MAGE-ML: MicroArray Gene Expression Markup Language
|
|
|
- Leon Curtis
- 5 years ago
- Views:
Transcription
1 MAGE-ML: MicroArray Gene Expression Markup Language Links: - Full MAGE specification: - MAGE-ML Document Type Definition (DTD): Microarray Gene Expression Markup Language (MAGE-ML) is a language designed to describe and communicate information about microarray based experiments. MAGE-ML is based on XML and can describe microarray designs, microarray manufacturing information, microarray experiment setup and execution information, gene expression data and data analysis results. This paper describes the basic MAGE-ML structure and its main concepts. The structure of MAGE-ML is not simple, therefore we envisage that typically MAGE-ML documents will not be created by hand (i.e., just using text editors), but by user-friendly tools providing web form interface and the ability to link external tab-delimited data, that are currently being developed. Therefore the purpose of this paper is twofold: 1. For microarray laboratory scientists it may be describing all they need to know about MAGE-ML; 2. For bioinformaticians that intend to develop tools for microarray data management or to run such tools for microarray laboratories, this paper can serve as the first introduction to MAGE-ML. MAGE-ML has been automatically derived from Microarray Gene Expression Object Model (MAGE-OM), which is developed and described using the Unified Modelling Language (UML) a standard language for describing object models. Descriptions using UML have an advantage over direct XML document type definitions (DTDs), in many respects. First they use graphical representation depicting the relationships between different entities in a way which is much easier to follow than DTDs. Second, the UML diagrams are primarily meant for humans, while DTDs are meant for computers. Therefore MAGE-OM should be considered as the primary model, and we will explain MAGE-ML by providing simplified fragments of MAGE-OM, rather then XML DTD or XML Schema. For full details of MAGE-OM, mapping rules used to derive MAGE-ML from MAGE-OM, as well as comprehensive glossary of terms see MAGE-OM is expressed in UML, however, for the purposes of this preliminary explanation knowledge of UML is not necessary. The main principle is that boxes represent classes of some objects and lines represent various relationships between the classes. Classes can represent various sets of entities: real-world physical objects (e.g., microarrays), real-world abstractions (e.g., microarray experiments), real-world actions (e.g., hybridization events) or information objects (e.g., microarray data matrices). Here
2 we will not analyse what kinds of relationships there are in UML and how they are represented on diagrams, for details, see, e.g., MAGE-OM is a bit two large to be represented on a single diagram in a readable way. In order to structure the model the UML notion of packages is used. Related classes are grouped together into packages, and quite often represented on the same diagrams. MAGE-OM will be explained package-by-package; a tool able to export MAGE-ML will probably have separate modules and/or user interface sections for separate packages, e.g., you can enter information about array designs in one UI section and information about steps of your microarray experiment using another UI section. On diagrams classes belonging to the package under discussion are coloured yellow, while classes belonging to other packages, therefore detailed elsewhere, but drawn on the current diagram for the purposes of showing inter-package relationships, are coloured grey. Experiment This package is for describing a microarray experiment as a unit. Note two parallel branches on the diagram. On the right-hand side we have experiment blueprint information experiment design and one or more experimental factors that are changed in the course of the experiment to explore whether and how gene expression levels change (e.g., time or drug concentration). On the left-hand side there is experiment execution information. An experiment consists of one or more bioassays (experiment steps), and each bioassay can test for gene expression with one or more experimental factor values fixed (e.g., time = 30min, drug concentration = 15ug/ml). BioAssay A bioassay is a single step within a microarray experiment. There are 3 types of bioassays. A physical bioassay correspond to wet-lab microarray experimental step. A
3 measured bioassays corresponds to a situation after feature extraction has been performed. A derived bioassay corresponds to data processing experimental steps. A physical bioassay is created by applying some amount of some biomaterial to a microarray. Bioassay treatment events (e.g., wash, apply blocking agent etc.) transform physical bioassays into new physical bioassays. A particular type of bioassay treatment is image acquisition. Measured bioassays can have corresponding MeasuredBioAssayData objects (raw data). Derived bioassays are obtained by data transformations, they are linked by BioAssayMap objects. ArrayDesign An array design consists of design element groups as well as information about element zone layout. Physical array design has been made as a subclass of array design, to allow virtual array designs with element groups but no zone layout information; such virtual designs can be used, e.g., to define different reporter-composite sequence mappings for the same physical array design. Zones can be grouped together to form zone groups; zones within the same zone group would have the same spacing between them, whereas zones from different zone groups can have different spacing. Zones/zone groups sometimes are referred to as blocks/metablocks.
4 DesignElement There are three types of design elements. A feature has a position on the array (within some zone), it can be a control feature for other features or controlled by other features. A reporter corresponds to the physical substance synthesized/printed on the array, it can be characterized by one or more biosequence objects which in turn can be characterized by database entries. There can be many features for the same reporter, and features can have one or several mismatches compared to reporter s reference sequence. The third, most abstract kind of design element is a composite sequence. It can have more than one reporter on the same array (e.g., different splice variants) and is characterized by biological characteristics, which are actually again sequences with corresponding database entries. The mapping between reporters and composite sequences is not shown on this diagram, but this is similar to the model of feature-reporter mapping. Also, composite sequences can be aggregated into more abstract composite sequences (also not shown here), e.g., genes of the same functional group etc.
5 BioMaterial BioMaterial is an abstraction of various states of biology-based materials used in various stages of the microarray experiment. Biosource refers to the initial source of material used in hybridization (e.g., cell line or tissue). Biosample is what is extracted from the biosource, and labeled extract is the last state of the biomaterial before hybridization. A biomaterial can be a result of a chain of treatments, each treatment involving one or more biomaterials in some amounts. A special kind of treatment is treatment with some amount of a compound. A simple way to model compounds consisting of other compounds is provided.
6 BioAssayData One of the central principles of MAGE is that data objects are regarded as 3-dimensional matrices, where there are bioassays (experimental steps or conditions) along one dimension, design elements (spots) along the other dimension and quantitation types (e.g., signal intensity, background intensity) along the 3 rd dimension. Bioassay data objects can be represented in one of two ways: as a set of vectors in the form (value, dimension1, dimension2, dimension3) (useful for small amounts of data), or as a 3-D matrix (BioDataCube). Transformations (e.g., filtering, normalization) can be applied to one or more bioassay data objects, resulting in derived data objects. A transformation involves computing values of the resulting 3-D matrix from the values of source matrices, and it also transforms dimensions. On this diagram just the mapping of quantitation types into new quantitation types has been shown; DesignElementMapping and BioAssayMapping are modeled similarly. A quantitation type mapping transforms a list of quantitation types into another list of quantitation types, and it consists of maps that deal with single target quantitation types.
7 QuantitationType A quantitation type can be either a standard quantitation type (a list of these is provided within MAGE) or a specialized quantitation type which should be described in detail. A quantitation type may reference a channel (e.g., Cy3 green signal intensity). Array An array is a physical array which corresponds to some array design. There are three types of information that can be captured about individual arrays and array production process. An individual array can have deviations from the design, either zone defects (e.g., a whole zone of spots is shifted) or individual feature defects. An array group can
8 consist of more than one array printed on the same slide, and fiducials (markings on the surface of the slide that can be used to identify arrays origins) can be printed to facilitate feature detection software accuracy. Array manufacture information also can refer to more than one individual array (sometimes referred to as an array batch), and it can contain protocol information (how the arrays were manufactured) as well as some limited LIMS information (what was printed on the array, on a feature-by-feature basis). BioEvent This diagram is provided to summarize what kinds of events are possible to describe in MAGE. Each event can have a sequence of protocol applications. On the left-hand side there are pysical events (biomaterial treatment, bioassay creation as a generalization for hybridization, and bioassay treatment and image acquisition as a special case), while on the right-hand side there are information processing events (feature extraction, data transformation, maps of data dimensions).
9 Protocol There are two parts for this package. On the upper part there are Protocol, Hardware and Software classes, representing abstract entities. All the parameterizable objects can have parameters, a protocol can involve usage of specific hardware and software, specific hardware might be needed to run some software, and software objects can be composed of other software objects (modules). On the lower part there are classes representing application of abstract entities at a given time point, with parameters filled in by some parameter values. AuditAndSecurity A contact can be either an organization or a person. A person can work for an organization, and an organization can consist of other sub-organizations.
10 Description Many MAGE objects can be further chracterized by attaching Descriptions to them, if there is some important information that cannot be recorded using provided attributes and relations. A Description can consist of a piece of free text, references to database entries, references to ontology entries (annotations) and one or more bibliographic references. A generic NameValueType class also is provided, objects of which can be attached to every MAGE object if even the Description functionality is not sufficient. HigherLevelAnalysis Experimental data (BioAssayData) can be clustered, obtaining one or more top level clusters. Each cluster consists of nodes, where each node in turn can contain subnodes (in the case of hierarchical clustering). A node can be characterized by its values (e.g., some metric of cluster quality), and a node groups together design elements (e.g., spots, genes) or bioassays (i.e., experimental conditions). BioAssayDimension is just an ordered list of bioassays, and DesignElementDimension is an ordered list of design elements.
11
Overview. Experiment Specifications. This tutorial will enable you to
 Defining a protocol in BioAssay Overview BioAssay provides an interface to store, manipulate, and retrieve biological assay data. The application allows users to define customized protocol tables representing
Defining a protocol in BioAssay Overview BioAssay provides an interface to store, manipulate, and retrieve biological assay data. The application allows users to define customized protocol tables representing
SEEK User Manual. Introduction
 SEEK User Manual Introduction SEEK is a computational gene co-expression search engine. It utilizes a vast human gene expression compendium to deliver fast, integrative, cross-platform co-expression analyses.
SEEK User Manual Introduction SEEK is a computational gene co-expression search engine. It utilizes a vast human gene expression compendium to deliver fast, integrative, cross-platform co-expression analyses.
ISAcreator V User Guide. User Guide: V1.3.2 February2011 Contact: Download:
 ISAcreator V 1.3.2 User Guide User Guide: V1.3.2 February2011 Contact: isatools@googlegroups.com Download: http://isa-tools.org 1 USER GUIDE LIST OF IMPROVEMENTS Improved user interface - addition of pure
ISAcreator V 1.3.2 User Guide User Guide: V1.3.2 February2011 Contact: isatools@googlegroups.com Download: http://isa-tools.org 1 USER GUIDE LIST OF IMPROVEMENTS Improved user interface - addition of pure
MAGE-TAB Specification Version 1.1
 MAGE-TAB Specification Version 1.1 July 24, 2009 Abstract This is a specification for a microarray data acquisition and communication standard MAGE-TAB, considerably simpler than MAGE-ML, but powerful
MAGE-TAB Specification Version 1.1 July 24, 2009 Abstract This is a specification for a microarray data acquisition and communication standard MAGE-TAB, considerably simpler than MAGE-ML, but powerful
Extracting Experiment Data from MAGE Files for Analysis
 Extracting Experiment Data from MAGE Files for Analysis Ian Fore 17 March 2005 TABLE OF CONTENTS SUMMARY... 3 INTRODUCTION... 3 Scenario... 3 Overall findings... 4 SOME EXAMPLES... 5 E-MEXP-3... 5 Discovering
Extracting Experiment Data from MAGE Files for Analysis Ian Fore 17 March 2005 TABLE OF CONTENTS SUMMARY... 3 INTRODUCTION... 3 Scenario... 3 Overall findings... 4 SOME EXAMPLES... 5 E-MEXP-3... 5 Discovering
Mining the Biomedical Research Literature. Ken Baclawski
 Mining the Biomedical Research Literature Ken Baclawski Data Formats Flat files Spreadsheets Relational databases Web sites XML Documents Flexible very popular text format Self-describing records XML Documents
Mining the Biomedical Research Literature Ken Baclawski Data Formats Flat files Spreadsheets Relational databases Web sites XML Documents Flexible very popular text format Self-describing records XML Documents
Agilent CytoGenomics 2.0 Feature Extraction for CytoGenomics
 Agilent CytoGenomics 2.0 Feature Extraction for CytoGenomics Quick Start Guide What is Agilent Feature Extraction for CytoGenomics software? 2 Getting Help 4 Starting the program 6 Setting up a Standard
Agilent CytoGenomics 2.0 Feature Extraction for CytoGenomics Quick Start Guide What is Agilent Feature Extraction for CytoGenomics software? 2 Getting Help 4 Starting the program 6 Setting up a Standard
Goal-oriented Schema in Biological Database Design
 Goal-oriented Schema in Biological Database Design Ping Chen Department of Computer Science University of Helsinki Helsinki, Finland 00014 EMAIL: pchen@cs.helsinki.fi Abstract In this paper, I reviewed
Goal-oriented Schema in Biological Database Design Ping Chen Department of Computer Science University of Helsinki Helsinki, Finland 00014 EMAIL: pchen@cs.helsinki.fi Abstract In this paper, I reviewed
Data management: databases and standards. Practical Bioinformatics
 Data management: databases and standards 1 Motivation to submit your data into microarray database 2 Why would you publish your gene expression data in a standard form? Peer-review: review: If the data
Data management: databases and standards 1 Motivation to submit your data into microarray database 2 Why would you publish your gene expression data in a standard form? Peer-review: review: If the data
Gene Clustering & Classification
 BINF, Introduction to Computational Biology Gene Clustering & Classification Young-Rae Cho Associate Professor Department of Computer Science Baylor University Overview Introduction to Gene Clustering
BINF, Introduction to Computational Biology Gene Clustering & Classification Young-Rae Cho Associate Professor Department of Computer Science Baylor University Overview Introduction to Gene Clustering
CEN/ISSS WS/eCAT. Terminology for ecatalogues and Product Description and Classification
 CEN/ISSS WS/eCAT Terminology for ecatalogues and Product Description and Classification Report Final Version This report has been written for WS/eCAT by Mrs. Bodil Nistrup Madsen (bnm.danterm@cbs.dk) and
CEN/ISSS WS/eCAT Terminology for ecatalogues and Product Description and Classification Report Final Version This report has been written for WS/eCAT by Mrs. Bodil Nistrup Madsen (bnm.danterm@cbs.dk) and
How to store and visualize RNA-seq data
 How to store and visualize RNA-seq data Gabriella Rustici Functional Genomics Group gabry@ebi.ac.uk EBI is an Outstation of the European Molecular Biology Laboratory. Talk summary How do we archive RNA-seq
How to store and visualize RNA-seq data Gabriella Rustici Functional Genomics Group gabry@ebi.ac.uk EBI is an Outstation of the European Molecular Biology Laboratory. Talk summary How do we archive RNA-seq
Standardization of Microarray Data and Public Databases
 Standardization of Microarray Data and Public Databases Philippe Rocca-Serra Scientific Data Manager Microarray Informatics Team @ EMBL-EBI European Bioinformatics Institute, Cambridge, UK TIGR DNA Microarray
Standardization of Microarray Data and Public Databases Philippe Rocca-Serra Scientific Data Manager Microarray Informatics Team @ EMBL-EBI European Bioinformatics Institute, Cambridge, UK TIGR DNA Microarray
Organizing, cleaning, and normalizing (smoothing) cdna microarray data
 Organizing, cleaning, and normalizing (smoothing) cdna microarray data All product names are given as examples only and they are not endorsed by the USDA or the University of Illinois. INTRODUCTION The
Organizing, cleaning, and normalizing (smoothing) cdna microarray data All product names are given as examples only and they are not endorsed by the USDA or the University of Illinois. INTRODUCTION The
A Reliable and Distributed LIMS for Efficient Management of the Microarray Experiment Environment
 A Reliable and Distributed LIMS for Efficient Management of the Microarray Experiment Environment Hee-Jeong Jin BK Center for U-Port IT Research Education, Pusan National University, Busan, South Korea,
A Reliable and Distributed LIMS for Efficient Management of the Microarray Experiment Environment Hee-Jeong Jin BK Center for U-Port IT Research Education, Pusan National University, Busan, South Korea,
An Intelligent Agents Architecture for DNA-microarray Data Integration
 An Intelligent Agents Architecture for DNA-microarray Data Integration Mauro Angeletti, Rosario Culmone and Emanuela Merelli Università di Camerino NETTAB Genova, 17 May 2001 NETTAB 2001 University of
An Intelligent Agents Architecture for DNA-microarray Data Integration Mauro Angeletti, Rosario Culmone and Emanuela Merelli Università di Camerino NETTAB Genova, 17 May 2001 NETTAB 2001 University of
VISDA. Installation Guide
 VISDA Installation Guide Table of Contents Introduction...1 Overview of VISDA...1 VISDA Minimal System Requirements...1 Minimal System Requirements...1 VISDA Software and Technology Requirements...2 Software
VISDA Installation Guide Table of Contents Introduction...1 Overview of VISDA...1 VISDA Minimal System Requirements...1 Minimal System Requirements...1 VISDA Software and Technology Requirements...2 Software
Giri Narasimhan. CAP 5510: Introduction to Bioinformatics. ECS 254; Phone: x3748
 CAP 5510: Introduction to Bioinformatics Giri Narasimhan ECS 254; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs07.html 3/3/08 CAP5510 1 Gene g Probe 1 Probe 2 Probe N 3/3/08 CAP5510
CAP 5510: Introduction to Bioinformatics Giri Narasimhan ECS 254; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs07.html 3/3/08 CAP5510 1 Gene g Probe 1 Probe 2 Probe N 3/3/08 CAP5510
Towards Interactive Exploration of Images, Meta-Data, and Analytic Results in the Open Microscopy Environment
 Towards Interactive Exploration of Images, Meta-Data, and Analytic Results in the Open Microscopy Environment Harry Hochheiser and Ilya G. Goldberg Image Informatics and Computational Biology Unit, Laboratory
Towards Interactive Exploration of Images, Meta-Data, and Analytic Results in the Open Microscopy Environment Harry Hochheiser and Ilya G. Goldberg Image Informatics and Computational Biology Unit, Laboratory
A Quick Guide to Using Cerebral in InnateDB
 A Quick Guide to Using Cerebral in InnateDB Cerebral can be used to visualize interaction networks from a set of interactions from InnateDB. Cerebral uses subcellular localization annotations to provide
A Quick Guide to Using Cerebral in InnateDB Cerebral can be used to visualize interaction networks from a set of interactions from InnateDB. Cerebral uses subcellular localization annotations to provide
XML in the bipharmaceutical
 XML in the bipharmaceutical sector XML holds out the opportunity to integrate data across both the enterprise and the network of biopharmaceutical alliances - with little technological dislocation and
XML in the bipharmaceutical sector XML holds out the opportunity to integrate data across both the enterprise and the network of biopharmaceutical alliances - with little technological dislocation and
Dendrogram export options
 BioNumerics Tutorial: Dendrogram export options 1 Introduction In this tutorial, the export options of a dendrogram, displayed in the Dendrogram panel of the Comparison window is covered. This tutorial
BioNumerics Tutorial: Dendrogram export options 1 Introduction In this tutorial, the export options of a dendrogram, displayed in the Dendrogram panel of the Comparison window is covered. This tutorial
Information Management (IM)
 1 2 3 4 5 6 7 8 9 Information Management (IM) Information Management (IM) is primarily concerned with the capture, digitization, representation, organization, transformation, and presentation of information;
1 2 3 4 5 6 7 8 9 Information Management (IM) Information Management (IM) is primarily concerned with the capture, digitization, representation, organization, transformation, and presentation of information;
Customisable Curation Workflows in Argo
 Customisable Curation Workflows in Argo Rafal Rak*, Riza Batista-Navarro, Andrew Rowley, Jacob Carter and Sophia Ananiadou National Centre for Text Mining, University of Manchester, UK *Corresponding author:
Customisable Curation Workflows in Argo Rafal Rak*, Riza Batista-Navarro, Andrew Rowley, Jacob Carter and Sophia Ananiadou National Centre for Text Mining, University of Manchester, UK *Corresponding author:
Clustering Techniques
 Clustering Techniques Bioinformatics: Issues and Algorithms CSE 308-408 Fall 2007 Lecture 16 Lopresti Fall 2007 Lecture 16-1 - Administrative notes Your final project / paper proposal is due on Friday,
Clustering Techniques Bioinformatics: Issues and Algorithms CSE 308-408 Fall 2007 Lecture 16 Lopresti Fall 2007 Lecture 16-1 - Administrative notes Your final project / paper proposal is due on Friday,
Kinematic Analysis dialog
 Dips version 6.0 has arrived, a major new upgrade to our popular stereonet analysis program. New features in Dips 6.0 include a comprehensive kinematic analysis toolkit for planar, wedge and toppling analysis;
Dips version 6.0 has arrived, a major new upgrade to our popular stereonet analysis program. New features in Dips 6.0 include a comprehensive kinematic analysis toolkit for planar, wedge and toppling analysis;
Gene expression & Clustering (Chapter 10)
 Gene expression & Clustering (Chapter 10) Determining gene function Sequence comparison tells us if a gene is similar to another gene, e.g., in a new species Dynamic programming Approximate pattern matching
Gene expression & Clustering (Chapter 10) Determining gene function Sequence comparison tells us if a gene is similar to another gene, e.g., in a new species Dynamic programming Approximate pattern matching
Lazy Big Data Integration
 Lazy Big Integration Prof. Dr. Andreas Thor Hochschule für Telekommunikation Leipzig (HfTL) Martin-Luther-Universität Halle-Wittenberg 16.12.2016 Agenda Integration analytics for domain-specific questions
Lazy Big Integration Prof. Dr. Andreas Thor Hochschule für Telekommunikation Leipzig (HfTL) Martin-Luther-Universität Halle-Wittenberg 16.12.2016 Agenda Integration analytics for domain-specific questions
Corra v2.0 User s Guide
 Corra v2.0 User s Guide Corra is an open source software Licensed under the Apache License, Version 2.0 and it s source code, demo data and this guide can be downloaded at the http://tools.proteomecenter.org/corra/corra.html.
Corra v2.0 User s Guide Corra is an open source software Licensed under the Apache License, Version 2.0 and it s source code, demo data and this guide can be downloaded at the http://tools.proteomecenter.org/corra/corra.html.
ViTraM: VIsualization of TRAnscriptional Modules
 ViTraM: VIsualization of TRAnscriptional Modules Version 2.0 October 1st, 2009 KULeuven, Belgium 1 Contents 1 INTRODUCTION AND INSTALLATION... 4 1.1 Introduction...4 1.2 Software structure...5 1.3 Requirements...5
ViTraM: VIsualization of TRAnscriptional Modules Version 2.0 October 1st, 2009 KULeuven, Belgium 1 Contents 1 INTRODUCTION AND INSTALLATION... 4 1.1 Introduction...4 1.2 Software structure...5 1.3 Requirements...5
D WSMO Data Grounding Component
 Project Number: 215219 Project Acronym: SOA4All Project Title: Instrument: Thematic Priority: Service Oriented Architectures for All Integrated Project Information and Communication Technologies Activity
Project Number: 215219 Project Acronym: SOA4All Project Title: Instrument: Thematic Priority: Service Oriented Architectures for All Integrated Project Information and Communication Technologies Activity
WBEM Web-based Enterprise Management
 1 WBEM Web-based Enterprise Management Outline What is Enterprise Management? What are the drivers in Enterprise Mgt.? Distributed Management Technology Forum (DMTF) Web Based Enterprise Management (WBEM)
1 WBEM Web-based Enterprise Management Outline What is Enterprise Management? What are the drivers in Enterprise Mgt.? Distributed Management Technology Forum (DMTF) Web Based Enterprise Management (WBEM)
ISO INTERNATIONAL STANDARD. Health informatics Genomic Sequence Variation Markup Language (GSVML)
 INTERNATIONAL STANDARD ISO 25720 First edition 2009-08-15 Health informatics Genomic Sequence Variation Markup Language (GSVML) Informatique de santé Langage de balisage de la variation de séquence génomique
INTERNATIONAL STANDARD ISO 25720 First edition 2009-08-15 Health informatics Genomic Sequence Variation Markup Language (GSVML) Informatique de santé Langage de balisage de la variation de séquence génomique
User guide to the Product Registry February 2018
 User guide to the Product Registry February 2018 Here is a guide on how to start to notify products to the web based system for the Danish Product Register. The user guide also describes all the other
User guide to the Product Registry February 2018 Here is a guide on how to start to notify products to the web based system for the Danish Product Register. The user guide also describes all the other
Mascot Insight is a new application designed to help you to organise and manage your Mascot search and quantitation results. Mascot Insight provides
 1 Mascot Insight is a new application designed to help you to organise and manage your Mascot search and quantitation results. Mascot Insight provides ways to flexibly merge your Mascot search and quantitation
1 Mascot Insight is a new application designed to help you to organise and manage your Mascot search and quantitation results. Mascot Insight provides ways to flexibly merge your Mascot search and quantitation
ViTraM: VIsualization of TRAnscriptional Modules
 ViTraM: VIsualization of TRAnscriptional Modules Version 1.0 June 1st, 2009 Hong Sun, Karen Lemmens, Tim Van den Bulcke, Kristof Engelen, Bart De Moor and Kathleen Marchal KULeuven, Belgium 1 Contents
ViTraM: VIsualization of TRAnscriptional Modules Version 1.0 June 1st, 2009 Hong Sun, Karen Lemmens, Tim Van den Bulcke, Kristof Engelen, Bart De Moor and Kathleen Marchal KULeuven, Belgium 1 Contents
STEM. Short Time-series Expression Miner (v1.1) User Manual
 STEM Short Time-series Expression Miner (v1.1) User Manual Jason Ernst (jernst@cs.cmu.edu) Ziv Bar-Joseph Center for Automated Learning and Discovery School of Computer Science Carnegie Mellon University
STEM Short Time-series Expression Miner (v1.1) User Manual Jason Ernst (jernst@cs.cmu.edu) Ziv Bar-Joseph Center for Automated Learning and Discovery School of Computer Science Carnegie Mellon University
GenViewer Tutorial / Manual
 GenViewer Tutorial / Manual Table of Contents Importing Data Files... 2 Configuration File... 2 Primary Data... 4 Primary Data Format:... 4 Connectivity Data... 5 Module Declaration File Format... 5 Module
GenViewer Tutorial / Manual Table of Contents Importing Data Files... 2 Configuration File... 2 Primary Data... 4 Primary Data Format:... 4 Connectivity Data... 5 Module Declaration File Format... 5 Module
Dimension reduction : PCA and Clustering
 Dimension reduction : PCA and Clustering By Hanne Jarmer Slides by Christopher Workman Center for Biological Sequence Analysis DTU The DNA Array Analysis Pipeline Array design Probe design Question Experimental
Dimension reduction : PCA and Clustering By Hanne Jarmer Slides by Christopher Workman Center for Biological Sequence Analysis DTU The DNA Array Analysis Pipeline Array design Probe design Question Experimental
Microarray Data Analysis (V) Preprocessing (i): two-color spotted arrays
 Microarray Data Analysis (V) Preprocessing (i): two-color spotted arrays Preprocessing Probe-level data: the intensities read for each of the components. Genomic-level data: the measures being used in
Microarray Data Analysis (V) Preprocessing (i): two-color spotted arrays Preprocessing Probe-level data: the intensities read for each of the components. Genomic-level data: the measures being used in
Analyzing ICAT Data. Analyzing ICAT Data
 Analyzing ICAT Data Gary Van Domselaar University of Alberta Analyzing ICAT Data ICAT: Isotope Coded Affinity Tag Introduced in 1999 by Ruedi Aebersold as a method for quantitative analysis of complex
Analyzing ICAT Data Gary Van Domselaar University of Alberta Analyzing ICAT Data ICAT: Isotope Coded Affinity Tag Introduced in 1999 by Ruedi Aebersold as a method for quantitative analysis of complex
Data Immersion : Providing Integrated Data to Infinity Scientists. Kevin Gilpin Principal Engineer Infinity Pharmaceuticals October 19, 2004
 Data Immersion : Providing Integrated Data to Infinity Scientists Kevin Gilpin Principal Engineer Infinity Pharmaceuticals October 19, 2004 Informatics at Infinity Understand the nature of the science
Data Immersion : Providing Integrated Data to Infinity Scientists Kevin Gilpin Principal Engineer Infinity Pharmaceuticals October 19, 2004 Informatics at Infinity Understand the nature of the science
Methodology for spot quality evaluation
 Methodology for spot quality evaluation Semi-automatic pipeline in MAIA The general workflow of the semi-automatic pipeline analysis in MAIA is shown in Figure 1A, Manuscript. In Block 1 raw data, i.e..tif
Methodology for spot quality evaluation Semi-automatic pipeline in MAIA The general workflow of the semi-automatic pipeline analysis in MAIA is shown in Figure 1A, Manuscript. In Block 1 raw data, i.e..tif
enanomapper database, search tools and templates Nina Jeliazkova, Nikolay Kochev IdeaConsult Ltd. Sofia, Bulgaria
 enanomapper database, search tools and templates Nina Jeliazkova, Nikolay Kochev IdeaConsult Ltd. Sofia, Bulgaria www.ideaconsult.net Ø enanomapper database: data model, technology; NANoREG data transfer
enanomapper database, search tools and templates Nina Jeliazkova, Nikolay Kochev IdeaConsult Ltd. Sofia, Bulgaria www.ideaconsult.net Ø enanomapper database: data model, technology; NANoREG data transfer
Introduction to BEST Viewpoints
 Introduction to BEST Viewpoints This is not all but just one of the documentation files included in BEST Viewpoints. Introduction BEST Viewpoints is a user friendly data manipulation and analysis application
Introduction to BEST Viewpoints This is not all but just one of the documentation files included in BEST Viewpoints. Introduction BEST Viewpoints is a user friendly data manipulation and analysis application
MAGE-TAB. Draft, 24 March 2006
 MAGE-TAB Draft, 24 March 2006 This is a proposal for a microarray data acquisition and communication standard MAGE-TAB, considerably simpler than MAGE-ML, but powerful enough to encode any microarray experiment
MAGE-TAB Draft, 24 March 2006 This is a proposal for a microarray data acquisition and communication standard MAGE-TAB, considerably simpler than MAGE-ML, but powerful enough to encode any microarray experiment
Network Analysis, Visualization, & Graphing TORonto (NAViGaTOR) User Documentation
 Network Analysis, Visualization, & Graphing TORonto (NAViGaTOR) User Documentation Jurisica Lab, Ontario Cancer Institute http://ophid.utoronto.ca/navigator/ November 10, 2006 Contents 1 Introduction 2
Network Analysis, Visualization, & Graphing TORonto (NAViGaTOR) User Documentation Jurisica Lab, Ontario Cancer Institute http://ophid.utoronto.ca/navigator/ November 10, 2006 Contents 1 Introduction 2
Domain Engineering And Variability In The Reuse-Driven Software Engineering Business.
 OBM 7 -draft 09/02/00 1 Domain Engineering And Variability In The Reuse-Driven Software Engineering Business. Martin L. Griss, Laboratory Scientist, Hewlett-Packard Laboratories, Palo Alto, CA. Effective
OBM 7 -draft 09/02/00 1 Domain Engineering And Variability In The Reuse-Driven Software Engineering Business. Martin L. Griss, Laboratory Scientist, Hewlett-Packard Laboratories, Palo Alto, CA. Effective
The basic arrangement of numeric data is called an ARRAY. Array is the derived data from fundamental data Example :- To store marks of 50 student
 Organizing data Learning Outcome 1. make an array 2. divide the array into class intervals 3. describe the characteristics of a table 4. construct a frequency distribution table 5. constructing a composite
Organizing data Learning Outcome 1. make an array 2. divide the array into class intervals 3. describe the characteristics of a table 4. construct a frequency distribution table 5. constructing a composite
University of Bath Seminar - Biology
 CambridgeSoft Solutions University of Bath Seminar - Biology ChemBioOffice Ultra ChemBioDraw ChemBio3D Ultra ChemBioFinder/ Ultra ChemBioViz Ultra E-Notebook Ultra Inventory Ultra BioAssay Ultra ChemDraw
CambridgeSoft Solutions University of Bath Seminar - Biology ChemBioOffice Ultra ChemBioDraw ChemBio3D Ultra ChemBioFinder/ Ultra ChemBioViz Ultra E-Notebook Ultra Inventory Ultra BioAssay Ultra ChemDraw
Software Service Engineering
 Software Service Engineering Lecture 4: Unified Modeling Language Doctor Guangyu Gao Some contents and notes selected from Fowler, M. UML Distilled, 3rd edition. Addison-Wesley Unified Modeling Language
Software Service Engineering Lecture 4: Unified Modeling Language Doctor Guangyu Gao Some contents and notes selected from Fowler, M. UML Distilled, 3rd edition. Addison-Wesley Unified Modeling Language
Overview of Reporting in the Business Information Warehouse
 Overview of Reporting in the Business Information Warehouse Contents What Is the Business Information Warehouse?...2 Business Information Warehouse Architecture: An Overview...2 Business Information Warehouse
Overview of Reporting in the Business Information Warehouse Contents What Is the Business Information Warehouse?...2 Business Information Warehouse Architecture: An Overview...2 Business Information Warehouse
SAS Data Integration Studio 3.3. User s Guide
 SAS Data Integration Studio 3.3 User s Guide The correct bibliographic citation for this manual is as follows: SAS Institute Inc. 2006. SAS Data Integration Studio 3.3: User s Guide. Cary, NC: SAS Institute
SAS Data Integration Studio 3.3 User s Guide The correct bibliographic citation for this manual is as follows: SAS Institute Inc. 2006. SAS Data Integration Studio 3.3: User s Guide. Cary, NC: SAS Institute
Experiment no 4 Study of Class Diagram in Rational Rose
 Experiment no 4 Study of Class Diagram in Rational Rose Objective-: To studyclass Diagram in Rational Rose. References-: www.developer.com The Unified Modeling Language User Guide by Grady Booch Mastering
Experiment no 4 Study of Class Diagram in Rational Rose Objective-: To studyclass Diagram in Rational Rose. References-: www.developer.com The Unified Modeling Language User Guide by Grady Booch Mastering
SOFTWARE DESIGN COSC 4353 / Dr. Raj Singh
 SOFTWARE DESIGN COSC 4353 / 6353 Dr. Raj Singh UML - History 2 The Unified Modeling Language (UML) is a general purpose modeling language designed to provide a standard way to visualize the design of a
SOFTWARE DESIGN COSC 4353 / 6353 Dr. Raj Singh UML - History 2 The Unified Modeling Language (UML) is a general purpose modeling language designed to provide a standard way to visualize the design of a
Agilent Feature Extraction Software (v10.5)
 Agilent Feature Extraction Software (v10.5) Quick Start Guide What is Agilent Feature Extraction software? Agilent Feature Extraction software extracts data from microarray images produced in two different
Agilent Feature Extraction Software (v10.5) Quick Start Guide What is Agilent Feature Extraction software? Agilent Feature Extraction software extracts data from microarray images produced in two different
Clustering. Lecture 6, 1/24/03 ECS289A
 Clustering Lecture 6, 1/24/03 What is Clustering? Given n objects, assign them to groups (clusters) based on their similarity Unsupervised Machine Learning Class Discovery Difficult, and maybe ill-posed
Clustering Lecture 6, 1/24/03 What is Clustering? Given n objects, assign them to groups (clusters) based on their similarity Unsupervised Machine Learning Class Discovery Difficult, and maybe ill-posed
Tutorial: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and Expression measures
 : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
An Experiment in Visual Clustering Using Star Glyph Displays
 An Experiment in Visual Clustering Using Star Glyph Displays by Hanna Kazhamiaka A Research Paper presented to the University of Waterloo in partial fulfillment of the requirements for the degree of Master
An Experiment in Visual Clustering Using Star Glyph Displays by Hanna Kazhamiaka A Research Paper presented to the University of Waterloo in partial fulfillment of the requirements for the degree of Master
The Arena View. Tutorial. November The Arena is an easy-to-use image management system for analysis of high volume data.
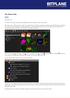 The Arena View Tutorial November 2014 The Arena is an easy-to-use image management system for analysis of high volume data. The Arena is your working centre in Imaris and an interface for visualizing,
The Arena View Tutorial November 2014 The Arena is an easy-to-use image management system for analysis of high volume data. The Arena is your working centre in Imaris and an interface for visualizing,
Comparative Sequencing
 Tutorial for Windows and Macintosh Comparative Sequencing 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Tutorial for Windows and Macintosh Comparative Sequencing 2017 Gene Codes Corporation Gene Codes Corporation 525 Avis Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074
Band matching and polymorphism analysis
 BioNumerics Tutorial: Band matching and polymorphism analysis 1 Aim Fingerprint patterns do not have well-defined characters. Band positions vary continuously, although they do tend to fall into categories,
BioNumerics Tutorial: Band matching and polymorphism analysis 1 Aim Fingerprint patterns do not have well-defined characters. Band positions vary continuously, although they do tend to fall into categories,
CompClustTk Manual & Tutorial
 CompClustTk Manual & Tutorial Brandon King Copyright c California Institute of Technology Version 0.1.10 May 13, 2004 Contents 1 Introduction 1 1.1 Purpose.............................................
CompClustTk Manual & Tutorial Brandon King Copyright c California Institute of Technology Version 0.1.10 May 13, 2004 Contents 1 Introduction 1 1.1 Purpose.............................................
Anchovy User Guide. Copyright Maxprograms
 Copyright 2009-2018 Maxprograms Table of Contents Introduction... 1 Anchovy... 1 Supported Platforms... 1 Supported Formats... 1 GlossML: Glossary Markup Language... 1 Comma Separated Values (CSV)... 1
Copyright 2009-2018 Maxprograms Table of Contents Introduction... 1 Anchovy... 1 Supported Platforms... 1 Supported Formats... 1 GlossML: Glossary Markup Language... 1 Comma Separated Values (CSV)... 1
TIGR MIDAS Version 2.19 TIGR MIDAS. Microarray Data Analysis System. Version 2.19 November Page 1 of 85
 TIGR MIDAS Microarray Data Analysis System Version 2.19 November 2004 Page 1 of 85 Table of Contents 1 General Information...4 1.1 Obtaining MIDAS... 4 1.2 Referencing MIDAS... 4 1.3 A note on non-windows
TIGR MIDAS Microarray Data Analysis System Version 2.19 November 2004 Page 1 of 85 Table of Contents 1 General Information...4 1.1 Obtaining MIDAS... 4 1.2 Referencing MIDAS... 4 1.3 A note on non-windows
Gene Survey: FAQ. Gene Survey: FAQ Tod Casasent DRAFT
 Gene Survey: FAQ Tod Casasent 2016-02-22-1245 DRAFT 1 What is this document? This document is intended for use by internal and external users of the Gene Survey package, results, and output. This document
Gene Survey: FAQ Tod Casasent 2016-02-22-1245 DRAFT 1 What is this document? This document is intended for use by internal and external users of the Gene Survey package, results, and output. This document
(Q)SAR APPLICATION TOOLBOX. Guidance on Importing Databases VERSION 1.1 JANUARY 2009 TABLE OF CONTENTS
 TABLE OF CONTENTS (Q)SAR APPLICATION TOOLBOX VERSION.. INTRODUCTION.... VERTICAL IMPORT.... HORIZONTAL IMPORT.... IMPORTING A DATABASE WITH A NEW ENDPOINT... 5. TIPS AND TRICKS... ANNEX : DIMENSIONS...
TABLE OF CONTENTS (Q)SAR APPLICATION TOOLBOX VERSION.. INTRODUCTION.... VERTICAL IMPORT.... HORIZONTAL IMPORT.... IMPORTING A DATABASE WITH A NEW ENDPOINT... 5. TIPS AND TRICKS... ANNEX : DIMENSIONS...
SlideReader V12. Software Manual. System Requirements. Required Privileges
 SlideReader V12 Software Manual System Requirements - 1.8 GHz Processor or higher - Operating System: WIN XP professional or WIN 7 professional, 64-Bit - 1 Gb RAM or more - Minimum 2 Gb free disk space,
SlideReader V12 Software Manual System Requirements - 1.8 GHz Processor or higher - Operating System: WIN XP professional or WIN 7 professional, 64-Bit - 1 Gb RAM or more - Minimum 2 Gb free disk space,
Genomics - Problem Set 2 Part 1 due Friday, 1/26/2018 by 9:00am Part 2 due Friday, 2/2/2018 by 9:00am
 Genomics - Part 1 due Friday, 1/26/2018 by 9:00am Part 2 due Friday, 2/2/2018 by 9:00am One major aspect of functional genomics is measuring the transcript abundance of all genes simultaneously. This was
Genomics - Part 1 due Friday, 1/26/2018 by 9:00am Part 2 due Friday, 2/2/2018 by 9:00am One major aspect of functional genomics is measuring the transcript abundance of all genes simultaneously. This was
ECS 234: Data Analysis: Clustering ECS 234
 : Data Analysis: Clustering What is Clustering? Given n objects, assign them to groups (clusters) based on their similarity Unsupervised Machine Learning Class Discovery Difficult, and maybe ill-posed
: Data Analysis: Clustering What is Clustering? Given n objects, assign them to groups (clusters) based on their similarity Unsupervised Machine Learning Class Discovery Difficult, and maybe ill-posed
A Mathematical Model For Treatment Selection Literature
 A Mathematical Model For Treatment Selection Literature G. Duncan and W. W. Koczkodaj Abstract Business Intelligence tools and techniques, when applied to a data store of bibliographical references, can
A Mathematical Model For Treatment Selection Literature G. Duncan and W. W. Koczkodaj Abstract Business Intelligence tools and techniques, when applied to a data store of bibliographical references, can
microarray systems VersArray Analyzer 5.0 Image Analysis Software User Manual
 microarray systems VersArray Analyzer 5.0 Image Analysis Software User Manual VersArray Analyzer 5.0 Software User s Guide The VersArray Analyzer Software is for research purposes only. It is not intended
microarray systems VersArray Analyzer 5.0 Image Analysis Software User Manual VersArray Analyzer 5.0 Software User s Guide The VersArray Analyzer Software is for research purposes only. It is not intended
Customizable information fields (or entries) linked to each database level may be replicated and summarized to upstream and downstream levels.
 Manage. Analyze. Discover. NEW FEATURES BioNumerics Seven comes with several fundamental improvements and a plethora of new analysis possibilities with a strong focus on user friendliness. Among the most
Manage. Analyze. Discover. NEW FEATURES BioNumerics Seven comes with several fundamental improvements and a plethora of new analysis possibilities with a strong focus on user friendliness. Among the most
UNIT 5 - UML STATE DIAGRAMS AND MODELING
 UNIT 5 - UML STATE DIAGRAMS AND MODELING UML state diagrams and modeling - Operation contracts- Mapping design to code UML deployment and component diagrams UML state diagrams: State diagrams are used
UNIT 5 - UML STATE DIAGRAMS AND MODELING UML state diagrams and modeling - Operation contracts- Mapping design to code UML deployment and component diagrams UML state diagrams: State diagrams are used
Contents. ! Data sets. ! Distance and similarity metrics. ! K-means clustering. ! Hierarchical clustering. ! Evaluation of clustering results
 Statistical Analysis of Microarray Data Contents Data sets Distance and similarity metrics K-means clustering Hierarchical clustering Evaluation of clustering results Clustering Jacques van Helden Jacques.van.Helden@ulb.ac.be
Statistical Analysis of Microarray Data Contents Data sets Distance and similarity metrics K-means clustering Hierarchical clustering Evaluation of clustering results Clustering Jacques van Helden Jacques.van.Helden@ulb.ac.be
Adaptable and Adaptive Web Information Systems. Lecture 1: Introduction
 Adaptable and Adaptive Web Information Systems School of Computer Science and Information Systems Birkbeck College University of London Lecture 1: Introduction George Magoulas gmagoulas@dcs.bbk.ac.uk October
Adaptable and Adaptive Web Information Systems School of Computer Science and Information Systems Birkbeck College University of London Lecture 1: Introduction George Magoulas gmagoulas@dcs.bbk.ac.uk October
Enterprise Data Catalog for Microsoft Azure Tutorial
 Enterprise Data Catalog for Microsoft Azure Tutorial VERSION 10.2 JANUARY 2018 Page 1 of 45 Contents Tutorial Objectives... 4 Enterprise Data Catalog Overview... 5 Overview... 5 Objectives... 5 Enterprise
Enterprise Data Catalog for Microsoft Azure Tutorial VERSION 10.2 JANUARY 2018 Page 1 of 45 Contents Tutorial Objectives... 4 Enterprise Data Catalog Overview... 5 Overview... 5 Objectives... 5 Enterprise
You can download missing data from the course website, together with the codes R and python that we will run in this exercise.
 Exercise 4: Samples Characterization Aim: Sampling environmental conditions Principal Component Analysis of environmental conditions Hierarchical clustering of sampling spots Interpretation of the environmental
Exercise 4: Samples Characterization Aim: Sampling environmental conditions Principal Component Analysis of environmental conditions Hierarchical clustering of sampling spots Interpretation of the environmental
GE Healthcare. Visualize Analyze. Realize. IN Cell Miner HCM Data management for high-content analysis and screening
 GE Healthcare Visualize Analyze Realize IN Cell Miner HCM Data management for high-content analysis and screening Managing the data mountain High-content analysis (HCA) provides quantitative insights in
GE Healthcare Visualize Analyze Realize IN Cell Miner HCM Data management for high-content analysis and screening Managing the data mountain High-content analysis (HCA) provides quantitative insights in
Clustering Jacques van Helden
 Statistical Analysis of Microarray Data Clustering Jacques van Helden Jacques.van.Helden@ulb.ac.be Contents Data sets Distance and similarity metrics K-means clustering Hierarchical clustering Evaluation
Statistical Analysis of Microarray Data Clustering Jacques van Helden Jacques.van.Helden@ulb.ac.be Contents Data sets Distance and similarity metrics K-means clustering Hierarchical clustering Evaluation
Introduction to GE Microarray data analysis Practical Course MolBio 2012
 Introduction to GE Microarray data analysis Practical Course MolBio 2012 Claudia Pommerenke Nov-2012 Transkriptomanalyselabor TAL Microarray and Deep Sequencing Core Facility Göttingen University Medical
Introduction to GE Microarray data analysis Practical Course MolBio 2012 Claudia Pommerenke Nov-2012 Transkriptomanalyselabor TAL Microarray and Deep Sequencing Core Facility Göttingen University Medical
Data Processing and Analysis in Systems Medicine. Milena Kraus Data Management for Digital Health Summer 2017
 Milena Kraus Digital Health Summer Agenda Real-world Use Cases Oncology Nephrology Heart Insufficiency Additional Topics Data Management & Foundations Biology Recap Data Sources Data Formats Business Processes
Milena Kraus Digital Health Summer Agenda Real-world Use Cases Oncology Nephrology Heart Insufficiency Additional Topics Data Management & Foundations Biology Recap Data Sources Data Formats Business Processes
Fuzzy C-means with Bi-dimensional Empirical Mode Decomposition for Segmentation of Microarray Image
 www.ijcsi.org 316 Fuzzy C-means with Bi-dimensional Empirical Mode Decomposition for Segmentation of Microarray Image J.Harikiran 1, D.RamaKrishna 2, M.L.Phanendra 3, Dr.P.V.Lakshmi 4, Dr.R.Kiran Kumar
www.ijcsi.org 316 Fuzzy C-means with Bi-dimensional Empirical Mode Decomposition for Segmentation of Microarray Image J.Harikiran 1, D.RamaKrishna 2, M.L.Phanendra 3, Dr.P.V.Lakshmi 4, Dr.R.Kiran Kumar
SOFTWARE ENGINEERING Prof.N.L.Sarda Computer Science & Engineering IIT Bombay. Lecture #10 Process Modelling DFD, Function Decomp (Part 2)
 SOFTWARE ENGINEERING Prof.N.L.Sarda Computer Science & Engineering IIT Bombay Lecture #10 Process Modelling DFD, Function Decomp (Part 2) Let us continue with the data modeling topic. So far we have seen
SOFTWARE ENGINEERING Prof.N.L.Sarda Computer Science & Engineering IIT Bombay Lecture #10 Process Modelling DFD, Function Decomp (Part 2) Let us continue with the data modeling topic. So far we have seen
MiChip. Jonathon Blake. October 30, Introduction 1. 5 Plotting Functions 3. 6 Normalization 3. 7 Writing Output Files 3
 MiChip Jonathon Blake October 30, 2018 Contents 1 Introduction 1 2 Reading the Hybridization Files 1 3 Removing Unwanted Rows and Correcting for Flags 2 4 Summarizing Intensities 3 5 Plotting Functions
MiChip Jonathon Blake October 30, 2018 Contents 1 Introduction 1 2 Reading the Hybridization Files 1 3 Removing Unwanted Rows and Correcting for Flags 2 4 Summarizing Intensities 3 5 Plotting Functions
Gegenees genome format...7. Gegenees comparisons...8 Creating a fragmented all-all comparison...9 The alignment The analysis...
 User Manual: Gegenees V 1.1.0 What is Gegenees?...1 Version system:...2 What's new...2 Installation:...2 Perspectives...4 The workspace...4 The local database...6 Populate the local database...7 Gegenees
User Manual: Gegenees V 1.1.0 What is Gegenees?...1 Version system:...2 What's new...2 Installation:...2 Perspectives...4 The workspace...4 The local database...6 Populate the local database...7 Gegenees
UBL Library Content Methodology
 UBL Library Content Methodology The purpose of this document is two-fold: 1. To explain how we got to where we are with the UBL vocabulary, we felt it necessary to provide a background to the rationale
UBL Library Content Methodology The purpose of this document is two-fold: 1. To explain how we got to where we are with the UBL vocabulary, we felt it necessary to provide a background to the rationale
Conceptual Model for a Software Maintenance Environment
 Conceptual Model for a Software Environment Miriam. A. M. Capretz Software Engineering Lab School of Computer Science & Engineering University of Aizu Aizu-Wakamatsu City Fukushima, 965-80 Japan phone:
Conceptual Model for a Software Environment Miriam. A. M. Capretz Software Engineering Lab School of Computer Science & Engineering University of Aizu Aizu-Wakamatsu City Fukushima, 965-80 Japan phone:
Unsupervised learning: Clustering & Dimensionality reduction. Theo Knijnenburg Jorma de Ronde
 Unsupervised learning: Clustering & Dimensionality reduction Theo Knijnenburg Jorma de Ronde Source of slides Marcel Reinders TU Delft Lodewyk Wessels NKI Bioalgorithms.info Jeffrey D. Ullman Stanford
Unsupervised learning: Clustering & Dimensionality reduction Theo Knijnenburg Jorma de Ronde Source of slides Marcel Reinders TU Delft Lodewyk Wessels NKI Bioalgorithms.info Jeffrey D. Ullman Stanford
Data Science. Data Analyst. Data Scientist. Data Architect
 Data Science Data Analyst Data Analysis in Excel Programming in R Introduction to Python/SQL/Tableau Data Visualization in R / Tableau Exploratory Data Analysis Data Scientist Inferential Statistics &
Data Science Data Analyst Data Analysis in Excel Programming in R Introduction to Python/SQL/Tableau Data Visualization in R / Tableau Exploratory Data Analysis Data Scientist Inferential Statistics &
Triclustering in Gene Expression Data Analysis: A Selected Survey
 Triclustering in Gene Expression Data Analysis: A Selected Survey P. Mahanta, H. A. Ahmed Dept of Comp Sc and Engg Tezpur University Napaam -784028, India Email: priyakshi@tezu.ernet.in, hasin@tezu.ernet.in
Triclustering in Gene Expression Data Analysis: A Selected Survey P. Mahanta, H. A. Ahmed Dept of Comp Sc and Engg Tezpur University Napaam -784028, India Email: priyakshi@tezu.ernet.in, hasin@tezu.ernet.in
Batch Processing (CasaXPS version and above)
 Copyright 2004, Casa Software Ltd. All Rights Reserved. 1 of 8 Batch Processing (CasaXPS version 2.2.50 and above) Batch processing is aimed at situations where a set of essentially equivalent samples
Copyright 2004, Casa Software Ltd. All Rights Reserved. 1 of 8 Batch Processing (CasaXPS version 2.2.50 and above) Batch processing is aimed at situations where a set of essentially equivalent samples
Biosphere: the interoperation of web services in microarray cluster analysis
 Biosphere: the interoperation of web services in microarray cluster analysis Kei-Hoi Cheung 1,2,*, Remko de Knikker 1, Youjun Guo 1, Guoneng Zhong 1, Janet Hager 3,4, Kevin Y. Yip 5, Albert K.H. Kwan 5,
Biosphere: the interoperation of web services in microarray cluster analysis Kei-Hoi Cheung 1,2,*, Remko de Knikker 1, Youjun Guo 1, Guoneng Zhong 1, Janet Hager 3,4, Kevin Y. Yip 5, Albert K.H. Kwan 5,
Database and R Interfacing for Annotated Microarray Data
 DSC 2003 Working Papers (Draft Versions) http://www.ci.tuwien.ac.at/conferences/dsc-2003/ Database and R Interfacing for Annotated Microarray Data Michael Mader, Werner Mewes GSF Research Center, Institute
DSC 2003 Working Papers (Draft Versions) http://www.ci.tuwien.ac.at/conferences/dsc-2003/ Database and R Interfacing for Annotated Microarray Data Michael Mader, Werner Mewes GSF Research Center, Institute
 GUIDELINES FOR DATABASES AS PUBLIC RECORDS PURPOSE... 1 OVERVIEW... 1 POLICY GUIDELINES... 2 OFFICIAL REQUEST... 2 EXEMPT RECORDS... 2 REQUESTS FOR SPECIFIC RECORDS... 3 REQUEST FOR ENTIRE DATABASES OR
GUIDELINES FOR DATABASES AS PUBLIC RECORDS PURPOSE... 1 OVERVIEW... 1 POLICY GUIDELINES... 2 OFFICIAL REQUEST... 2 EXEMPT RECORDS... 2 REQUESTS FOR SPECIFIC RECORDS... 3 REQUEST FOR ENTIRE DATABASES OR
Appendix A - Glossary(of OO software term s)
 Appendix A - Glossary(of OO software term s) Abstract Class A class that does not supply an implementation for its entire interface, and so consequently, cannot be instantiated. ActiveX Microsoft s component
Appendix A - Glossary(of OO software term s) Abstract Class A class that does not supply an implementation for its entire interface, and so consequently, cannot be instantiated. ActiveX Microsoft s component
Order Preserving Triclustering Algorithm. (Version1.0)
 Order Preserving Triclustering Algorithm User Manual (Version1.0) Alain B. Tchagang alain.tchagang@nrc-cnrc.gc.ca Ziying Liu ziying.liu@nrc-cnrc.gc.ca Sieu Phan sieu.phan@nrc-cnrc.gc.ca Fazel Famili fazel.famili@nrc-cnrc.gc.ca
Order Preserving Triclustering Algorithm User Manual (Version1.0) Alain B. Tchagang alain.tchagang@nrc-cnrc.gc.ca Ziying Liu ziying.liu@nrc-cnrc.gc.ca Sieu Phan sieu.phan@nrc-cnrc.gc.ca Fazel Famili fazel.famili@nrc-cnrc.gc.ca
EECS730: Introduction to Bioinformatics
 EECS730: Introduction to Bioinformatics Lecture 15: Microarray clustering http://compbio.pbworks.com/f/wood2.gif Some slides were adapted from Dr. Shaojie Zhang (University of Central Florida) Microarray
EECS730: Introduction to Bioinformatics Lecture 15: Microarray clustering http://compbio.pbworks.com/f/wood2.gif Some slides were adapted from Dr. Shaojie Zhang (University of Central Florida) Microarray
Agilent Genomic Workbench Feature Extraction 10.10
 Agilent Genomic Workbench Feature Extraction 10.10 Quick Start Guide Starting Feature Extraction on PCs with Agilent Scanners 2 Getting Help 6 What s new in version 10.10? 9 Starting the program and setting
Agilent Genomic Workbench Feature Extraction 10.10 Quick Start Guide Starting Feature Extraction on PCs with Agilent Scanners 2 Getting Help 6 What s new in version 10.10? 9 Starting the program and setting
21. Document Component Design
 Page 1 of 17 1. Plan for Today's Lecture Methods for identifying aggregate components 21. Document Component Design Bob Glushko (glushko@sims.berkeley.edu) Document Engineering (IS 243) - 11 April 2005
Page 1 of 17 1. Plan for Today's Lecture Methods for identifying aggregate components 21. Document Component Design Bob Glushko (glushko@sims.berkeley.edu) Document Engineering (IS 243) - 11 April 2005
