de Edda Klipp Wolfram Liebermeister klipp@molgen.mpg.de
SBMLmerge 1 SBMLmerge, a system for combining biochemical network models Marvin Schulz schulzma@molgen.mpg.de Jannis Uhlendorf uhlndorf@molgen.mpg.de Edda Klipp Wolfram Liebermeister klipp@molgen.mpg.de
A New Approach To Graph Based Object Classification On Images
 A New Approach To Graph Based Object Classification On Images Sandhya S Krishnan,Kavitha V K P.G Scholar, Dept of CSE, BMCE, Kollam, Kerala, India Sandhya4parvathy@gmail.com Abstract: The main idea of
A New Approach To Graph Based Object Classification On Images Sandhya S Krishnan,Kavitha V K P.G Scholar, Dept of CSE, BMCE, Kollam, Kerala, India Sandhya4parvathy@gmail.com Abstract: The main idea of
8/19/13. Computational problems. Introduction to Algorithm
 I519, Introduction to Introduction to Algorithm Yuzhen Ye (yye@indiana.edu) School of Informatics and Computing, IUB Computational problems A computational problem specifies an input-output relationship
I519, Introduction to Introduction to Algorithm Yuzhen Ye (yye@indiana.edu) School of Informatics and Computing, IUB Computational problems A computational problem specifies an input-output relationship
motifs In the context of networks, the term motif may refer to di erent notions. Subgraph motifs Coloured motifs { }
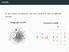 motifs In the context of networks, the term motif may refer to di erent notions. Subgraph motifs Coloured motifs G M { } 2 subgraph motifs 3 motifs Find interesting patterns in a network. 4 motifs Find
motifs In the context of networks, the term motif may refer to di erent notions. Subgraph motifs Coloured motifs G M { } 2 subgraph motifs 3 motifs Find interesting patterns in a network. 4 motifs Find
Structure of biological networks. Presentation by Atanas Kamburov
 Structure of biological networks Presentation by Atanas Kamburov Seminar Gute Ideen in der theoretischen Biologie / Systembiologie 08.05.2007 Overview Motivation Definitions Large-scale properties of cellular
Structure of biological networks Presentation by Atanas Kamburov Seminar Gute Ideen in der theoretischen Biologie / Systembiologie 08.05.2007 Overview Motivation Definitions Large-scale properties of cellular
Database and Knowledge-Base Systems: Data Mining. Martin Ester
 Database and Knowledge-Base Systems: Data Mining Martin Ester Simon Fraser University School of Computing Science Graduate Course Spring 2006 CMPT 843, SFU, Martin Ester, 1-06 1 Introduction [Fayyad, Piatetsky-Shapiro
Database and Knowledge-Base Systems: Data Mining Martin Ester Simon Fraser University School of Computing Science Graduate Course Spring 2006 CMPT 843, SFU, Martin Ester, 1-06 1 Introduction [Fayyad, Piatetsky-Shapiro
Biological Networks Analysis
 Biological Networks Analysis Introduction and Dijkstra s algorithm Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The clustering problem: partition genes into distinct
Biological Networks Analysis Introduction and Dijkstra s algorithm Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The clustering problem: partition genes into distinct
Graph Modeling and Analysis in Oracle
 Graph Modeling and Analysis in Oracle Susie Stephens Principal Product Manager, Life Sciences Oracle Corporation BioPathways, July 30, 2004 Access Distributed Data UltraSearch External Sites Distributed
Graph Modeling and Analysis in Oracle Susie Stephens Principal Product Manager, Life Sciences Oracle Corporation BioPathways, July 30, 2004 Access Distributed Data UltraSearch External Sites Distributed
Incoming, Outgoing Degree and Importance Analysis of Network Motifs
 Available Online at www.ijcsmc.com International Journal of Computer Science and Mobile Computing A Monthly Journal of Computer Science and Information Technology IJCSMC, Vol. 4, Issue. 6, June 2015, pg.758
Available Online at www.ijcsmc.com International Journal of Computer Science and Mobile Computing A Monthly Journal of Computer Science and Information Technology IJCSMC, Vol. 4, Issue. 6, June 2015, pg.758
The Weisfeiler-Lehman Kernel
 The Weisfeiler-Lehman Kernel Karsten Borgwardt and Nino Shervashidze Machine Learning and Computational Biology Research Group, Max Planck Institute for Biological Cybernetics and Max Planck Institute
The Weisfeiler-Lehman Kernel Karsten Borgwardt and Nino Shervashidze Machine Learning and Computational Biology Research Group, Max Planck Institute for Biological Cybernetics and Max Planck Institute
CyKEGGParser User Manual
 CyKEGGParser User Manual Table of Contents Introduction... 3 Development... 3 Citation... 3 License... 3 Getting started... 4 Pathway loading... 4 Laoding KEGG pathways from local KGML files... 4 Importing
CyKEGGParser User Manual Table of Contents Introduction... 3 Development... 3 Citation... 3 License... 3 Getting started... 4 Pathway loading... 4 Laoding KEGG pathways from local KGML files... 4 Importing
Learning Graph Grammars
 Learning Graph Grammars 1 Aly El Gamal ECE Department and Coordinated Science Laboratory University of Illinois at Urbana-Champaign Abstract Discovery of patterns in a given graph - in the form of repeated
Learning Graph Grammars 1 Aly El Gamal ECE Department and Coordinated Science Laboratory University of Illinois at Urbana-Champaign Abstract Discovery of patterns in a given graph - in the form of repeated
Important Example: Gene Sequence Matching. Corrigiendum. Central Dogma of Modern Biology. Genetics. How Nucleotides code for Amino Acids
 Important Example: Gene Sequence Matching Century of Biology Two views of computer science s relationship to biology: Bioinformatics: computational methods to help discover new biology from lots of data
Important Example: Gene Sequence Matching Century of Biology Two views of computer science s relationship to biology: Bioinformatics: computational methods to help discover new biology from lots of data
MARGIN: Maximal Frequent Subgraph Mining Λ
 MARGIN: Maximal Frequent Subgraph Mining Λ Lini T Thomas Satyanarayana R Valluri Kamalakar Karlapalem enter For Data Engineering, IIIT, Hyderabad flini,satyag@research.iiit.ac.in, kamal@iiit.ac.in Abstract
MARGIN: Maximal Frequent Subgraph Mining Λ Lini T Thomas Satyanarayana R Valluri Kamalakar Karlapalem enter For Data Engineering, IIIT, Hyderabad flini,satyag@research.iiit.ac.in, kamal@iiit.ac.in Abstract
EMPIRICAL COMPARISON OF GRAPH CLASSIFICATION AND REGRESSION ALGORITHMS. By NIKHIL S. KETKAR
 EMPIRICAL COMPARISON OF GRAPH CLASSIFICATION AND REGRESSION ALGORITHMS By NIKHIL S. KETKAR A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTORATE OF PHILOSOPHY
EMPIRICAL COMPARISON OF GRAPH CLASSIFICATION AND REGRESSION ALGORITHMS By NIKHIL S. KETKAR A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTORATE OF PHILOSOPHY
Optimal Linear Arrangement of Interval Graphs
 /5 Optimal Linear Arrangement of Interval Graphs Johanne Cohen Fedor Fomin Pinar Heggernes Dieter Kratsch Gregory Kucherov 4 LORIA/CNRS, Nancy, France. Department of Informatics, University of Bergen,
/5 Optimal Linear Arrangement of Interval Graphs Johanne Cohen Fedor Fomin Pinar Heggernes Dieter Kratsch Gregory Kucherov 4 LORIA/CNRS, Nancy, France. Department of Informatics, University of Bergen,
A Comparison of Global and Local Probabilistic Approximations in Mining Data with Many Missing Attribute Values
 A Comparison of Global and Local Probabilistic Approximations in Mining Data with Many Missing Attribute Values Patrick G. Clark Department of Electrical Eng. and Computer Sci. University of Kansas Lawrence,
A Comparison of Global and Local Probabilistic Approximations in Mining Data with Many Missing Attribute Values Patrick G. Clark Department of Electrical Eng. and Computer Sci. University of Kansas Lawrence,
Pathway Assistant: a web portal for metabolic modelling
 Pathway Assistant: a web portal for metabolic modelling Pekko Parikka, Esa Pitkänen, Ari Rantanen, Arto Åkerlund, Esko Ukkonen Firstname.Lastname@cs.Helsinki.Fi Department of Computer Science and HIIT
Pathway Assistant: a web portal for metabolic modelling Pekko Parikka, Esa Pitkänen, Ari Rantanen, Arto Åkerlund, Esko Ukkonen Firstname.Lastname@cs.Helsinki.Fi Department of Computer Science and HIIT
SEEK User Manual. Introduction
 SEEK User Manual Introduction SEEK is a computational gene co-expression search engine. It utilizes a vast human gene expression compendium to deliver fast, integrative, cross-platform co-expression analyses.
SEEK User Manual Introduction SEEK is a computational gene co-expression search engine. It utilizes a vast human gene expression compendium to deliver fast, integrative, cross-platform co-expression analyses.
An overview of Graph Categories and Graph Primitives
 An overview of Graph Categories and Graph Primitives Dino Ienco (dino.ienco@irstea.fr) https://sites.google.com/site/dinoienco/ Topics I m interested in: Graph Database and Graph Data Mining Social Network
An overview of Graph Categories and Graph Primitives Dino Ienco (dino.ienco@irstea.fr) https://sites.google.com/site/dinoienco/ Topics I m interested in: Graph Database and Graph Data Mining Social Network
Optimally Orienting Physical Networks
 Optimally Orienting Physical Networks Dana Silverbush,1, Michael Elberfeld,2, and Roded Sharan 1 1 Blavatnik School of Computer Science, Tel Aviv University, Tel Aviv 69978, Israel, {danasilv,roded}@post.tau.ac.il
Optimally Orienting Physical Networks Dana Silverbush,1, Michael Elberfeld,2, and Roded Sharan 1 1 Blavatnik School of Computer Science, Tel Aviv University, Tel Aviv 69978, Israel, {danasilv,roded}@post.tau.ac.il
Advanced Algorithms and Models for Computational Biology -- a machine learning approach
 Advanced Algorithms and Models for Computational Biology -- a machine learning approach Biological Networks & Network Evolution Eric Xing Lecture 22, April 10, 2006 Reading: Molecular Networks Interaction
Advanced Algorithms and Models for Computational Biology -- a machine learning approach Biological Networks & Network Evolution Eric Xing Lecture 22, April 10, 2006 Reading: Molecular Networks Interaction
Prognosis of Lung Cancer Using Data Mining Techniques
 Prognosis of Lung Cancer Using Data Mining Techniques 1 C. Saranya, M.Phil, Research Scholar, Dr.M.G.R.Chockalingam Arts College, Arni 2 K. R. Dillirani, Associate Professor, Department of Computer Science,
Prognosis of Lung Cancer Using Data Mining Techniques 1 C. Saranya, M.Phil, Research Scholar, Dr.M.G.R.Chockalingam Arts College, Arni 2 K. R. Dillirani, Associate Professor, Department of Computer Science,
GRaph theory is a field of mathematics with applications
 IAENG International Journal of Computer Science, 44:2, IJCS_44_2_3 Partitioning and Classification of RNA Secondary Structures into Pseudonotted and Pseudoknot-free Regions Using a Graph-Theoretical Approach
IAENG International Journal of Computer Science, 44:2, IJCS_44_2_3 Partitioning and Classification of RNA Secondary Structures into Pseudonotted and Pseudoknot-free Regions Using a Graph-Theoretical Approach
Genetic Algorithms Based Solution To Maximum Clique Problem
 Genetic Algorithms Based Solution To Maximum Clique Problem Harsh Bhasin computergrad.com Faridabad, India i_harsh_bhasin@yahoo.com Rohan Mahajan Lingaya s University Faridabad, India mahajanr28@gmail.com
Genetic Algorithms Based Solution To Maximum Clique Problem Harsh Bhasin computergrad.com Faridabad, India i_harsh_bhasin@yahoo.com Rohan Mahajan Lingaya s University Faridabad, India mahajanr28@gmail.com
MetScape User Manual
 MetScape 2.3.2 User Manual A Plugin for Cytoscape National Center for Integrative Biomedical Informatics July 2012 2011 University of Michigan This work is supported by the National Center for Integrative
MetScape 2.3.2 User Manual A Plugin for Cytoscape National Center for Integrative Biomedical Informatics July 2012 2011 University of Michigan This work is supported by the National Center for Integrative
A quick review. Which molecular processes/functions are involved in a certain phenotype (e.g., disease, stress response, etc.)
 Gene expression profiling A quick review Which molecular processes/functions are involved in a certain phenotype (e.g., disease, stress response, etc.) The Gene Ontology (GO) Project Provides shared vocabulary/annotation
Gene expression profiling A quick review Which molecular processes/functions are involved in a certain phenotype (e.g., disease, stress response, etc.) The Gene Ontology (GO) Project Provides shared vocabulary/annotation
Boolean Network Modeling
 Boolean Network Modeling Bioinformatics: Sequence Analysis COMP 571 - Spring 2015 Luay Nakhleh, Rice University Gene Regulatory Networks Gene regulatory networks describe the molecules involved in gene
Boolean Network Modeling Bioinformatics: Sequence Analysis COMP 571 - Spring 2015 Luay Nakhleh, Rice University Gene Regulatory Networks Gene regulatory networks describe the molecules involved in gene
Locality-sensitive hashing and biological network alignment
 Locality-sensitive hashing and biological network alignment Laura LeGault - University of Wisconsin, Madison 12 May 2008 Abstract Large biological networks contain much information about the functionality
Locality-sensitive hashing and biological network alignment Laura LeGault - University of Wisconsin, Madison 12 May 2008 Abstract Large biological networks contain much information about the functionality
Coupling Two Complementary Knowledge Discovery Systems *
 From: Proceedings of the Eleventh nternational FLARS Conference. Copyright 1998, AAA (www.aaai.org). All rights reserved. Coupling Two Complementary Knowledge Discovery Systems * Lawrence B. Holder and
From: Proceedings of the Eleventh nternational FLARS Conference. Copyright 1998, AAA (www.aaai.org). All rights reserved. Coupling Two Complementary Knowledge Discovery Systems * Lawrence B. Holder and
Innovative Study to the Graph-based Data Mining: Application of the Data Mining
 Innovative Study to the Graph-based Data Mining: Application of the Data Mining Amit Kr. Mishra, Pradeep Gupta, Ashutosh Bhatt, Jainendra Singh Rana Abstract Graph-based data mining represents a collection
Innovative Study to the Graph-based Data Mining: Application of the Data Mining Amit Kr. Mishra, Pradeep Gupta, Ashutosh Bhatt, Jainendra Singh Rana Abstract Graph-based data mining represents a collection
Langara College Spring archived
 Instructor: Office: Anoush Dadgar A303k Office Phone: 604-323-5236 Email: Office Hours: Text: GENERAL BIOLOGY I adadgar@langara.bc.ca 10:30 M, T, W & Th or by appointment Biology. Campbell, Neil A. and
Instructor: Office: Anoush Dadgar A303k Office Phone: 604-323-5236 Email: Office Hours: Text: GENERAL BIOLOGY I adadgar@langara.bc.ca 10:30 M, T, W & Th or by appointment Biology. Campbell, Neil A. and
Graph Theory. Graph Theory. COURSE: Introduction to Biological Networks. Euler s Solution LECTURE 1: INTRODUCTION TO NETWORKS.
 Graph Theory COURSE: Introduction to Biological Networks LECTURE 1: INTRODUCTION TO NETWORKS Arun Krishnan Koenigsberg, Russia Is it possible to walk with a route that crosses each bridge exactly once,
Graph Theory COURSE: Introduction to Biological Networks LECTURE 1: INTRODUCTION TO NETWORKS Arun Krishnan Koenigsberg, Russia Is it possible to walk with a route that crosses each bridge exactly once,
An Edge-Swap Heuristic for Finding Dense Spanning Trees
 Theory and Applications of Graphs Volume 3 Issue 1 Article 1 2016 An Edge-Swap Heuristic for Finding Dense Spanning Trees Mustafa Ozen Bogazici University, mustafa.ozen@boun.edu.tr Hua Wang Georgia Southern
Theory and Applications of Graphs Volume 3 Issue 1 Article 1 2016 An Edge-Swap Heuristic for Finding Dense Spanning Trees Mustafa Ozen Bogazici University, mustafa.ozen@boun.edu.tr Hua Wang Georgia Southern
Metabolic Network Visualization Using Constraint Planar Graph Drawing Algorithm.
 Metabolic Network Visualization Using Constraint Planar Graph Drawing Algorithm. Romain Bourqui, Université Bordeaux I LaBRI In Collaboration with : David Auber (LaBRI), Vincent Lacroix (INRIA) and Fabien
Metabolic Network Visualization Using Constraint Planar Graph Drawing Algorithm. Romain Bourqui, Université Bordeaux I LaBRI In Collaboration with : David Auber (LaBRI), Vincent Lacroix (INRIA) and Fabien
A Study of Random Duplication Graphs and Degree Distribution Pattern of Protein-Protein Interaction Networks
 A Study of Random Duplication Graphs and Degree Distribution Pattern of Protein-Protein Interaction Networks by Zheng Ma A thesis presented to the University of Waterloo in fulfillment of the thesis requirement
A Study of Random Duplication Graphs and Degree Distribution Pattern of Protein-Protein Interaction Networks by Zheng Ma A thesis presented to the University of Waterloo in fulfillment of the thesis requirement
REDUCING GRAPH COLORING TO CLIQUE SEARCH
 Asia Pacific Journal of Mathematics, Vol. 3, No. 1 (2016), 64-85 ISSN 2357-2205 REDUCING GRAPH COLORING TO CLIQUE SEARCH SÁNDOR SZABÓ AND BOGDÁN ZAVÁLNIJ Institute of Mathematics and Informatics, University
Asia Pacific Journal of Mathematics, Vol. 3, No. 1 (2016), 64-85 ISSN 2357-2205 REDUCING GRAPH COLORING TO CLIQUE SEARCH SÁNDOR SZABÓ AND BOGDÁN ZAVÁLNIJ Institute of Mathematics and Informatics, University
Discovering Frequent Geometric Subgraphs
 Discovering Frequent Geometric Subgraphs Michihiro Kuramochi and George Karypis Department of Computer Science/Army HPC Research Center University of Minnesota 4-192 EE/CS Building, 200 Union St SE Minneapolis,
Discovering Frequent Geometric Subgraphs Michihiro Kuramochi and George Karypis Department of Computer Science/Army HPC Research Center University of Minnesota 4-192 EE/CS Building, 200 Union St SE Minneapolis,
Data Mining in Bioinformatics Day 3: Graph Mining
 Graph Mining and Graph Kernels Data Mining in Bioinformatics Day 3: Graph Mining Karsten Borgwardt & Chloé-Agathe Azencott February 6 to February 17, 2012 Machine Learning and Computational Biology Research
Graph Mining and Graph Kernels Data Mining in Bioinformatics Day 3: Graph Mining Karsten Borgwardt & Chloé-Agathe Azencott February 6 to February 17, 2012 Machine Learning and Computational Biology Research
Parallelization of Graph Isomorphism using OpenMP
 Parallelization of Graph Isomorphism using OpenMP Vijaya Balpande Research Scholar GHRCE, Nagpur Priyadarshini J L College of Engineering, Nagpur ABSTRACT Advancement in computer architecture leads to
Parallelization of Graph Isomorphism using OpenMP Vijaya Balpande Research Scholar GHRCE, Nagpur Priyadarshini J L College of Engineering, Nagpur ABSTRACT Advancement in computer architecture leads to
APD tool: Mining Anomalous Patterns from Event Logs
 APD tool: Mining Anomalous Patterns from Event Logs Laura Genga 1, Mahdi Alizadeh 1, Domenico Potena 2, Claudia Diamantini 2, and Nicola Zannone 1 1 Eindhoven University of Technology 2 Università Politecnica
APD tool: Mining Anomalous Patterns from Event Logs Laura Genga 1, Mahdi Alizadeh 1, Domenico Potena 2, Claudia Diamantini 2, and Nicola Zannone 1 1 Eindhoven University of Technology 2 Università Politecnica
Web Data mining-a Research area in Web usage mining
 IOSR Journal of Computer Engineering (IOSR-JCE) e-issn: 2278-0661, p- ISSN: 2278-8727Volume 13, Issue 1 (Jul. - Aug. 2013), PP 22-26 Web Data mining-a Research area in Web usage mining 1 V.S.Thiyagarajan,
IOSR Journal of Computer Engineering (IOSR-JCE) e-issn: 2278-0661, p- ISSN: 2278-8727Volume 13, Issue 1 (Jul. - Aug. 2013), PP 22-26 Web Data mining-a Research area in Web usage mining 1 V.S.Thiyagarajan,
Rearrangement of DNA fragments: a branch-and-cut algorithm Abstract. In this paper we consider a problem that arises in the process of reconstruction
 Rearrangement of DNA fragments: a branch-and-cut algorithm 1 C. E. Ferreira 1 C. C. de Souza 2 Y. Wakabayashi 1 1 Instituto de Mat. e Estatstica 2 Instituto de Computac~ao Universidade de S~ao Paulo e-mail:
Rearrangement of DNA fragments: a branch-and-cut algorithm 1 C. E. Ferreira 1 C. C. de Souza 2 Y. Wakabayashi 1 1 Instituto de Mat. e Estatstica 2 Instituto de Computac~ao Universidade de S~ao Paulo e-mail:
Graphs and Trees. An example. Graphs. Example 2
 Graphs and Trees An example How would you describe this network? What kind of model would you write for it? What kind of information would you expect to obtain? Relationship between some of the apoptotic
Graphs and Trees An example How would you describe this network? What kind of model would you write for it? What kind of information would you expect to obtain? Relationship between some of the apoptotic
MICROARRAY IMAGE SEGMENTATION USING CLUSTERING METHODS
 Mathematical and Computational Applications, Vol. 5, No. 2, pp. 240-247, 200. Association for Scientific Research MICROARRAY IMAGE SEGMENTATION USING CLUSTERING METHODS Volkan Uslan and Đhsan Ömür Bucak
Mathematical and Computational Applications, Vol. 5, No. 2, pp. 240-247, 200. Association for Scientific Research MICROARRAY IMAGE SEGMENTATION USING CLUSTERING METHODS Volkan Uslan and Đhsan Ömür Bucak
Sequences Modeling and Analysis Based on Complex Network
 Sequences Modeling and Analysis Based on Complex Network Li Wan 1, Kai Shu 1, and Yu Guo 2 1 Chongqing University, China 2 Institute of Chemical Defence People Libration Army {wanli,shukai}@cqu.edu.cn
Sequences Modeling and Analysis Based on Complex Network Li Wan 1, Kai Shu 1, and Yu Guo 2 1 Chongqing University, China 2 Institute of Chemical Defence People Libration Army {wanli,shukai}@cqu.edu.cn
Relational Learning. Jan Struyf and Hendrik Blockeel
 Relational Learning Jan Struyf and Hendrik Blockeel Dept. of Computer Science, Katholieke Universiteit Leuven Celestijnenlaan 200A, 3001 Leuven, Belgium 1 Problem definition Relational learning refers
Relational Learning Jan Struyf and Hendrik Blockeel Dept. of Computer Science, Katholieke Universiteit Leuven Celestijnenlaan 200A, 3001 Leuven, Belgium 1 Problem definition Relational learning refers
Ph.D. in Computer Science (
 Computer Science 1 COMPUTER SCIENCE http://www.cs.miami.edu Dept. Code: CSC Introduction The Department of Computer Science offers undergraduate and graduate education in Computer Science, and performs
Computer Science 1 COMPUTER SCIENCE http://www.cs.miami.edu Dept. Code: CSC Introduction The Department of Computer Science offers undergraduate and graduate education in Computer Science, and performs
A new Approach for Handling Numeric Ranges for Graph-Based. Knowledge Discovery.
 A new Approach for Handling Numeric Ranges for Graph-Based Knowledge Discovery Oscar E. Romero A. 1, Lawrence B. Holder 2, and Jesus A. Gonzalez B. 1 1 Computer Science Department, INAOE, San Andres Cholula,
A new Approach for Handling Numeric Ranges for Graph-Based Knowledge Discovery Oscar E. Romero A. 1, Lawrence B. Holder 2, and Jesus A. Gonzalez B. 1 1 Computer Science Department, INAOE, San Andres Cholula,
A quick review. The clustering problem: Hierarchical clustering algorithm: Many possible distance metrics K-mean clustering algorithm:
 The clustering problem: partition genes into distinct sets with high homogeneity and high separation Hierarchical clustering algorithm: 1. Assign each object to a separate cluster.. Regroup the pair of
The clustering problem: partition genes into distinct sets with high homogeneity and high separation Hierarchical clustering algorithm: 1. Assign each object to a separate cluster.. Regroup the pair of
Example for calculation of clustering coefficient Node N 1 has 8 neighbors (red arrows) There are 12 connectivities among neighbors (blue arrows)
 Example for calculation of clustering coefficient Node N 1 has 8 neighbors (red arrows) There are 12 connectivities among neighbors (blue arrows) Average clustering coefficient of a graph Overall measure
Example for calculation of clustering coefficient Node N 1 has 8 neighbors (red arrows) There are 12 connectivities among neighbors (blue arrows) Average clustering coefficient of a graph Overall measure
Molecular Distance Geometry and Atomic Orderings
 Molecular Distance Geometry and Atomic Orderings Antonio Mucherino IRISA, University of Rennes 1 Workshop Set Computation for Control ENSTA Bretagne, Brest, France December 5 th 2013 The Distance Geometry
Molecular Distance Geometry and Atomic Orderings Antonio Mucherino IRISA, University of Rennes 1 Workshop Set Computation for Control ENSTA Bretagne, Brest, France December 5 th 2013 The Distance Geometry
31 PathDB: a second generation metabolic database
 31 PathDB: a second generation metabolic database P. Mendes, D.L. Bulmore, A.D. Farmer, P.A. Steadman, M.E. Waugh and S.T. Wlodek National Center for Genome Resources 1800A Old Pecos Trail, Santa Fé, NM
31 PathDB: a second generation metabolic database P. Mendes, D.L. Bulmore, A.D. Farmer, P.A. Steadman, M.E. Waugh and S.T. Wlodek National Center for Genome Resources 1800A Old Pecos Trail, Santa Fé, NM
The Generalized Topological Overlap Matrix in Biological Network Analysis
 The Generalized Topological Overlap Matrix in Biological Network Analysis Andy Yip, Steve Horvath Email: shorvath@mednet.ucla.edu Depts Human Genetics and Biostatistics, University of California, Los Angeles
The Generalized Topological Overlap Matrix in Biological Network Analysis Andy Yip, Steve Horvath Email: shorvath@mednet.ucla.edu Depts Human Genetics and Biostatistics, University of California, Los Angeles
Chapter 28. Outline. Definitions of Data Mining. Data Mining Concepts
 Chapter 28 Data Mining Concepts Outline Data Mining Data Warehousing Knowledge Discovery in Databases (KDD) Goals of Data Mining and Knowledge Discovery Association Rules Additional Data Mining Algorithms
Chapter 28 Data Mining Concepts Outline Data Mining Data Warehousing Knowledge Discovery in Databases (KDD) Goals of Data Mining and Knowledge Discovery Association Rules Additional Data Mining Algorithms
HIDDEN MARKOV MODELS AND SEQUENCE ALIGNMENT
 HIDDEN MARKOV MODELS AND SEQUENCE ALIGNMENT - Swarbhanu Chatterjee. Hidden Markov models are a sophisticated and flexible statistical tool for the study of protein models. Using HMMs to analyze proteins
HIDDEN MARKOV MODELS AND SEQUENCE ALIGNMENT - Swarbhanu Chatterjee. Hidden Markov models are a sophisticated and flexible statistical tool for the study of protein models. Using HMMs to analyze proteins
Rapid Application Development using InforSense Open Workflow and Oracle Chemistry Cartridge Technologies
 Rapid Application Development using InforSense Open Workflow and Oracle Chemistry Cartridge Technologies Anthony C. Arvanites Lead Discovery Informatics Company Introduction Founded: 1999 Platform: Combining
Rapid Application Development using InforSense Open Workflow and Oracle Chemistry Cartridge Technologies Anthony C. Arvanites Lead Discovery Informatics Company Introduction Founded: 1999 Platform: Combining
BIOE 198MI Biomedical Data Analysis. Spring Semester Dynamic programming: finding the shortest path
 BIOE 98MI Biomedical Data Analysis. Spring Semester 09. Dynamic programming: finding the shortest path Page Problem Statement: we re going to learn how to convert real life problem into a graphical diagram
BIOE 98MI Biomedical Data Analysis. Spring Semester 09. Dynamic programming: finding the shortest path Page Problem Statement: we re going to learn how to convert real life problem into a graphical diagram
The Betweenness Centrality Of Biological Networks
 The Betweenness Centrality Of Biological Networks Shivaram Narayanan Thesis submitted to the Faculty of the Virginia Polytechnic Institute and State University in partial fulfillment of the requirements
The Betweenness Centrality Of Biological Networks Shivaram Narayanan Thesis submitted to the Faculty of the Virginia Polytechnic Institute and State University in partial fulfillment of the requirements
Extraction of Frequent Subgraph from Graph Database
 Extraction of Frequent Subgraph from Graph Database Sakshi S. Mandke, Sheetal S. Sonawane Deparment of Computer Engineering Pune Institute of Computer Engineering, Pune, India. sakshi.mandke@cumminscollege.in;
Extraction of Frequent Subgraph from Graph Database Sakshi S. Mandke, Sheetal S. Sonawane Deparment of Computer Engineering Pune Institute of Computer Engineering, Pune, India. sakshi.mandke@cumminscollege.in;
Theory of Computation. Prof. Kamala Krithivasan. Department of Computer Science and Engineering. Indian Institute of Technology, Madras
 Theory of Computation Prof. Kamala Krithivasan Department of Computer Science and Engineering Indian Institute of Technology, Madras Lecture No. # 42 Membrane Computing Today, we shall consider a new paradigm
Theory of Computation Prof. Kamala Krithivasan Department of Computer Science and Engineering Indian Institute of Technology, Madras Lecture No. # 42 Membrane Computing Today, we shall consider a new paradigm
Contents. ! Data sets. ! Distance and similarity metrics. ! K-means clustering. ! Hierarchical clustering. ! Evaluation of clustering results
 Statistical Analysis of Microarray Data Contents Data sets Distance and similarity metrics K-means clustering Hierarchical clustering Evaluation of clustering results Clustering Jacques van Helden Jacques.van.Helden@ulb.ac.be
Statistical Analysis of Microarray Data Contents Data sets Distance and similarity metrics K-means clustering Hierarchical clustering Evaluation of clustering results Clustering Jacques van Helden Jacques.van.Helden@ulb.ac.be
Complexity Results on Graphs with Few Cliques
 Discrete Mathematics and Theoretical Computer Science DMTCS vol. 9, 2007, 127 136 Complexity Results on Graphs with Few Cliques Bill Rosgen 1 and Lorna Stewart 2 1 Institute for Quantum Computing and School
Discrete Mathematics and Theoretical Computer Science DMTCS vol. 9, 2007, 127 136 Complexity Results on Graphs with Few Cliques Bill Rosgen 1 and Lorna Stewart 2 1 Institute for Quantum Computing and School
Algorithms for biological graphs: analysis and enumeration
 Algorithms for biological graphs: analysis and enumeration Andrea Marino Dipartimento di Informatica, Università di Milano, Milano, Italy The aim of enumeration is to list all the feasible solutions of
Algorithms for biological graphs: analysis and enumeration Andrea Marino Dipartimento di Informatica, Università di Milano, Milano, Italy The aim of enumeration is to list all the feasible solutions of
Polynomial-time Algorithm for Determining the Graph Isomorphism
 American Journal of Information Science and Computer Engineering Vol. 3, No. 6, 2017, pp. 71-76 http://www.aiscience.org/journal/ajisce ISSN: 2381-7488 (Print); ISSN: 2381-7496 (Online) Polynomial-time
American Journal of Information Science and Computer Engineering Vol. 3, No. 6, 2017, pp. 71-76 http://www.aiscience.org/journal/ajisce ISSN: 2381-7488 (Print); ISSN: 2381-7496 (Online) Polynomial-time
Clustering Jacques van Helden
 Statistical Analysis of Microarray Data Clustering Jacques van Helden Jacques.van.Helden@ulb.ac.be Contents Data sets Distance and similarity metrics K-means clustering Hierarchical clustering Evaluation
Statistical Analysis of Microarray Data Clustering Jacques van Helden Jacques.van.Helden@ulb.ac.be Contents Data sets Distance and similarity metrics K-means clustering Hierarchical clustering Evaluation
Interval Graphs. Joyce C. Yang. Nicholas Pippenger, Advisor. Arthur T. Benjamin, Reader. Department of Mathematics
 Interval Graphs Joyce C. Yang Nicholas Pippenger, Advisor Arthur T. Benjamin, Reader Department of Mathematics May, 2016 Copyright c 2016 Joyce C. Yang. The author grants Harvey Mudd College and the Claremont
Interval Graphs Joyce C. Yang Nicholas Pippenger, Advisor Arthur T. Benjamin, Reader Department of Mathematics May, 2016 Copyright c 2016 Joyce C. Yang. The author grants Harvey Mudd College and the Claremont
Vector PathBlazer 2.0. User s Manual
 TM Vector PathBlazer 2.0 User s Manual Vector PathBlazer 2.0 User s Manual Published by: Invitrogen 7305 Executive Way Frederick, MD 21704 www.informaxinc.com Copyright 2004 Invitrogen. All rights reserved.
TM Vector PathBlazer 2.0 User s Manual Vector PathBlazer 2.0 User s Manual Published by: Invitrogen 7305 Executive Way Frederick, MD 21704 www.informaxinc.com Copyright 2004 Invitrogen. All rights reserved.
Brief Survey on DNA Sequence Mining
 Available Online at www.ijcsmc.com International Journal of Computer Science and Mobile Computing A Monthly Journal of Computer Science and Information Technology IJCSMC, Vol. 2, Issue. 11, November 2013,
Available Online at www.ijcsmc.com International Journal of Computer Science and Mobile Computing A Monthly Journal of Computer Science and Information Technology IJCSMC, Vol. 2, Issue. 11, November 2013,
CONTENTS 1. Contents
 BIANA Tutorial CONTENTS 1 Contents 1 Getting Started 6 1.1 Starting BIANA......................... 6 1.2 Creating a new BIANA Database................ 8 1.3 Parsing External Databases...................
BIANA Tutorial CONTENTS 1 Contents 1 Getting Started 6 1.1 Starting BIANA......................... 6 1.2 Creating a new BIANA Database................ 8 1.3 Parsing External Databases...................
Enhancing Structure Discovery for Data Mining in Graphical Databases Using Evolutionary Programming
 From: FLAIRS-02 Proceedings. Copyright 2002, AAAI (www.aaai.org). All rights reserved. Enhancing Structure Discovery for Data Mining in Graphical Databases Using Evolutionary Programming Sanghamitra Bandyopadhyay,
From: FLAIRS-02 Proceedings. Copyright 2002, AAAI (www.aaai.org). All rights reserved. Enhancing Structure Discovery for Data Mining in Graphical Databases Using Evolutionary Programming Sanghamitra Bandyopadhyay,
Figure : Example Precedence Graph
 CS787: Advanced Algorithms Topic: Scheduling with Precedence Constraints Presenter(s): James Jolly, Pratima Kolan 17.5.1 Motivation 17.5.1.1 Objective Consider the problem of scheduling a collection of
CS787: Advanced Algorithms Topic: Scheduling with Precedence Constraints Presenter(s): James Jolly, Pratima Kolan 17.5.1 Motivation 17.5.1.1 Objective Consider the problem of scheduling a collection of
Towards a Weighted-Tree Similarity Algorithm for RNA Secondary Structure Comparison
 Towards a Weighted-Tree Similarity Algorithm for RNA Secondary Structure Comparison Jing Jin, Biplab K. Sarker, Virendra C. Bhavsar, Harold Boley 2, Lu Yang Faculty of Computer Science, University of New
Towards a Weighted-Tree Similarity Algorithm for RNA Secondary Structure Comparison Jing Jin, Biplab K. Sarker, Virendra C. Bhavsar, Harold Boley 2, Lu Yang Faculty of Computer Science, University of New
Subgraph Isomorphism. Artem Maksov, Yong Li, Reese Butler 03/04/2015
 Subgraph Isomorphism Artem Maksov, Yong Li, Reese Butler 03/04/2015 Similar Graphs The two graphs below look different but are structurally the same. Definition What is Graph Isomorphism? An isomorphism
Subgraph Isomorphism Artem Maksov, Yong Li, Reese Butler 03/04/2015 Similar Graphs The two graphs below look different but are structurally the same. Definition What is Graph Isomorphism? An isomorphism
Eliminating Annotations by Automatic Flow Analysis of Real-Time Programs
 Eliminating Annotations by Automatic Flow Analysis of Real-Time Programs Jan Gustafsson Department of Computer Engineering, Mälardalen University Box 883, S-721 23 Västerås, Sweden jangustafsson@mdhse
Eliminating Annotations by Automatic Flow Analysis of Real-Time Programs Jan Gustafsson Department of Computer Engineering, Mälardalen University Box 883, S-721 23 Västerås, Sweden jangustafsson@mdhse
Data Mining Technologies for Bioinformatics Sequences
 Data Mining Technologies for Bioinformatics Sequences Deepak Garg Computer Science and Engineering Department Thapar Institute of Engineering & Tecnology, Patiala Abstract Main tool used for sequence alignment
Data Mining Technologies for Bioinformatics Sequences Deepak Garg Computer Science and Engineering Department Thapar Institute of Engineering & Tecnology, Patiala Abstract Main tool used for sequence alignment
Instruction for Reaxys database
 Instruction for Reaxys database Reaxys database allow for easy literature search in the area of chemistry, biology and related Literature search can be performed on the independent ways: - literature;
Instruction for Reaxys database Reaxys database allow for easy literature search in the area of chemistry, biology and related Literature search can be performed on the independent ways: - literature;
Graph-Based Hierarchical Conceptual Clustering
 From: FLAIRS-00 Proceedings. opyright 2000, AAAI (www.aaai.org). All rights reserved. Graph-Based Hierarchical onceptual lustering Istvan Jonyer, Lawrence B. Holder and Diane J. ook Department of omputer
From: FLAIRS-00 Proceedings. opyright 2000, AAAI (www.aaai.org). All rights reserved. Graph-Based Hierarchical onceptual lustering Istvan Jonyer, Lawrence B. Holder and Diane J. ook Department of omputer
OPEN MP-BASED PARALLEL AND SCALABLE GENETIC SEQUENCE ALIGNMENT
 OPEN MP-BASED PARALLEL AND SCALABLE GENETIC SEQUENCE ALIGNMENT Asif Ali Khan*, Laiq Hassan*, Salim Ullah* ABSTRACT: In bioinformatics, sequence alignment is a common and insistent task. Biologists align
OPEN MP-BASED PARALLEL AND SCALABLE GENETIC SEQUENCE ALIGNMENT Asif Ali Khan*, Laiq Hassan*, Salim Ullah* ABSTRACT: In bioinformatics, sequence alignment is a common and insistent task. Biologists align
Dynamic Programming Course: A structure based flexible search method for motifs in RNA. By: Veksler, I., Ziv-Ukelson, M., Barash, D.
 Dynamic Programming Course: A structure based flexible search method for motifs in RNA By: Veksler, I., Ziv-Ukelson, M., Barash, D., Kedem, K Outline Background Motivation RNA s structure representations
Dynamic Programming Course: A structure based flexible search method for motifs in RNA By: Veksler, I., Ziv-Ukelson, M., Barash, D., Kedem, K Outline Background Motivation RNA s structure representations
Efficient SQL-Querying Method for Data Mining in Large Data Bases
 Efficient SQL-Querying Method for Data Mining in Large Data Bases Nguyen Hung Son Institute of Mathematics Warsaw University Banacha 2, 02095, Warsaw, Poland Abstract Data mining can be understood as a
Efficient SQL-Querying Method for Data Mining in Large Data Bases Nguyen Hung Son Institute of Mathematics Warsaw University Banacha 2, 02095, Warsaw, Poland Abstract Data mining can be understood as a
A 2k-Kernelization Algorithm for Vertex Cover Based on Crown Decomposition
 A 2k-Kernelization Algorithm for Vertex Cover Based on Crown Decomposition Wenjun Li a, Binhai Zhu b, a Hunan Provincial Key Laboratory of Intelligent Processing of Big Data on Transportation, Changsha
A 2k-Kernelization Algorithm for Vertex Cover Based on Crown Decomposition Wenjun Li a, Binhai Zhu b, a Hunan Provincial Key Laboratory of Intelligent Processing of Big Data on Transportation, Changsha
Configuration Management for Component-based Systems
 Configuration Management for Component-based Systems Magnus Larsson Ivica Crnkovic Development and Research Department of Computer Science ABB Automation Products AB Mälardalen University 721 59 Västerås,
Configuration Management for Component-based Systems Magnus Larsson Ivica Crnkovic Development and Research Department of Computer Science ABB Automation Products AB Mälardalen University 721 59 Västerås,
Static Analysis for Systems Biology
 Static Analysis for Systems Biology Flemming Nielson, Hanne Riis Nielson Technical University of enmark Corrado Priami, ebora Schuch da Rosa University of Trento Abstract This paper shows how static analysis
Static Analysis for Systems Biology Flemming Nielson, Hanne Riis Nielson Technical University of enmark Corrado Priami, ebora Schuch da Rosa University of Trento Abstract This paper shows how static analysis
Genomic pathways database and biological data management
 SHORT COMMUNICATION Genomic pathways database and biological data management Z. M. Ozsoyoglu*,, G. Ozsoyoglu*, and J. Nadeau*, *Center for Computational Genomics, Case Western Reserve University (CWRU),
SHORT COMMUNICATION Genomic pathways database and biological data management Z. M. Ozsoyoglu*,, G. Ozsoyoglu*, and J. Nadeau*, *Center for Computational Genomics, Case Western Reserve University (CWRU),
