Biomedical Paper AnatomyBrowser: A Novel Approach to Visualization and Integration of Medical Information
|
|
|
- Maximillian Simmons
- 5 years ago
- Views:
Transcription
1 Computer Aided Surgery 4: (1999) Biomedical Paper AnatomyBrowser: A Novel Approach to Visualization and Integration of Medical Information P. Golland, M.Sc., R. Kikinis, M.D., M. Halle, Ph.D., C. Umans, B.A., W.E.L. Grimson, Ph.D., M.E. Shenton, Ph.D., and J.A. Richolt, M.D. Artificial Intelligence Laboratory, Massachusetts Institute of Technology (P.G., W.E.L.G.), Cambridge, Massachusetts and Surgical Planning Laboratory, Brigham and Women s Hospital (R.K., M.H., C.U., M.E.S., J.A.R.), Boston, Massachusetts ABSTRACT In this article, we present a novel technique for visualization of three-dimensional (3D) surface models, as well as its implementation in a system called AnatomyBrowser. Using our approach, visualization of 3D surface models is performed in two separate steps: a pre-rendering step, in which the models are rendered and saved in a special format, and an actual display step, in which the final result of rendering is generated using information from the prerendering step. Whereas prerendering requires high-end graphics hardware, the final image generation and display can be implemented efficiently in software. Moreover, our current implementation of Anatomy- Browser interface uses the Java programming language and can therefore be readily run on a wide range of systems, including low-end computers with no special graphics hardware. In addition to visualization of 3D models and 2D slices, AnatomyBrowser provides a rich set of annotation and cross-referencing capabilities. We demonstrate several possible applications for AnatomyBrowser, including interactive anatomy atlases, surgery planning, and assistance in segmentation. Comp Aid Surg 4: (1999) Wiley-Liss, Inc. Key words: Key links: AnatomyBrowser, multilayer images, 3D surface models, visualization OBJECTIVE With recent developments in magnetic resonance imaging (MRI) technology, it has become possible to obtain high-quality medical images that are extremely useful for clinical studies, surgical planning, and other applications. This has spurred the rapid development of a family of information extraction and analysis algorithms using medical imagery. Examples include segmentation, registration, and shape and deformation modeling. With the introduction of such images and methods of analysis, the importance of visualization of the results, especially three-dimensional (3D) data, has also become apparent. In this article, we introduce Anatomy- Browser, a visualization and integration package for medical applications. A novel approach to 3D surface model visualization has been developed and implemented in this system. Visualization is Received September 8, 1998; accepted July 6, Address correspondence/reprint requests: Polina Golland, Artificial Intelligence Laboratory, 545 Technology Square, No. 737, Cambridge, MA polina@ai.mit.edu Wiley-Liss, Inc.
2 130 Golland et al.: AnatomyBrowser performed by two separate components of the system: the back-end component and the user-end interface. The back-end component uses special graphics hardware to render the models and save both intensity and depth information in a special representation called multilayer images. The userend interface reads the multilayer images of the scene and generates an image to be displayed for the user. Since multilayer images contain depth information, the user-end interface supports 3D model manipulation, such as partially transparent surfaces, changes in surface color, and depth queries. The image generation process in the user-end interface requires few computational resources and can be run on a low-end computer. The user-end interface is implemented as a Java applet, and as a consequence is platform independent. It can be readily run using any Web browser that supports Java, and in fact is available on-line. 1 Possible applications of AnatomyBrowser include interactive anatomy atlases (which was an original inspiration for this work), assistance in model-based segmentation, surgery planning, and others. Related Work Work in digital anatomy atlases was pioneered by Höhne et al., 6 followed by other research groups, 7,11 in an effort to develop both extensive medical image databases and visualization software suitable for display and manipulation of medical data. Clinical systems for surgical planning, surgical simulation, and image guided surgery 5,14 are other examples of applications that depend greatly on visualization tools. These systems usually provide features specific for the type of surgery, whereas the atlas-oriented visualization systems tend to support fairly general visualization and annotation capabilities. A common feature among these systems is a need for high-end graphics hardware. To the best of our knowledge, Anatomy- Browser is the first system to provide an extensive set of visualization and cross-referencing capabilities without using specialized graphics support at the user end of the system. Because of its design, AnatomyBrowser is better suited for visualization of atlases, but the same approach to visualization can be used in a surgical tool. A later section of this article contains examples of AnatomyBrowser being used as a general visualization tool as well as a reference tool for surgical applications. Digital atlases are commonly used for modeldriven segmentation. In this application, the atlas is treated as a template that is to be warped by the algorithm to match the input gray-scale data set (McInerney and Tersopoulos 10 contains a survey of deformable model methods). Another approach to segmentation that uses atlases is based on registration of gray-scale images. The atlas gray-scale data set is registered to the input gray-scale data set, and the segmentation is mapped from the atlas onto the input data using the results of registration (see Maintz and Viergever 9 for a comprehensive survey on registration methods). In both cases, Anatomy- Browser can provide visualization of intermediate results in the algorithms. Before proceeding with the system description, we would like to stress that AnatomyBrowser is a visualization tool for displaying segmentation results combined with other types of medical information, but it is not directly a part of a segmentation process. A segmented data set can be an output of an automatic segmentation algorithm or a result of manual segmentation. It is processed by AnatomyBrowser in exactly the same manner, independently of the way it was obtained. This article is organized as follows. Our approach to 3D visualization is introduced and discussed in the next section, and the two subsequent sections describe implementation of the Anatomy- Browser back-end components and its user-end interface, respectively. Next comes a section presenting several examples of AnatomyBrowser being used on various data sets, followed by the system performance evaluation and concluding remarks. MATERIALS AND METHODS Novel Approach to 3D Model Visualization It has traditionally been accepted that special hardware and software are required for visualization of 3D surface models. Rendering of 3D surface models is a computationally intensive operation that requires special hardware to achieve acceptable update rates, and since rendering is considered an integral part of the visualization process, the visualization itself is thought to be feasible only on high-end computers with hardware graphics acceleration. In this article, we propose a change in this paradigm. We separate the visualization process into two steps. First, the models are pre-rendered using a high-end graphics system, and the resulting images are saved in an intermediate format called multilayer images. Then, multilayer images are processed by the user-end interface to produce an image of a 3D scene. Multilayer images are orga-
3 Golland et al.: AnatomyBrowser 131 Fig. 1. Multilayer image. Three-dimensional scene consists of three surfaces (A, B, C). The view rays and resulting stacks in the multilayer image are shown for six different pixels. Intensity and depth values are omitted for simplicity; only the structure indices are shown. nized in such a way that the image generation by the user-end component requires only modest computational resources. This approach can be considered a compromise between dynamic rendering of a scene and a set of static images of the same scene. Multilayer images contain depth information, and therefore allow partial manipulation of the scene, such as removal of models, changing surface transparency, changing surface color, etc., but they are not equivalent to dynamic rendering, as they provide only the views that were selected and rendered in the pre-processing step. The input to the pre-rendering step is a set of surface models of the anatomical structures of interest. These surface models are represented using polygonal meshes. During the pre-rendering step, every model is rendered in isolation with all other models removed from the image. Two images are computed and saved for every model: an intensity image generated by the renderer, and a depth image. To compute the depth values, every vertex of the model mesh is projected onto the optical axis of the imaginary camera, and the depth of the mesh faces is obtained by interpolating between the depth values of the vertices. Intensity and depth images of all models are then combined in the following way: for every pixel, a stack of k,i,d tuples is saved, where k is a unique index associated with a surface model (i.e., an anatomical structure that this model represents), I is the surface intensity from the intensity image, and d is the depth value from the corresponding depth image. The tuples are sorted in order of increasing depth (Fig. 1). To obtain the resulting image intensity for a particular pixel, intensity values in the corresponding stack are treated as colored (and perhaps partially transparent) layers that are piled on top of each other; hence, the name multilayer image. This approach is reminiscent of z-buffering techniques, 4 with one significant difference: For a particular pixel, a z-buffer stores a single intensity value which is overwritten every time a surface with a smaller depth value is encountered, whereas in a multilayer image the previous intensity value is stored in a stack when a new intensity with a smaller depth value is found for the pixel. At rendering time, all models are assigned the color white and are treated as fully opaque. Surface colors and opacities are set at display time and are used to compute the resulting image intensity. For modeling colored surfaces, we assume that the reflectivity properties of a surface are independent of the light wavelength. 4 Consider a white surface that produces an image of intensity (color) I w at a certain pixel. Strictly speaking, the color of the image of the white surface is a threevector [I w I w I w ], but we treat it as if it were a scalar value because its three components are always equal under white illumination. Suppose we change the color of the surface to C [R GB] without
4 132 Golland et al.: AnatomyBrowser changing its reflectivity properties, such as diffuse and specular reflectivity coefficients. According to our model, the image intensity at the same pixel will change to c [r gb] such that c CI w N r gb RI w GI w BI w. (1) This implies that the color of a layer in a multilayer image can be obtained by simply multiplying the layer intensity, which is computed during the prerendering step, by the color of the corresponding surface, which is set by a user at display time. To handle partially transparent surfaces, every surface model is assigned an opacity value,, ranging between 0 and 1 ( 0 corresponds to a fully transparent surface, 1 corresponds to a fully opaque surface). We use a premultiplied color representation with a front-to-back compositing algorithm 2 for layer compositing. For a layer of color c (r, g, b) and opacity, a premultiplied color is defined as a four-vector ĉ c r g b (2) Combining with Equation (1), we obtain ĉ RI GI BI (3) where I is the intensity of the layer computed in the prerendering step, and C [R GB] is the userdefined surface color. Premultiplied colors provide a very efficient way to composite color layers. It can be shown 2 that a layer of (premultiplied) color fˆ composited on top of a layer of color bˆ is equivalent to a single layer of (premultiplied) color fˆ 1 bˆ (4) where is the opacity of top layer fˆ. This is a very simple compositing rule that requires only a few additions and multiplications and can be implemented efficiently using lookup tables for surface colors and opacities. Additional time savings come from the fact that for many pixels, it is not necessary to composite all the layers in their stacks, as the first visible surface is usually opaque. A simple test on the opacity of the resulting layer can be used to detect this situation and stop the compositing process. To summarize, our approach to visualization of 3D surface models consists of two separate stages. First, the models are rendered, and for every pixel in the image a set of surface intensities and depth values is saved in a multilayer image. This operation requires high-end graphics hardware for rendering. Then, at the display time, the intensity values are multiplied by the user-defined colors and opacities and composited using the front-to-back compositing rule. This process can be efficiently implemented in software, as it does not require a lot of computation. The next two sections describe AnatomyBrowser, a system that employs the technique presented in this section for visualization of medical images. AnatomyBrowser Back-End Components AnatomyBrowser consists of two parts: the backend system for data preprocessing, and the user-end interface for data visualization. Fig. 2 shows the data flow in the system. The input to the back-end system includes gray-scale and segmented data sets, a list of anatomical structures with the label values used for segmentation of the structures, and an optional hierarchy file that defines organization of the structures. At the pre-processing stage, 3D surface models are generated, rendered, and saved as multilayer images; the slice data sets are resampled and compressed; and the text information is processed to establish correspondence between different types of information available to the system. An anatomical structure can have information of a different nature associated with it: a name, a 3D surface model, a set of 2D slices, or text notes. To tie them together and provide efficient cross-referencing between them, AnatomyBrowser automatically assigns a unique index (id) to every anatomical structure in the input data set, i.e., in the list of structures provided by the user. The index is then used by the back-end components for tagging the data associated with the structure and by the user-end components for looking up information on the structure. The current implementation of the model generation and the model-rendering components is based on the Visualization Toolkit (VTK), a software package that uses available GL libraries to take advantage of graphics resources of a particular system. 13 Any other rendering library that supports scene manipulation (e.g., Inventor by SGI) could also be used for this purpose. 3D Model Generation Surface models are constructed from segmented data sets using the Marching Cubes algorithm. 8 First, every border voxel in the segmented slice set is considered with its neighboring voxels, and a
5 Golland et al.: AnatomyBrowser 133 Fig. 2. Data flow in AnatomyBrowser. Arrows indicate flow of data between different components of the system. single patch of the surface is constructed based on the local behavior of the boundary. Next, patches from neighboring voxels are merged to form a single mesh that approximates the surface of the structure. To decrease the number of polygons (triangles) in the representation and improve the surface appearance, a certain amount of decimation and smoothing is performed on the mesh under constraints on the accuracy of representation. Each surface model is saved in a separate file (one file per model) to be read by the pre-rendering component. We used an implementation of the algorithm that is publicly available as a part of VTK. 13 Note that the models are specified in the coordinate system that is naturally defined by the slice volume. This simplifies implementation of cross-referencing between 3D models and crosssectional views. 3D Image Pre-rendering The main difference between AnatomyBrowser and other visualization packages lies in this component. It is in charge of model rendering and multilayer image generation, and is essentially an implementation of the prerendering step described earlier. The current implementation of this component is very similar to other 3D model viewers used in the field. 5,6,14 The viewer provides controls for adjusting global scene parameters, such as a viewpoint and lighting, as well as surface properties of individual models (color, transparency, and reflectivity). Fig. 3(a) shows the viewer interface with the 3D surface models for the brain atlas data set. The menu buttons correspond to control panels for scene manipulation. The viewer also maintains a list of views to be saved as multilayer images: The default is the six standard views (front, back, left, right, top, and bottom), but other views can be added to the list by adjusting the scene parameters until the image is satisfactory and clicking on the Add View button on the viewer control panel. Fig. 3(b) shows additional examples for the brain atlas data set generated by AnatomyBrowser. After selection of views is completed, the rendering process is activated. For each view, a separate multilayer image is generated and stored. Slice Reformatting The slice data sets undergo a set of transformations in the back-end component. The goal of this component is to provide the best representation of slice data for the user-end interface. Gray-scale slice intensities are scaled from a
6 134 Golland et al.: AnatomyBrowser Fig. 3. Prerendering component of AnatomyBrowser. (a) Viewer interface with the brain atlas data set. The view list contains the six standard views. (b) Additional examples of the 3D views generated for the brain atlas data set. 16-bit representation to an 8-bit representation based on the user-defined window width w and window level l parameters. Window level l defines the gray-scale level that will be mapped to grayscale level 128, and window width w defines the width of the gray-scale range to be mapped to 256 gray-scale levels of the display: Î I 0, I l w/ w I l, l w/2 I l w/2 255, I l w/2 (5) Segmented slice intensities are remapped using the set of unique indices associated with the anatomical structures. For example, if a certain model is segmented using the value 15 and is assigned the index 8, all the pixels in the segmented data set with values of 15 will be assigned the value 8. After remapping is performed, the pixel value can be used as an index into a lookup table for the structure name, color, transparency, etc. Both gray-scale and segmented slice volumes are resampled along three standard directions (axial, coronal, and sagittal) using an isotropic voxel size. The size of the new voxels is determined by the smallest dimension of the original voxels. This operation is needed to speed up the display of slices, trading storage space for performance time. We have also implemented and tested a version that resamples the volume online in the user-end inter-
7 Fig. 4. AnatomyBrowser user-end interface. (a) Example of the interface for the brain data set. The cross-hair positions correspond to the same 3D point. Some of the structures are annotated on the 3D display. The sagittal slice is slightly zoomed in on. (b) AnatomyBrowser for the brain data set with the frontal, temporal, and parietal lobes collapsed into single nodes. Compare to (a) for the differences in the hierarchy panel, the colors of the structures, and annotation labels.
8 136 Golland et al.: AnatomyBrowser face, but it proved to be significantly slower than the current implementation that retrieves resampled slices off a disk. The system can switch between the two implementations in a manner transparent to the user. After resampling, gray-scale slices are stored as gif files and segmented slices are compressed using run-length encoding. Structure Hierarchy AnatomyBrowser supports hierarchical organization of anatomical structures. A simple text file format is used to specify the nodes of the hierarchical tree. For example, [Head]? Brain x Skin x Skull [Brain] x Temporal lobe x Frontal lobe x Parietal lobe defines a hierarchy tree with a root named Head that has three children: two leaves, Skin and Skull, and the node Brain, which has three children of its own: Temporal lobe, Frontal lobe, and Parietal lobe. A name can be declared to be a single structure (x) or a group of structures (?). The group of structures is specified by listing all the elements (structures or subgroups) right after the declaration of the group name ([...]). Hierarchy files can be as simple as a list of structures, or as complicated as a tree of several levels with more than a hundred structures in it (e.g., the brain atlas in Fig. 4). The back-end system reads the hierarchy file, verifies that there exists a surface model for every name that was specified as a single structure, and attaches the structure index to the name in the hierarchy. A new, augmented hierarchy file is then written out for the user-end interface. In addition to the hierarchy file, an HTML file is generated by this component. The HTML file contains all the parameters required by the interface, such as file names, slice volume size, and voxel size. In summary, after processing the input data, the back-end part of the system generates the following files: an HTML file that defines all the necessary parameters for the interface a hierarchy file that specifies organization of the anatomical structures multilayer images (one file per view) for the 3D display resampled and compressed slices (two files per slice) for the slice displays. These files are used by the user-end interface at display time. The next section describes the interface components in detail. AnatomyBrowser User-End Interface AnatomyBrowser integrates three main types of information: 3D surface models, slice data sets, and text. Accordingly, the user-end interface consists of three main components: a 3D display, three slice displays, and a hierarchy panel (Fig. 4(a)). In addition to visualization capabilities, Anatomy- Browser provides cross-referencing among all types of displayed information. The user-end interface is implemented as a Java applet, and is therefore platform independent. Interface Components 3D Display. The 3D display uses multilayer images to generate and display images of the 3D models using Equations (3) and (4). It provides the user with controls for view selection, zoom in/out, and manipulation of surface colors and opacities. Cross-Sectional Slices. Axial, coronal and sagittal cross-sections are displayed on the three slice displays. The cross-sectional slices are computed from the original slice volume by resampling it along the three orthogonal directions. Similarly to the 3D display, zoom in/out functionality is implemented for the slice displays. AnatomyBrowser provides four different ways to view the slices. A user can choose to view just the gray-scale images, or just the segmented set, or blend them together, or view the gray-scale images with the outlines of the segmentation. Fig. 5 demonstrates all four choices for one slice from the brain atlas data set. Hierarchy Panel. A hierarchy tree (graph) is a natural, systematic way to classify and present anatomical structures. AnatomyBrowser uses a directed acyclic graph (DAG) to model anatomical hierarchy. The hierarchy file generated by the backend system is parsed by the interface and a corresponding graph structure is constructed. However, we use a tree model to display the structure hierarchy, as it is more suitable than a DAG for visualization purposes. If a particular structure (or a
9 Golland et al.: AnatomyBrowser 137 Fig. 5. Slice display. The same coronal slice from the brain data set is displayed using four different ways of combining gray-scale and segmented images: (a) gray-scale, (b) segmented, (c) blended, (d) outline. group of structures) is shared by several parents, several instances of the corresponding node (or a subgraph) are generated and displayed under each of the parents. Since the internal representation uses a DAG, and all the instances of a group or a structure point back to the same element in the representation, the element properties can be changed using any of the instances. The hierarchy panel allows the user to set a desirable level of detail for displayed images. Any subtree in the hierarchy panel can be collapsed into a single node. This causes all the structures in the subtree to be rerendered using the color of the new collapsed node and to be treated as one structure from that point forward (Fig. 4(b)). This feature can be useful for studying anatomy with the help of AnatomyBrowser. A user can start with a very coarse hierarchy tree and expand group nodes gradually, rather than try to learn the whole tree at once. It can also be used to change properties of all elements in a subtree in one step. For example, if we wish to remove all muscles from the knee atlas in Fig. 7(a), collapsing the muscle group and removing it as one element is easier than removing the muscles one by one. Cross-Referencing between Displays An important capability provided by Anatomy- Browser is cross-referencing among the displays. If a user clicks on one of the display areas (Shift-Click), a cross-hair appears on all four dislays at locations corresponding to the same 3D point. Although all four displays exhibit very similar behavior, the cross-referencing definition and implementation are different for the 3D display and the slice displays. Any pixel in the slice display is characterized by three numbers: the slice index and the pixel coordinates in the slice (Fig. 6(a)). These three numbers are sufficient to compute the 3D coordinates of the corresponding physical point in the volume defined by the slice set. Therefore, the mapping between pixels in the slice set and physical points in the 3D volume is well-defined and can be readily computed from the physical dimensions of the voxel. This is not true for the 3D display. Each pixel in the 3D display is described by only two coordinates, and a user has no means to specify or obtain depth values directly in the 3D display. A natural oneto-one correspondence between pixels in the 3D display and entities in the 3D space is the one from pixels to view rays, and vice versa (Fig. 6(b)). This construction provides us with a useful interpretation of the cross-referencing for the 3D display. If a cross-reference is initiated from the slice display, the corresponding 3D point can be uniquely computed. For the 3D display, we place the cross-hair on the pixel corresponding to the selected 3D point, but we might not see the point itself on the display. Instead, we will see a surface that occludes the selected 3D point. The cross-hair on the 3D display should be interpreted as a view ray along which the selected 3D point lies. In a similar way, by clicking on the 3D display, a user specifies a view ray rather than a single 3D point. AnatomyBrowser uses depth information available to it from the multilayer image to disambiguate this point referenced by the ray. The as-
10 138 Golland et al.: AnatomyBrowser Fig. 6. Coordinate correspondence between the volumes and the physical world: (a) A point in the slice display defines a 3D point in the physical volume. (b) A point in the 3D display defines a view ray in the physical world; the depth value can be determined by identifying the intersection point of the view ray with the nearest surface. (c) The depth value can be looked up in the multilayer image of the scene. sumption used by the system is that the point of interest lies on the intersection of the view ray with the closest visible surface (Fig. 6(b,c)). This decision is justified by a natural notion of pointing: when interpreting somebody s pointing, an observer follows an imaginary ray defined by the pointer, until it meets an object that is not fully transparent. Text and Annotation AnatomyBrowser also provides annotation capabilities, which can be considered as cross-referencing between text and images. This feature can be invoked from any of the main components of the AnatomyBrowser interface. To use the hierarchy panel for annotation of a particular structure, a user clicks on the name of the structure on the panel. If the structure is visible on the 3D display, a pointer to the structure will appear in the 3D image. The name of the structure will be displayed on the title bar between the 3D display and the slice displays. The implementation of this feature is fairly straightforward: A multilayer image contains information on the structures currently visible on the 3D display. If a user clicks on any of the displays, the structure index is extracted from the corresponding segmented slice for the slice displays, or from the multilayer image for the 3D display. Again, the name of the structure is located in the hierarchy graph and a pointer with the structure name is displayed on the 3D image. As mentioned before, AnatomyBrowser allows text notes to be attached to a particular structure. The text notes are saved in a file with the structure name, which is used by the interface to locate the notes. We use a title bar with the name of the structure as a hypertext link to a collection of text notes. Clicking on it brings up a text window with all the text information available on the structure. Both annotation and text documentation can be invoked for all leaves in the hierarchy panel, whether they represent real structures or groups of structures that have been collapsed into a single node. RESULTS AND DISCUSSION Example Applications AnatomyBrowser provides a framework for visualization of medical images and their integration with other information. Anatomy studies are the most obvious application for such a system. AnatomyBrowser can effectively replace a conventional anatomical atlas book, as it provides at least as much information as the book. Students can see anatomical structures, view an MR or computed tomographic (CT) scan of those structures, read an attached description, and study their hierarchy. Visualization and full cross-referencing make AnatomyBrowser a useful interactive teaching tool, and the fact that the user-end interface can run essentially on any computer makes AnatomyBrowser particularly well-suited for this purpose. We have generated interactive atlases using AnatomyBrowser for brain (Fig. 4), knee (Fig. 7(a)), abdomen, inner ear, and other areas. Building digital atlases is an ongoing effort in our group, and they become available on-line 1 as new data sets are segmented. All the atlas cases are based on scans of individual healthy subjects, but the system can po-
11 Golland et al.: AnatomyBrowser 139 Fig. 7. Additional examples of AnatomyBrowser. Both examples demonstrate cross-reference and label correspondence. (a) Atlas example: knee atlas. (b) Clinical case example: pelvic tumor.
12 140 Golland et al.: AnatomyBrowser tentially be used on phantom data as well as on atlases derived from a group of subjects. Most of the cases are based on a single segmented scan, but some examples were generated by merging several different modalities using registration. These include the brain atlas that was generated from two different MRI scans that covered overlapping but different parts of the subject head and were merged together using rigid registration techniques, and the inner ear atlas that was generated by merging bone segmentation from a CT scan and vessel segmentation from an MR angiogram. Once on the Web site, the brain atlas has been extensively used as a reference tool by the researchers in our laboratories. In addition to anatomy studies, digital atlases can also be used for model-driven segmentation. We can view the process of model-based segmentation as a loop in which the results of the segmentation of the previous scans are used in segmentation of the view data sets. The digital atlas is an intermediate step in this loop, where knowledge about anatomy is accumulated in a particular representation, visualized for the user, and possibly processed for future use by the segmentation algorithm. In a way, its purpose is to close the loop from one stage of the segmentation to the next one, which uses the result of the first stage. Anatomy- Browser can be helpful for visualization of both the atlas and the intermediate results in this process. As we pointed out in the Introduction, AnatomyBrowser is different from a digital atlas. It is a visualization system that can be used on any scan, not necessarily the reference (atlas) data. This naturally brings up another application for Anatomy- Browser. It can be employed as a visualization and annotation tool for clinical cases, when the analysis must be performed on a per-case basis. We have used AnatomyBrowser for visualization of several tumor cases in different anatomical areas: pelvis (Fig. 7(b)), sacrum, brain, and kidney. These cases are also available on-line. 1 The tool was tested for both surgical planning and as a reference tool during surgery. Before the surgery, an MRI (or/and CT) scan of a patient was taken and segmented in the Surgical Planning Laboratory at Brigham and Women s Hospital. Then, the 3D surface models were generated and presented to the surgeons in the back-end viewer. The surgeons used the viewer to familiarize themselves with the 3D shape and location of a tumor and structures around it and to plan the surgery. They were also asked to select about a dozen views that would be useful during surgery. During surgery, the user-end interface with the selected views and the slice data were made available to the surgeons for general reference on a personal computer (a laptop) in the operating room. The surgeons response to the system was positive in all cases. It provided them with much greater visualization capabilities than they had had before, while establishing clear correspondences between the 3D models and the more commonly used cross-sectional slice sets. We would like to mention two cases in which AnatomyBrowser allowed the surgeons to improve their performance significantly. In a case involving a sacral tumor, the visualization system made it obvious that certain vertebrae were covered by the tumor on the right side of the sacrum, but not on the left, which was of great help in planning the surgery. More information on this case can be found in Richolt. 12 In the case of a pelvic tumor, an initial surgery plan was changed after the analysis of the 3D models using AnatomyBrowser had demonstrated that the cuts originally planned would not be feasible. Then, in the operating room, the surgeons consulted the user-end interface to establish correspondence between different structures on the slices and on the 3D display. The surgeons reported that the information on spatial relationships between different structures provided by AnatomyBrowser allowed them to reduce the time of surgery substantially. Although AnatomyBrowser cannot provide as much functionality as the specialized surgical systems, 5,14 it can definitely be useful for surgeons who do not have access to high-level computational resources and specialized surgical systems. In particular, AnatomyBrowser can be considered a testing tool, allowing the surgeons to assess the value of 3D image visualization before committing to a certain surgical system. Another interesting (future) application in which AnatomyBrowser can be helpful is in remote data processing. Consider an imaging center that receives medical scans from other hospitals or imaging facilities that are not equipped to perform segmentation and visualization onsite. The scans are processed in the imaging center and sent back to the users, who can then employ Anatomy- Browser interface on a workstation or personal computer to visualize the data. This model of medical image processing follows the same principle of uneven distribution of computational resources between the processing site and visualization site that was used in AnatomyBrowser design and implementation, so it can naturally provide visualization services in such a scenario.
13 Golland et al.: AnatomyBrowser 141 Table 1. Comparison Between the Original Data Set Size and the Size of the Data Set Produced By the Back-End Component for 12 Different Cases Case number Input size (MB) New size (MB) Current Implementation Evaluation In this section, we report statistics collected on 12 cases that were processed and visualized using AnatomyBrowser. They range from tumor cases with a small number of structures to atlas cases with over 100 structures. We note that the current system was implemented as a proof of concept for the new visualization approach we presented in the report. It can be improved to decrease significantly the size of the intermediate data used by Anatomy- Browser, as well as the running time of all the components. Table 1 compares the sizes of slice data sets generated in the preprocessing step and the original data sets. In most cases, the output size is comparable to the input size, but for some cases (for example, case 5), the size of the output is significantly higher than that of the input data set. These are statistics reported for the implementation that resamples a slice volume off-line and stores all resampled slices. In this implementation, if an aspect ratio between slice thickness and slice resolution is high, the number of slices in the output data set is significantly higher than the number of slices in the input data set. In the future, when it becomes feasible to resample slices online, the intermediate data sets will be significantly smaller than the input data sets. Table 2 reports the average size of a multilayer image generated and stored by the back-end system. There is some correlation between the number of models and the size of a multilayer image, but it is obvious that this is not the only factor affecting the size of the images. Another important parameter (that is difficult to quantify) is the density of a scene i.e., an average number of models per pixel in the image but since the size of the multilayer images is two orders of magnitude smaller than the size of the slice data set, it is clearly not a bottleneck of the system. The amount of time required by the back-end system to process the data varies significantly depending on the number, size, and complexity of surface models. Table 3 summarizes our experience with the system. In general, it takes under two hours to process the data on a Sun Ultra Sparc10 workstation with graphics acceleration. Most of the time is taken up by rendering and file I/O. Although the processing time for the slice data sets does not change as the number of models increases, the time for surface mode generation and model rendering has a high correlation with the number of models in the system. In the process of system development and use, several different modes of running the userend interface emerged. The first and the simplest one is when the data files and the user-interface reside in the same file system, and the files are accessed through that file system. In this case, the update speed is sufficient for most applications. Once the applet is loaded, the interface reacts to user commands almost instantaneously. Later in the project, we put a few sample cases on our Web site, and users started accessing the system through an http connection. The system performance dropped, but was still acceptable if the access was through the local network (within the same domain). However, it proved too slow for remote access from other sites. Analysis of the data size above provides an important insight into this problem: The amount of data that needs to be transferred over the network makes remote use difficult. That is why we Table 2. Number of Surface Models (Anatomical Structures) and an Average Size of Multi-Layer Images Generated by the Pre-Rendering Component for 12 Different Cases Case number Num. of models Image size (MB)
14 142 Golland et al.: AnatomyBrowser Table 3. Running Time for Different Back- End Components Reported Separately for Clinical Cases (Fewer Than 20 Structures) and Atlas Cases (More Than 20 Structures) Surface Component model generation Model pre-rendering Slice pre-processing Clinical cases 5 10 min min 2 3 min Atlas cases min min 2 3 min encourage our users to obtain a local copy of the data and the interface. Improving the system performance for remote access is an exciting and promising direction for future work for this project. The current implementation, however, is not suited for remote use, as it is not optimized with respect to the amount of transmitted data, which is known to be a bottleneck for many distributed applications. CONCLUSIONS We have presented a novel framework for visualization of 3D models of anatomical structures. In this framework, the visualization process is divided between the back-end system that renders the images and saves them in a special format, and the user-end interface that reads prerendered images and displays 3D models, while providing a set of 3D scene manipulation capabilities similar to visualization packages based on true dynamic rendering. The main advantage of this method is that the computational resources can be allocated unevenly in the system. In particular, the back-end components require graphics acceleration hardware, but the user-end interface does not need special hardware or software support. We demonstrated an implementation of this approach in a system called AnatomyBrowser. The user-end interface of AnatomyBrowser is implemented in Java and can run on any platform. The current implementation proved to be efficient if the data resided in the same file system with the interface. Remote access through an http connection does not yield sufficient update speed for the system to be used over the network. Future optimizations of the interface implementation can help reduce the amount of information transmitted over the network and thus make remote use of the system feasible. We demonstrated several example applications for AnatomyBrowser as a visualization system. They included studies of anatomy, or visualization of anatomical atlases in general, as well as visualization of clinical cases for medical research or surgery. ACKNOWLEDGMENTS This article presents research that was supported in part by Mitsubishi Electric Research Laboratories and NSF Grant IIS REFERENCES 1. AnatomyBrowser URL. edu:8000/pages/papers/browser-update/index.html. Mirror site: 2. Blinn JF. Jim Blinn s corner: compositing, part I. IEEE Comput Graphics Appl 1994;14: Blinn JF. Jim Blinn s corner: compositing, part II. IEEE Comput Graphics Appl 1994;14: Foley JD, Van Dam A, Feiner SK, Hughes JF. Computer graphics: principles and practice. 2nd edition. New York: Addison-Wesley, Grimson WEL, Leventon ME, Ettinger GJ, Chabrerie A, Ozlen F, Nakajima S, Atsumi H, Kikinis R, Black PML. Clinical experience with a high precision image-guided neurosurgery system. In: Wells WM, Colchester A, Delp S, editors. Medical image computing and computer-assisted intervention: MICCAI 98. Proceedings of the First International Conference, Cambridge, MA, October Lecture Notes in Computer Science Berlin: Springer, p Höhne KH, Bomans M, Riemer M Schubert R, Tiede U, Lierse W. A 3D anatomical atlas based on a volume model. IEEE Comput Graphics Appl 1992;2: Kikinis R, Shenton ME, Iosifescu DV, McCarley RW, Saiviroonporn P, Hokama HH, Robatino A, Metcalf D, Wible CG, Portas CM, Donnino R, Jolesz FA. A digital brain atlas for surgical planning, model driven segmentation and teaching. IEEE Trans Vis Comput Graphics 1996;2: Lorensen WE, Cline HE. Marching cubes: a high resolution 3D surface construction algorithm. Comput Graphics 1987;21: Maintz JBA, Viergever MA. A survey of medical image registration. Med Image Anal 1998;2: McInerney T, Tersopoulos D. Deformable models in medical image analysis: a survey. Med Image Anal 1996;1: Nowinski WL, Fang A, Nguyen BT, Raphel JK, Jagannathan L, Raghaven R, Bryan RN, Miller GA. Multiple brain atlas database and atlas-based neuroimaging system. Comput Aid Surg 1997;2: Richolt JA. Case of the month report on the sacral tumor case. pages/comonth/97/may2/com.html. Surgical Planning Lab, Brigham and Women s Hospital, Boston, MA, Schroeder W, Martin K, Lorensen WE. The visualiza-
15 Golland et al.: AnatomyBrowser 143 tion toolkit: an object-oriented approach to 3D graphics. Princeton, NJ: Prentice-Hall, Serra L, Nowinski WL, Poston T, Chua BC, Ng H, Lee CM, Chua GG, Pillay PK. The brain bench: virtual tools for neurosurgery. Med Image Anal 1997; 1: Umans C, Halle M, Kikinis R. Multilayer images for interactive 3D visualization on the World Wide Web. papers/mli/paper.html. Tech. Rep. 51. Surgical Planning Lab, Brigham and Women s Hospital, Boston, MA, 1997.
2 Michael E. Leventon and Sarah F. F. Gibson a b c d Fig. 1. (a, b) Two MR scans of a person's knee. Both images have high resolution in-plane, but ha
 Model Generation from Multiple Volumes using Constrained Elastic SurfaceNets Michael E. Leventon and Sarah F. F. Gibson 1 MIT Artificial Intelligence Laboratory, Cambridge, MA 02139, USA leventon@ai.mit.edu
Model Generation from Multiple Volumes using Constrained Elastic SurfaceNets Michael E. Leventon and Sarah F. F. Gibson 1 MIT Artificial Intelligence Laboratory, Cambridge, MA 02139, USA leventon@ai.mit.edu
Calculating the Distance Map for Binary Sampled Data
 MITSUBISHI ELECTRIC RESEARCH LABORATORIES http://www.merl.com Calculating the Distance Map for Binary Sampled Data Sarah F. Frisken Gibson TR99-6 December 999 Abstract High quality rendering and physics-based
MITSUBISHI ELECTRIC RESEARCH LABORATORIES http://www.merl.com Calculating the Distance Map for Binary Sampled Data Sarah F. Frisken Gibson TR99-6 December 999 Abstract High quality rendering and physics-based
An Anatomical Atlas to Support the Virtual Planning of Hip Operations
 An Anatomical Atlas to Support the Virtual Planning of Hip Operations J. Ehrhardt a, H. Handels a, T. Malina a, B. Strathmann b, W. Plötz b, S. J. Pöppl a a Institute for Medical Informatics and b Department
An Anatomical Atlas to Support the Virtual Planning of Hip Operations J. Ehrhardt a, H. Handels a, T. Malina a, B. Strathmann b, W. Plötz b, S. J. Pöppl a a Institute for Medical Informatics and b Department
A Study of Medical Image Analysis System
 Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
3D Volume Mesh Generation of Human Organs Using Surface Geometries Created from the Visible Human Data Set
 3D Volume Mesh Generation of Human Organs Using Surface Geometries Created from the Visible Human Data Set John M. Sullivan, Jr., Ziji Wu, and Anand Kulkarni Worcester Polytechnic Institute Worcester,
3D Volume Mesh Generation of Human Organs Using Surface Geometries Created from the Visible Human Data Set John M. Sullivan, Jr., Ziji Wu, and Anand Kulkarni Worcester Polytechnic Institute Worcester,
Scene-Based Segmentation of Multiple Muscles from MRI in MITK
 Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
Previously... contour or image rendering in 2D
 Volume Rendering Visualisation Lecture 10 Taku Komura Institute for Perception, Action & Behaviour School of Informatics Volume Rendering 1 Previously... contour or image rendering in 2D 2D Contour line
Volume Rendering Visualisation Lecture 10 Taku Komura Institute for Perception, Action & Behaviour School of Informatics Volume Rendering 1 Previously... contour or image rendering in 2D 2D Contour line
First Steps in Hardware Two-Level Volume Rendering
 First Steps in Hardware Two-Level Volume Rendering Markus Hadwiger, Helwig Hauser Abstract We describe first steps toward implementing two-level volume rendering (abbreviated as 2lVR) on consumer PC graphics
First Steps in Hardware Two-Level Volume Rendering Markus Hadwiger, Helwig Hauser Abstract We describe first steps toward implementing two-level volume rendering (abbreviated as 2lVR) on consumer PC graphics
Volume Rendering. Computer Animation and Visualisation Lecture 9. Taku Komura. Institute for Perception, Action & Behaviour School of Informatics
 Volume Rendering Computer Animation and Visualisation Lecture 9 Taku Komura Institute for Perception, Action & Behaviour School of Informatics Volume Rendering 1 Volume Data Usually, a data uniformly distributed
Volume Rendering Computer Animation and Visualisation Lecture 9 Taku Komura Institute for Perception, Action & Behaviour School of Informatics Volume Rendering 1 Volume Data Usually, a data uniformly distributed
11/1/13. Visualization. Scientific Visualization. Types of Data. Height Field. Contour Curves. Meshes
 CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 2.11] Jernej Barbic University of Southern California Scientific Visualization
CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 2.11] Jernej Barbic University of Southern California Scientific Visualization
Visualization. CSCI 420 Computer Graphics Lecture 26
 CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 11] Jernej Barbic University of Southern California 1 Scientific Visualization
CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 11] Jernej Barbic University of Southern California 1 Scientific Visualization
Efficient Volumetric Ray Casting for Isosurface Rendering
 Efficient Volumetric Ray Casting for Isosurface Rendering Jae Jeong Choi 1 Byeong-Seok Shin 2 Yeong Gil Shin 1 Kevin Cleary 3 1. Department of Computer Science, Seoul National University, Seoul, Republic
Efficient Volumetric Ray Casting for Isosurface Rendering Jae Jeong Choi 1 Byeong-Seok Shin 2 Yeong Gil Shin 1 Kevin Cleary 3 1. Department of Computer Science, Seoul National University, Seoul, Republic
Copyright 2017 Medical IP - Tutorial Medip v /2018, Revision
 Copyright 2017 Medical IP - Tutorial Medip v.1.0.0.9 01/2018, Revision 1.0.0.2 List of Contents 1. Introduction......................................................... 2 2. Overview..............................................................
Copyright 2017 Medical IP - Tutorial Medip v.1.0.0.9 01/2018, Revision 1.0.0.2 List of Contents 1. Introduction......................................................... 2 2. Overview..............................................................
Computational Medical Imaging Analysis Chapter 4: Image Visualization
 Computational Medical Imaging Analysis Chapter 4: Image Visualization Jun Zhang Laboratory for Computational Medical Imaging & Data Analysis Department of Computer Science University of Kentucky Lexington,
Computational Medical Imaging Analysis Chapter 4: Image Visualization Jun Zhang Laboratory for Computational Medical Imaging & Data Analysis Department of Computer Science University of Kentucky Lexington,
Fast Interactive Region of Interest Selection for Volume Visualization
 Fast Interactive Region of Interest Selection for Volume Visualization Dominik Sibbing and Leif Kobbelt Lehrstuhl für Informatik 8, RWTH Aachen, 20 Aachen Email: {sibbing,kobbelt}@informatik.rwth-aachen.de
Fast Interactive Region of Interest Selection for Volume Visualization Dominik Sibbing and Leif Kobbelt Lehrstuhl für Informatik 8, RWTH Aachen, 20 Aachen Email: {sibbing,kobbelt}@informatik.rwth-aachen.de
Volume Illumination and Segmentation
 Volume Illumination and Segmentation Computer Animation and Visualisation Lecture 13 Institute for Perception, Action & Behaviour School of Informatics Overview Volume illumination Segmentation Volume
Volume Illumination and Segmentation Computer Animation and Visualisation Lecture 13 Institute for Perception, Action & Behaviour School of Informatics Overview Volume illumination Segmentation Volume
Saturn User Manual. Rubén Cárdenes. 29th January 2010 Image Processing Laboratory, University of Valladolid. Abstract
 Saturn User Manual Rubén Cárdenes 29th January 2010 Image Processing Laboratory, University of Valladolid Abstract Saturn is a software package for DTI processing and visualization, provided with a graphic
Saturn User Manual Rubén Cárdenes 29th January 2010 Image Processing Laboratory, University of Valladolid Abstract Saturn is a software package for DTI processing and visualization, provided with a graphic
Lecture 17: Recursive Ray Tracing. Where is the way where light dwelleth? Job 38:19
 Lecture 17: Recursive Ray Tracing Where is the way where light dwelleth? Job 38:19 1. Raster Graphics Typical graphics terminals today are raster displays. A raster display renders a picture scan line
Lecture 17: Recursive Ray Tracing Where is the way where light dwelleth? Job 38:19 1. Raster Graphics Typical graphics terminals today are raster displays. A raster display renders a picture scan line
Visualization Computer Graphics I Lecture 20
 15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] November 20, 2003 Doug James Carnegie Mellon University http://www.cs.cmu.edu/~djames/15-462/fall03
15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] November 20, 2003 Doug James Carnegie Mellon University http://www.cs.cmu.edu/~djames/15-462/fall03
Course Title: Computer Graphics Course no: CSC209
 Course Title: Computer Graphics Course no: CSC209 Nature of the Course: Theory + Lab Semester: III Full Marks: 60+20+20 Pass Marks: 24 +8+8 Credit Hrs: 3 Course Description: The course coversconcepts of
Course Title: Computer Graphics Course no: CSC209 Nature of the Course: Theory + Lab Semester: III Full Marks: 60+20+20 Pass Marks: 24 +8+8 Credit Hrs: 3 Course Description: The course coversconcepts of
3D Voxel-Based Volumetric Image Registration with Volume-View Guidance
 3D Voxel-Based Volumetric Image Registration with Volume-View Guidance Guang Li*, Huchen Xie, Holly Ning, Deborah Citrin, Jacek Copala, Barbara Arora, Norman Coleman, Kevin Camphausen, and Robert Miller
3D Voxel-Based Volumetric Image Registration with Volume-View Guidance Guang Li*, Huchen Xie, Holly Ning, Deborah Citrin, Jacek Copala, Barbara Arora, Norman Coleman, Kevin Camphausen, and Robert Miller
Modelling a Lamb Hind Leg
 Modelling a Lamb Hind Leg Joanne P. Crocombe Ross D. Clarke MIRINZ Food Technology & Research East Street (Ruakura Campus), PO Box 617 HAMILTON, NEW ZEALAND Andrew J. Pullan Department of Engineering Science
Modelling a Lamb Hind Leg Joanne P. Crocombe Ross D. Clarke MIRINZ Food Technology & Research East Street (Ruakura Campus), PO Box 617 HAMILTON, NEW ZEALAND Andrew J. Pullan Department of Engineering Science
A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images
 A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images Jianhua Yao 1, Russell Taylor 2 1. Diagnostic Radiology Department, Clinical Center,
A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images Jianhua Yao 1, Russell Taylor 2 1. Diagnostic Radiology Department, Clinical Center,
Visualization Computer Graphics I Lecture 20
 15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 15, 2003 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 15, 2003 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University
![Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University](/thumbs/90/102611276.jpg) 15-462 Computer Graphics I Lecture 21 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
15-462 Computer Graphics I Lecture 21 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
Multi-Modal Volume Registration Using Joint Intensity Distributions
 Multi-Modal Volume Registration Using Joint Intensity Distributions Michael E. Leventon and W. Eric L. Grimson Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA leventon@ai.mit.edu
Multi-Modal Volume Registration Using Joint Intensity Distributions Michael E. Leventon and W. Eric L. Grimson Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA leventon@ai.mit.edu
Fiber Selection from Diffusion Tensor Data based on Boolean Operators
 Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Using Probability Maps for Multi organ Automatic Segmentation
 Using Probability Maps for Multi organ Automatic Segmentation Ranveer Joyseeree 1,2, Óscar Jiménez del Toro1, and Henning Müller 1,3 1 University of Applied Sciences Western Switzerland (HES SO), Sierre,
Using Probability Maps for Multi organ Automatic Segmentation Ranveer Joyseeree 1,2, Óscar Jiménez del Toro1, and Henning Müller 1,3 1 University of Applied Sciences Western Switzerland (HES SO), Sierre,
Visualization. Images are used to aid in understanding of data. Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26]
![Visualization. Images are used to aid in understanding of data. Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26] Visualization. Images are used to aid in understanding of data. Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26]](/thumbs/74/70771954.jpg) Visualization Images are used to aid in understanding of data Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26] Tumor SCI, Utah Scientific Visualization Visualize large
Visualization Images are used to aid in understanding of data Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26] Tumor SCI, Utah Scientific Visualization Visualize large
Open Topology: A Toolkit for Brain Isosurface Correction
 Open Topology: A Toolkit for Brain Isosurface Correction Sylvain Jaume 1, Patrice Rondao 2, and Benoît Macq 2 1 National Institute of Research in Computer Science and Control, INRIA, France, sylvain@mit.edu,
Open Topology: A Toolkit for Brain Isosurface Correction Sylvain Jaume 1, Patrice Rondao 2, and Benoît Macq 2 1 National Institute of Research in Computer Science and Control, INRIA, France, sylvain@mit.edu,
Volume Illumination, Contouring
 Volume Illumination, Contouring Computer Animation and Visualisation Lecture 0 tkomura@inf.ed.ac.uk Institute for Perception, Action & Behaviour School of Informatics Contouring Scaler Data Overview -
Volume Illumination, Contouring Computer Animation and Visualisation Lecture 0 tkomura@inf.ed.ac.uk Institute for Perception, Action & Behaviour School of Informatics Contouring Scaler Data Overview -
Introduction. Welcome to 3D Slicer! Surgical Planning Laboratory -1- Brigham and Women s Hospital
 Introduction Welcome to 3D Slicer! -1- Overview of Training What is Slicer? Uses of Slicer Getting Slicer How to Use Slicer Loading Data Viewing Data Modifying Data Saving Data -2- What is Slicer? -3-
Introduction Welcome to 3D Slicer! -1- Overview of Training What is Slicer? Uses of Slicer Getting Slicer How to Use Slicer Loading Data Viewing Data Modifying Data Saving Data -2- What is Slicer? -3-
3DMMVR REFERENCE MANUAL V 0.81
 3DMMVR REFERENCE MANUAL V 0.81 Page 1 of 30 Index: 1.0 System Requirements...5 1.1 System Processor...5 1.2 System RAM...5 1.3 Graphics Card...5 1.4 Operating System...5 2.0 Conventions...6 2.1 Typographic
3DMMVR REFERENCE MANUAL V 0.81 Page 1 of 30 Index: 1.0 System Requirements...5 1.1 System Processor...5 1.2 System RAM...5 1.3 Graphics Card...5 1.4 Operating System...5 2.0 Conventions...6 2.1 Typographic
Applications of Explicit Early-Z Culling
 Applications of Explicit Early-Z Culling Jason L. Mitchell ATI Research Pedro V. Sander ATI Research Introduction In past years, in the SIGGRAPH Real-Time Shading course, we have covered the details of
Applications of Explicit Early-Z Culling Jason L. Mitchell ATI Research Pedro V. Sander ATI Research Introduction In past years, in the SIGGRAPH Real-Time Shading course, we have covered the details of
CSC Computer Graphics
 // CSC. Computer Graphics Lecture Kasun@dscs.sjp.ac.lk Department of Computer Science University of Sri Jayewardanepura Polygon Filling Scan-Line Polygon Fill Algorithm Span Flood-Fill Algorithm Inside-outside
// CSC. Computer Graphics Lecture Kasun@dscs.sjp.ac.lk Department of Computer Science University of Sri Jayewardanepura Polygon Filling Scan-Line Polygon Fill Algorithm Span Flood-Fill Algorithm Inside-outside
Queen s University CISC 454 Final Exam. April 19, :00pm Duration: 3 hours. One two sided aid sheet allowed. Initial of Family Name:
 Page 1 of 11 Queen s University CISC 454 Final Exam April 19, 2005 2:00pm Duration: 3 hours One two sided aid sheet allowed. Initial of Family Name: Student Number: (Write this at the top of every page.)
Page 1 of 11 Queen s University CISC 454 Final Exam April 19, 2005 2:00pm Duration: 3 hours One two sided aid sheet allowed. Initial of Family Name: Student Number: (Write this at the top of every page.)
ECE1778 Final Report MRI Visualizer
 ECE1778 Final Report MRI Visualizer David Qixiang Chen Alex Rodionov Word Count: 2408 Introduction We aim to develop a mobile phone/tablet based neurosurgical MRI visualization application with the goal
ECE1778 Final Report MRI Visualizer David Qixiang Chen Alex Rodionov Word Count: 2408 Introduction We aim to develop a mobile phone/tablet based neurosurgical MRI visualization application with the goal
CHAPTER 2. Morphometry on rodent brains. A.E.H. Scheenstra J. Dijkstra L. van der Weerd
 CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
Visualisation : Lecture 1. So what is visualisation? Visualisation
 So what is visualisation? UG4 / M.Sc. Course 2006 toby.breckon@ed.ac.uk Computer Vision Lab. Institute for Perception, Action & Behaviour Introducing 1 Application of interactive 3D computer graphics to
So what is visualisation? UG4 / M.Sc. Course 2006 toby.breckon@ed.ac.uk Computer Vision Lab. Institute for Perception, Action & Behaviour Introducing 1 Application of interactive 3D computer graphics to
Image Registration I
 Image Registration I Comp 254 Spring 2002 Guido Gerig Image Registration: Motivation Motivation for Image Registration Combine images from different modalities (multi-modality registration), e.g. CT&MRI,
Image Registration I Comp 254 Spring 2002 Guido Gerig Image Registration: Motivation Motivation for Image Registration Combine images from different modalities (multi-modality registration), e.g. CT&MRI,
A MORPHOLOGY-BASED FILTER STRUCTURE FOR EDGE-ENHANCING SMOOTHING
 Proceedings of the 1994 IEEE International Conference on Image Processing (ICIP-94), pp. 530-534. (Austin, Texas, 13-16 November 1994.) A MORPHOLOGY-BASED FILTER STRUCTURE FOR EDGE-ENHANCING SMOOTHING
Proceedings of the 1994 IEEE International Conference on Image Processing (ICIP-94), pp. 530-534. (Austin, Texas, 13-16 November 1994.) A MORPHOLOGY-BASED FILTER STRUCTURE FOR EDGE-ENHANCING SMOOTHING
Level Set Extraction from Gridded 2D and 3D Data
 Level Set Extraction from Gridded 2D and 3D Data David Eberly, Geometric Tools, Redmond WA 98052 https://www.geometrictools.com/ This work is licensed under the Creative Commons Attribution 4.0 International
Level Set Extraction from Gridded 2D and 3D Data David Eberly, Geometric Tools, Redmond WA 98052 https://www.geometrictools.com/ This work is licensed under the Creative Commons Attribution 4.0 International
Stereoscopic Atlas of Intrinsic Brain Networks (SAIBN)
 Stereoscopic Atlas of Intrinsic Brain Networks (SAIBN) Gonzalo M. Rojas, Marcelo Gálvez, Daniel Margulies, Cameron Craddock, Xavier Castellanos, Michael Milham 1.- Introduction Stereoscopic Atlas of Intrinsic
Stereoscopic Atlas of Intrinsic Brain Networks (SAIBN) Gonzalo M. Rojas, Marcelo Gálvez, Daniel Margulies, Cameron Craddock, Xavier Castellanos, Michael Milham 1.- Introduction Stereoscopic Atlas of Intrinsic
Synthesis of Textures with Intricate Geometries using BTF and Large Number of Textured Micropolygons. Abstract. 2. Related studies. 1.
 Synthesis of Textures with Intricate Geometries using BTF and Large Number of Textured Micropolygons sub047 Abstract BTF has been studied extensively and much progress has been done for measurements, compression
Synthesis of Textures with Intricate Geometries using BTF and Large Number of Textured Micropolygons sub047 Abstract BTF has been studied extensively and much progress has been done for measurements, compression
3D VISUALIZATION OF SEGMENTED CRUCIATE LIGAMENTS 1. INTRODUCTION
 JOURNAL OF MEDICAL INFORMATICS & TECHNOLOGIES Vol. 10/006, ISSN 164-6037 Paweł BADURA * cruciate ligament, segmentation, fuzzy connectedness,3d visualization 3D VISUALIZATION OF SEGMENTED CRUCIATE LIGAMENTS
JOURNAL OF MEDICAL INFORMATICS & TECHNOLOGIES Vol. 10/006, ISSN 164-6037 Paweł BADURA * cruciate ligament, segmentation, fuzzy connectedness,3d visualization 3D VISUALIZATION OF SEGMENTED CRUCIATE LIGAMENTS
Lecture 3: Art Gallery Problems and Polygon Triangulation
 EECS 396/496: Computational Geometry Fall 2017 Lecture 3: Art Gallery Problems and Polygon Triangulation Lecturer: Huck Bennett In this lecture, we study the problem of guarding an art gallery (specified
EECS 396/496: Computational Geometry Fall 2017 Lecture 3: Art Gallery Problems and Polygon Triangulation Lecturer: Huck Bennett In this lecture, we study the problem of guarding an art gallery (specified
3D Slicer. NA-MIC National Alliance for Medical Image Computing 4 February 2011
 NA-MIC http://na-mic.org 3D Slicer 4 February 2011 Andrey Fedorov, PhD Steve Pieper, PhD Ron Kikinis, MD Surgical Planning Lab Brigham and Women's Hospital Acknowledgments Picture courtesy Kapur, Jakab,
NA-MIC http://na-mic.org 3D Slicer 4 February 2011 Andrey Fedorov, PhD Steve Pieper, PhD Ron Kikinis, MD Surgical Planning Lab Brigham and Women's Hospital Acknowledgments Picture courtesy Kapur, Jakab,
Chapter 3 Set Redundancy in Magnetic Resonance Brain Images
 16 Chapter 3 Set Redundancy in Magnetic Resonance Brain Images 3.1 MRI (magnetic resonance imaging) MRI is a technique of measuring physical structure within the human anatomy. Our proposed research focuses
16 Chapter 3 Set Redundancy in Magnetic Resonance Brain Images 3.1 MRI (magnetic resonance imaging) MRI is a technique of measuring physical structure within the human anatomy. Our proposed research focuses
Whole Body MRI Intensity Standardization
 Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Volume Rendering. Lecture 21
 Volume Rendering Lecture 21 Acknowledgements These slides are collected from many sources. A particularly valuable source is the IEEE Visualization conference tutorials. Sources from: Roger Crawfis, Klaus
Volume Rendering Lecture 21 Acknowledgements These slides are collected from many sources. A particularly valuable source is the IEEE Visualization conference tutorials. Sources from: Roger Crawfis, Klaus
3D Surface Reconstruction of the Brain based on Level Set Method
 3D Surface Reconstruction of the Brain based on Level Set Method Shijun Tang, Bill P. Buckles, and Kamesh Namuduri Department of Computer Science & Engineering Department of Electrical Engineering University
3D Surface Reconstruction of the Brain based on Level Set Method Shijun Tang, Bill P. Buckles, and Kamesh Namuduri Department of Computer Science & Engineering Department of Electrical Engineering University
Improved Navigated Spine Surgery Utilizing Augmented Reality Visualization
 Improved Navigated Spine Surgery Utilizing Augmented Reality Visualization Zein Salah 1,2, Bernhard Preim 1, Erck Elolf 3, Jörg Franke 4, Georg Rose 2 1Department of Simulation and Graphics, University
Improved Navigated Spine Surgery Utilizing Augmented Reality Visualization Zein Salah 1,2, Bernhard Preim 1, Erck Elolf 3, Jörg Franke 4, Georg Rose 2 1Department of Simulation and Graphics, University
COMP30019 Graphics and Interaction Scan Converting Polygons and Lines
 COMP30019 Graphics and Interaction Scan Converting Polygons and Lines Department of Computer Science and Software Engineering The Lecture outline Introduction Scan conversion Scan-line algorithm Edge coherence
COMP30019 Graphics and Interaction Scan Converting Polygons and Lines Department of Computer Science and Software Engineering The Lecture outline Introduction Scan conversion Scan-line algorithm Edge coherence
Automatic Optimization of Segmentation Algorithms Through Simultaneous Truth and Performance Level Estimation (STAPLE)
 Automatic Optimization of Segmentation Algorithms Through Simultaneous Truth and Performance Level Estimation (STAPLE) Mahnaz Maddah, Kelly H. Zou, William M. Wells, Ron Kikinis, and Simon K. Warfield
Automatic Optimization of Segmentation Algorithms Through Simultaneous Truth and Performance Level Estimation (STAPLE) Mahnaz Maddah, Kelly H. Zou, William M. Wells, Ron Kikinis, and Simon K. Warfield
New Technology Allows Multiple Image Contrasts in a Single Scan
 These images were acquired with an investigational device. PD T2 T2 FLAIR T1 MAP T1 FLAIR PSIR T1 New Technology Allows Multiple Image Contrasts in a Single Scan MR exams can be time consuming. A typical
These images were acquired with an investigational device. PD T2 T2 FLAIR T1 MAP T1 FLAIR PSIR T1 New Technology Allows Multiple Image Contrasts in a Single Scan MR exams can be time consuming. A typical
Comparison of Different Metrics for Appearance-model-based 2D/3D-registration with X-ray Images
 Comparison of Different Metrics for Appearance-model-based 2D/3D-registration with X-ray Images Philipp Steininger 1, Karl D. Fritscher 1, Gregor Kofler 1, Benedikt Schuler 1, Markus Hänni 2, Karsten Schwieger
Comparison of Different Metrics for Appearance-model-based 2D/3D-registration with X-ray Images Philipp Steininger 1, Karl D. Fritscher 1, Gregor Kofler 1, Benedikt Schuler 1, Markus Hänni 2, Karsten Schwieger
Lecture 17: Solid Modeling.... a cubit on the one side, and a cubit on the other side Exodus 26:13
 Lecture 17: Solid Modeling... a cubit on the one side, and a cubit on the other side Exodus 26:13 Who is on the LORD's side? Exodus 32:26 1. Solid Representations A solid is a 3-dimensional shape with
Lecture 17: Solid Modeling... a cubit on the one side, and a cubit on the other side Exodus 26:13 Who is on the LORD's side? Exodus 32:26 1. Solid Representations A solid is a 3-dimensional shape with
Digital Volume Correlation for Materials Characterization
 19 th World Conference on Non-Destructive Testing 2016 Digital Volume Correlation for Materials Characterization Enrico QUINTANA, Phillip REU, Edward JIMENEZ, Kyle THOMPSON, Sharlotte KRAMER Sandia National
19 th World Conference on Non-Destructive Testing 2016 Digital Volume Correlation for Materials Characterization Enrico QUINTANA, Phillip REU, Edward JIMENEZ, Kyle THOMPSON, Sharlotte KRAMER Sandia National
Brain Surface Conformal Spherical Mapping
 Brain Surface Conformal Spherical Mapping Min Zhang Department of Industrial Engineering, Arizona State University mzhang33@asu.edu Abstract It is well known and proved that any genus zero surface can
Brain Surface Conformal Spherical Mapping Min Zhang Department of Industrial Engineering, Arizona State University mzhang33@asu.edu Abstract It is well known and proved that any genus zero surface can
Fast Image Registration via Joint Gradient Maximization: Application to Multi-Modal Data
 MITSUBISHI ELECTRIC RESEARCH LABORATORIES http://www.merl.com Fast Image Registration via Joint Gradient Maximization: Application to Multi-Modal Data Xue Mei, Fatih Porikli TR-19 September Abstract We
MITSUBISHI ELECTRIC RESEARCH LABORATORIES http://www.merl.com Fast Image Registration via Joint Gradient Maximization: Application to Multi-Modal Data Xue Mei, Fatih Porikli TR-19 September Abstract We
S U N G - E U I YO O N, K A I S T R E N D E R I N G F R E E LY A VA I L A B L E O N T H E I N T E R N E T
 S U N G - E U I YO O N, K A I S T R E N D E R I N G F R E E LY A VA I L A B L E O N T H E I N T E R N E T Copyright 2018 Sung-eui Yoon, KAIST freely available on the internet http://sglab.kaist.ac.kr/~sungeui/render
S U N G - E U I YO O N, K A I S T R E N D E R I N G F R E E LY A VA I L A B L E O N T H E I N T E R N E T Copyright 2018 Sung-eui Yoon, KAIST freely available on the internet http://sglab.kaist.ac.kr/~sungeui/render
Adaptive Point Cloud Rendering
 1 Adaptive Point Cloud Rendering Project Plan Final Group: May13-11 Christopher Jeffers Eric Jensen Joel Rausch Client: Siemens PLM Software Client Contact: Michael Carter Adviser: Simanta Mitra 4/29/13
1 Adaptive Point Cloud Rendering Project Plan Final Group: May13-11 Christopher Jeffers Eric Jensen Joel Rausch Client: Siemens PLM Software Client Contact: Michael Carter Adviser: Simanta Mitra 4/29/13
Assessing Accuracy Factors in Deformable 2D/3D Medical Image Registration Using a Statistical Pelvis Model
 Assessing Accuracy Factors in Deformable 2D/3D Medical Image Registration Using a Statistical Pelvis Model Jianhua Yao National Institute of Health Bethesda, MD USA jyao@cc.nih.gov Russell Taylor The Johns
Assessing Accuracy Factors in Deformable 2D/3D Medical Image Registration Using a Statistical Pelvis Model Jianhua Yao National Institute of Health Bethesda, MD USA jyao@cc.nih.gov Russell Taylor The Johns
Conformal flattening maps for the visualization of vessels
 Conformal flattening maps for the visualization of vessels Lei Zhua, Steven Hakerb, and Allen Tannenbauma adept of Biomedical Engineering, Georgia Tech, Atlanta bdept of Radiology, Brigham and Women's
Conformal flattening maps for the visualization of vessels Lei Zhua, Steven Hakerb, and Allen Tannenbauma adept of Biomedical Engineering, Georgia Tech, Atlanta bdept of Radiology, Brigham and Women's
Scalar Visualization
 Scalar Visualization Visualizing scalar data Popular scalar visualization techniques Color mapping Contouring Height plots outline Recap of Chap 4: Visualization Pipeline 1. Data Importing 2. Data Filtering
Scalar Visualization Visualizing scalar data Popular scalar visualization techniques Color mapping Contouring Height plots outline Recap of Chap 4: Visualization Pipeline 1. Data Importing 2. Data Filtering
Chapter 8 Visualization and Optimization
 Chapter 8 Visualization and Optimization Recommended reference books: [1] Edited by R. S. Gallagher: Computer Visualization, Graphics Techniques for Scientific and Engineering Analysis by CRC, 1994 [2]
Chapter 8 Visualization and Optimization Recommended reference books: [1] Edited by R. S. Gallagher: Computer Visualization, Graphics Techniques for Scientific and Engineering Analysis by CRC, 1994 [2]
A Method of Automated Landmark Generation for Automated 3D PDM Construction
 A Method of Automated Landmark Generation for Automated 3D PDM Construction A. D. Brett and C. J. Taylor Department of Medical Biophysics University of Manchester Manchester M13 9PT, Uk adb@sv1.smb.man.ac.uk
A Method of Automated Landmark Generation for Automated 3D PDM Construction A. D. Brett and C. J. Taylor Department of Medical Biophysics University of Manchester Manchester M13 9PT, Uk adb@sv1.smb.man.ac.uk
Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry
 Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry Nivedita Agarwal, MD Nivedita.agarwal@apss.tn.it Nivedita.agarwal@unitn.it Volume and surface morphometry Brain volume White matter
Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry Nivedita Agarwal, MD Nivedita.agarwal@apss.tn.it Nivedita.agarwal@unitn.it Volume and surface morphometry Brain volume White matter
Scalar Data. Visualization Torsten Möller. Weiskopf/Machiraju/Möller
 Scalar Data Visualization Torsten Möller Weiskopf/Machiraju/Möller Overview Basic strategies Function plots and height fields Isolines Color coding Volume visualization (overview) Classification Segmentation
Scalar Data Visualization Torsten Möller Weiskopf/Machiraju/Möller Overview Basic strategies Function plots and height fields Isolines Color coding Volume visualization (overview) Classification Segmentation
Data Partitioning. Figure 1-31: Communication Topologies. Regular Partitions
 Data In single-program multiple-data (SPMD) parallel programs, global data is partitioned, with a portion of the data assigned to each processing node. Issues relevant to choosing a partitioning strategy
Data In single-program multiple-data (SPMD) parallel programs, global data is partitioned, with a portion of the data assigned to each processing node. Issues relevant to choosing a partitioning strategy
Volume Illumination. Visualisation Lecture 11. Taku Komura. Institute for Perception, Action & Behaviour School of Informatics
 Volume Illumination Visualisation Lecture 11 Taku Komura Institute for Perception, Action & Behaviour School of Informatics Taku Komura Volume Illumination & Vector Vis. 1 Previously : Volume Rendering
Volume Illumination Visualisation Lecture 11 Taku Komura Institute for Perception, Action & Behaviour School of Informatics Taku Komura Volume Illumination & Vector Vis. 1 Previously : Volume Rendering
EMSegment Tutorial. How to Define and Fine-Tune Automatic Brain Compartment Segmentation and the Detection of White Matter Hyperintensities
 EMSegment Tutorial How to Define and Fine-Tune Automatic Brain Compartment Segmentation and the Detection of White Matter Hyperintensities This documentation serves as a tutorial to learn to customize
EMSegment Tutorial How to Define and Fine-Tune Automatic Brain Compartment Segmentation and the Detection of White Matter Hyperintensities This documentation serves as a tutorial to learn to customize
Multipass GPU Surface Rendering in 4D Ultrasound
 2012 Cairo International Biomedical Engineering Conference (CIBEC) Cairo, Egypt, December 20-21, 2012 Multipass GPU Surface Rendering in 4D Ultrasound Ahmed F. Elnokrashy 1,2, Marwan Hassan 1, Tamer Hosny
2012 Cairo International Biomedical Engineering Conference (CIBEC) Cairo, Egypt, December 20-21, 2012 Multipass GPU Surface Rendering in 4D Ultrasound Ahmed F. Elnokrashy 1,2, Marwan Hassan 1, Tamer Hosny
Scalar Visualization
 Scalar Visualization 5-1 Motivation Visualizing scalar data is frequently encountered in science, engineering, and medicine, but also in daily life. Recalling from earlier, scalar datasets, or scalar fields,
Scalar Visualization 5-1 Motivation Visualizing scalar data is frequently encountered in science, engineering, and medicine, but also in daily life. Recalling from earlier, scalar datasets, or scalar fields,
The Allen Human Brain Atlas offers three types of searches to allow a user to: (1) obtain gene expression data for specific genes (or probes) of
 Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
RT_Image v0.2β User s Guide
 RT_Image v0.2β User s Guide RT_Image is a three-dimensional image display and analysis suite developed in IDL (ITT, Boulder, CO). It offers a range of flexible tools for the visualization and quantitation
RT_Image v0.2β User s Guide RT_Image is a three-dimensional image display and analysis suite developed in IDL (ITT, Boulder, CO). It offers a range of flexible tools for the visualization and quantitation
Video Registration Virtual Reality for Non-linkage Stereotactic Surgery
 Video Registration Virtual Reality for Non-linkage Stereotactic Surgery P.L. Gleason, Ron Kikinis, David Altobelli, William Wells, Eben Alexander III, Peter McL. Black, Ferenc Jolesz Surgical Planning
Video Registration Virtual Reality for Non-linkage Stereotactic Surgery P.L. Gleason, Ron Kikinis, David Altobelli, William Wells, Eben Alexander III, Peter McL. Black, Ferenc Jolesz Surgical Planning
Chapter IV Fragment Processing and Output Merging. 3D Graphics for Game Programming
 Chapter IV Fragment Processing and Output Merging Fragment Processing The per-fragment attributes may include a normal vector, a set of texture coordinates, a set of color values, a depth, etc. Using these
Chapter IV Fragment Processing and Output Merging Fragment Processing The per-fragment attributes may include a normal vector, a set of texture coordinates, a set of color values, a depth, etc. Using these
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy
 The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
A Survey of Volumetric Visualization Techniques for Medical Images
 International Journal of Research Studies in Computer Science and Engineering (IJRSCSE) Volume 2, Issue 4, April 2015, PP 34-39 ISSN 2349-4840 (Print) & ISSN 2349-4859 (Online) www.arcjournals.org A Survey
International Journal of Research Studies in Computer Science and Engineering (IJRSCSE) Volume 2, Issue 4, April 2015, PP 34-39 ISSN 2349-4840 (Print) & ISSN 2349-4859 (Online) www.arcjournals.org A Survey
(12) Patent Application Publication (10) Pub. No.: US 2016/ A1
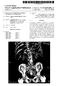 US 20160364902A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2016/0364902 A1 Hong et al. (43) Pub. Date: (54) HIGH QUALITY EMBEDDED GRAPHICS (52) U.S. Cl. FOR REMOTE VISUALIZATION
US 20160364902A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2016/0364902 A1 Hong et al. (43) Pub. Date: (54) HIGH QUALITY EMBEDDED GRAPHICS (52) U.S. Cl. FOR REMOTE VISUALIZATION
Machine Learning for Medical Image Analysis. A. Criminisi
 Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Reconstruction of complete 3D object model from multi-view range images.
 Header for SPIE use Reconstruction of complete 3D object model from multi-view range images. Yi-Ping Hung *, Chu-Song Chen, Ing-Bor Hsieh, Chiou-Shann Fuh Institute of Information Science, Academia Sinica,
Header for SPIE use Reconstruction of complete 3D object model from multi-view range images. Yi-Ping Hung *, Chu-Song Chen, Ing-Bor Hsieh, Chiou-Shann Fuh Institute of Information Science, Academia Sinica,
Creating a Vision Channel for Observing Deep-Seated Anatomy in Medical Augmented Reality
 Creating a Vision Channel for Observing Deep-Seated Anatomy in Medical Augmented Reality A Cut-Away Technique for In-Situ Visualization Felix Wimmer 1, Christoph Bichlmeier 1, Sandro M. Heining 2, Nassir
Creating a Vision Channel for Observing Deep-Seated Anatomy in Medical Augmented Reality A Cut-Away Technique for In-Situ Visualization Felix Wimmer 1, Christoph Bichlmeier 1, Sandro M. Heining 2, Nassir
Contours & Implicit Modelling 4
 Brief Recap Contouring & Implicit Modelling Contouring Implicit Functions Visualisation Lecture 8 lecture 6 Marching Cubes lecture 3 visualisation of a Quadric toby.breckon@ed.ac.uk Computer Vision Lab.
Brief Recap Contouring & Implicit Modelling Contouring Implicit Functions Visualisation Lecture 8 lecture 6 Marching Cubes lecture 3 visualisation of a Quadric toby.breckon@ed.ac.uk Computer Vision Lab.
Integrating patient-oriented data processing into the PREPaRe virtual hospital using XML technology
 Integrating patient-oriented data processing into the PREPaRe virtual hospital using XML technology René Tschirley, Kai Köchy, Steffen Märkle Dept. for Computer Science and Computer Assisted Medicine,
Integrating patient-oriented data processing into the PREPaRe virtual hospital using XML technology René Tschirley, Kai Köchy, Steffen Märkle Dept. for Computer Science and Computer Assisted Medicine,
Structural and Syntactic Pattern Recognition
 Structural and Syntactic Pattern Recognition Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr CS 551, Fall 2017 CS 551, Fall 2017 c 2017, Selim Aksoy (Bilkent
Structural and Syntactic Pattern Recognition Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr CS 551, Fall 2017 CS 551, Fall 2017 c 2017, Selim Aksoy (Bilkent
Isosurface Rendering. CSC 7443: Scientific Information Visualization
 Isosurface Rendering What is Isosurfacing? An isosurface is the 3D surface representing the locations of a constant scalar value within a volume A surface with the same scalar field value Isosurfaces form
Isosurface Rendering What is Isosurfacing? An isosurface is the 3D surface representing the locations of a constant scalar value within a volume A surface with the same scalar field value Isosurfaces form
Computer-Aided Tracking of MS lesions
 Computer-Aided Tracking of MS lesions Deborah Sturm* 1, Devorah Gurwitz Kletenik 2 Phillip Koshy 3 1 Department of Computer Science, College of Staten Island, City University of New York, 2800 Victory
Computer-Aided Tracking of MS lesions Deborah Sturm* 1, Devorah Gurwitz Kletenik 2 Phillip Koshy 3 1 Department of Computer Science, College of Staten Island, City University of New York, 2800 Victory
Simon K. Warfield, Michael Kaus, Ferenc A. Jolesz and Ron Kikinis
 Medical Image Analysis (2000) volume 4, number 1, pp 43 55 c Elsevier Science BV Adaptive, Template Moderated, Spatially Varying Statistical Classification Simon K. Warfield, Michael Kaus, Ferenc A. Jolesz
Medical Image Analysis (2000) volume 4, number 1, pp 43 55 c Elsevier Science BV Adaptive, Template Moderated, Spatially Varying Statistical Classification Simon K. Warfield, Michael Kaus, Ferenc A. Jolesz
Emissive Clip Planes for Volume Rendering Supplement.
 Emissive Clip Planes for Volume Rendering Supplement. More material than fit on the one page version for the SIGGRAPH 2003 Sketch by Jan Hardenbergh & Yin Wu of TeraRecon, Inc. Left Image: The clipped
Emissive Clip Planes for Volume Rendering Supplement. More material than fit on the one page version for the SIGGRAPH 2003 Sketch by Jan Hardenbergh & Yin Wu of TeraRecon, Inc. Left Image: The clipped
Volumetric Deformable Models for Simulation of Laparoscopic Surgery
 Volumetric Deformable Models for Simulation of Laparoscopic Surgery S. Cotin y, H. Delingette y, J.M. Clément z V. Tassetti z, J. Marescaux z, N. Ayache y y INRIA, Epidaure Project 2004, route des Lucioles,
Volumetric Deformable Models for Simulation of Laparoscopic Surgery S. Cotin y, H. Delingette y, J.M. Clément z V. Tassetti z, J. Marescaux z, N. Ayache y y INRIA, Epidaure Project 2004, route des Lucioles,
Introduction to Visualization and Computer Graphics
 Introduction to Visualization and Computer Graphics DH2320, Fall 2015 Prof. Dr. Tino Weinkauf Introduction to Visualization and Computer Graphics Visibility Shading 3D Rendering Geometric Model Color Perspective
Introduction to Visualization and Computer Graphics DH2320, Fall 2015 Prof. Dr. Tino Weinkauf Introduction to Visualization and Computer Graphics Visibility Shading 3D Rendering Geometric Model Color Perspective
MODELING AND HIERARCHY
 MODELING AND HIERARCHY Introduction Models are abstractions of the world both of the real world in which we live and of virtual worlds that we create with computers. We are all familiar with mathematical
MODELING AND HIERARCHY Introduction Models are abstractions of the world both of the real world in which we live and of virtual worlds that we create with computers. We are all familiar with mathematical
K-Means Clustering Using Localized Histogram Analysis
 K-Means Clustering Using Localized Histogram Analysis Michael Bryson University of South Carolina, Department of Computer Science Columbia, SC brysonm@cse.sc.edu Abstract. The first step required for many
K-Means Clustering Using Localized Histogram Analysis Michael Bryson University of South Carolina, Department of Computer Science Columbia, SC brysonm@cse.sc.edu Abstract. The first step required for many
white paper EPC Tag Data Specification abstract David Brock, Chris Cummins
 Published June 1, 2003. Distribution restricted to Sponsors until September 1, 2003. white paper EPC Tag Data Specification David Brock, Chris Cummins auto-id center massachusetts institute of technology,
Published June 1, 2003. Distribution restricted to Sponsors until September 1, 2003. white paper EPC Tag Data Specification David Brock, Chris Cummins auto-id center massachusetts institute of technology,
Hierarchical Multi structure Segmentation Guided by Anatomical Correlations
 Hierarchical Multi structure Segmentation Guided by Anatomical Correlations Oscar Alfonso Jiménez del Toro oscar.jimenez@hevs.ch Henning Müller henningmueller@hevs.ch University of Applied Sciences Western
Hierarchical Multi structure Segmentation Guided by Anatomical Correlations Oscar Alfonso Jiménez del Toro oscar.jimenez@hevs.ch Henning Müller henningmueller@hevs.ch University of Applied Sciences Western
CHAPTER 1 Graphics Systems and Models 3
 ?????? 1 CHAPTER 1 Graphics Systems and Models 3 1.1 Applications of Computer Graphics 4 1.1.1 Display of Information............. 4 1.1.2 Design.................... 5 1.1.3 Simulation and Animation...........
?????? 1 CHAPTER 1 Graphics Systems and Models 3 1.1 Applications of Computer Graphics 4 1.1.1 Display of Information............. 4 1.1.2 Design.................... 5 1.1.3 Simulation and Animation...........
Method of Background Subtraction for Medical Image Segmentation
 Method of Background Subtraction for Medical Image Segmentation Seongjai Kim Department of Mathematics and Statistics, Mississippi State University Mississippi State, MS 39762, USA and Hyeona Lim Department
Method of Background Subtraction for Medical Image Segmentation Seongjai Kim Department of Mathematics and Statistics, Mississippi State University Mississippi State, MS 39762, USA and Hyeona Lim Department
Dominic Filion, Senior Engineer Blizzard Entertainment. Rob McNaughton, Lead Technical Artist Blizzard Entertainment
 Dominic Filion, Senior Engineer Blizzard Entertainment Rob McNaughton, Lead Technical Artist Blizzard Entertainment Screen-space techniques Deferred rendering Screen-space ambient occlusion Depth of Field
Dominic Filion, Senior Engineer Blizzard Entertainment Rob McNaughton, Lead Technical Artist Blizzard Entertainment Screen-space techniques Deferred rendering Screen-space ambient occlusion Depth of Field
