An Anatomical Atlas to Support the Virtual Planning of Hip Operations
|
|
|
- Sheryl Copeland
- 6 years ago
- Views:
Transcription
1 An Anatomical Atlas to Support the Virtual Planning of Hip Operations J. Ehrhardt a, H. Handels a, T. Malina a, B. Strathmann b, W. Plötz b, S. J. Pöppl a a Institute for Medical Informatics and b Department of Orthopedic Surgery Medical University of Lübeck, Ratzeburger Allee 160, 3538 Lübeck, Germany. ehrhardt@medinf.mu-luebeck.de Abstract. Two three-dimensional digitized atlases of a female and a male pelvis were generated to support the virtual 3D-planning of hip operations. Beside the anatomical labeling of bone structures the determination of orthopedic measures, like angles, distances or sizes of contact areas, is important for the planning of hip operations. Thus, each atlas consists of labeled reference CT data sets, a set of landmarks as well as definitions of orthopedic measures and methods for their automatic computation. Furthermore, methods for the automatic transfer of anatomical labels from the atlas to an individual data set are presented resulting in three-dimensional models of the patient s bone structures. The anatomical atlases are designed to replace the interactive, time-consuming pre-processing steps for the virtual 3D operation planning. 1. Introduction In orthopedic surgery computer assisted systems are getting more and more important. Due to the complex geometry of the pelvis t he exact planning of the hip operation is essential for the operation result. The application of virtual planning procedures opens up new possibilities with regards to effort of time and costs and to an improved quality. During the computer assisted planning anatomical knowledge is required in the virtual environment. This is usually achieved by an interactive time-consuming segmentation step. The main idea of the presented approach is to represent the needed knowledge in an digital anatomical atlas and to use automatic algorithms to transfer this knowledge to the patient data. In the past digital atlases have been used for several purposes [1-4]. Especially atlases of the brain have been designed for teaching purposes, automatic segmentation and neurosurgical planning. In the considered application, a three-dimensional atlas of the human pelvis based on the CT data of the Visible Human Data Set [5] has been built up to facilitate virtual planning procedures in hip surgery. The atlases consists of labeled CT data sets, anatomical point landmarks and computation instructions for orthopedic measures.. Motivation The segmentation of different bone structures is necessary to address or analyze these structures in a virtual planning environment. Furthermore, the localization of landmarks is needed
2 to derive orthopedic measures, like distances or angles. For instance the virtual planning of an endoprosthetic reconstruction in bone tumor surgery requires at least the segmentation of the left and right hip bone, sacrum, left and right femur as well as a series of landmarks, e.g. the center of the femoral head or points on the border of the acetabulum [6]. The segmentation of the mentioned structures requires several hours using semi-automatic volume growing algorithms and interactive post-processing tools. If further measures are needed the number of labeled structures and landmarks increases. For example measuring of the contact area between acetabulum and femoral head requires the segmentation of both structures, the computation of the antetorsion requires the localization of the centers of the femoral head and the femoral leg as well as the condyles. This may increase the needed pre-processing time essentially. The goal of the presented approach is to avoid these time-consuming steps and to enable an automatic recognition of hip structures by using a digital atlas of the hip. 3. Digital anatomical atlas of the hip The virtual planning of hip operations is based on bone structures extracted from CT data. Starting point to built up the two atlases are high resolution CT image sequences of a woman and a man taken from the Visible Human Data Set. In a first step, threshold based volume growing algorithms are used for a rough segmentation of the bones. In the next interactive step the different bone structures are separated and insignificant structures, like vessels or sinews, are removed. Incomplete bone contours are closed with an semi-automatic "live-wire" [7] algorithm in a post-processing step. Finally, the segmentation result was reviewed and corrected by an expert. The separated structures are hip bones, acetabuli, sacrum, os coccygis and femuri composed by femoral head and shaft. A set of landmarks and measures for hip interventions are gathered by the surgeons. During the implementation two main problems arise: the verbal definitions of landmarks and measures are unprecise and most of them are based on x-rays. So the verbal descriptions are transformed to exact definitions of localization and computation instructions and the two-dimensional x-ray based measures are transferred into the three-dimensional space. Anatomical landmarks, e.g. promotorium or spina iliaca anterior superior, are selected interactively on the bone surface and methods for the computation of orthopedic measures, e.g. the center of femoral head, the antetorsion or the center-edge-angle, are implemented (fig. 1). Fig. 1: The centers of the femoral head (left) and the acetabulum (right) can be determined by an approximation with a sphere. In the right figure the center of femoral head (blue) and acetabulum (red) are visualized.
3 4. Matching of atlas and patient data For the transfer of the atlas information to the patient data set a registration of atlas and patient data is necessary. Due to the anatomical variations of the hip local transformations are needed in order to obtain correct results. A number of methods of varying complexity have been developed for the inter-patient 3D-3D registration of medical image volumes. Collins et al. has applied a multi-resolution approach with piecewise linear transformations [5]. Bajcsy et al. used an elastic model approach [6,7]. Algorithms based on viscous fluid models are developed by Christensen et al. [8] and Bro- Nielsen et al. [9]. Thirion presented an algorithm, which trades the strict physical modeling to simplify the implementation and speed up the execution of the registration [10]. In our application the applied registration method is divided into two parts: a global alignment of the two data sets and the local matching of corresponding structures. In the first step, an affine alignment of the two data sets is performed using the software tool AIR provided by Woods et al. [11]. Here, the affine Transformation T opt is determined, which minimizes the least squares distance T = arg min I ( T ( s )) I, opt T s where s are the coordinates of a point in the patient volume I P (s) and I A ( T ) is the intensity value of the atlas volume at the transformed coordinates T (s). Afterwards the atlas volume is resampled using the transformation T opt and trilinear interpolation. Due to the high resolution of the atlas data set the introduced interpolation errors are low. For the local matching of corresponding structures a totally freeform registration process based on Thirion s concept of demons is used. The basic idea of this algorithm is to consider the object contours in one static image as hemipermeable membrans and to consider the other image as a deformable model diffusing through these membranes. The demons are located onto the contours of the static image and push the deformable model in the normal direction of the object contour, but the orientation of the movement depends on the nature of the corresponding point in the model image. If the point is inside the model, the direction of the movement is inward, and if the point is outside, the direction is outward. In this way, the inside points of the model image are diffusing into the inside of the object contours of the static image, and vice versa for outside points. In an iterative process this strategy leads to a matching of static and model image. The iso-intensity contours in medical images are closely related to object contours. The demons are associated to each voxel s of the static image I 1, where the gradient norm I(s) is greater zero. The gradient I 1( s ) can be treated as the normal vector of the iso-intensity contour in s. The demon pushes the model image I according to I 1 if I 1 < I and according to I 1( s ) if I ( ) ( ) 1 s > I s. Precisely, the pushing force f of the demon is computed by ( I I1( s)) I1( s) f =. I1( s) + ( I I1( s)) The equation is derived from the optical flow formulation and has the desired behavior of a demon, for details see [11]. By the force vectors a displacement field is defined, i. e. each voxel could be moved by the computed vector. In order to achieve smooth deformations a regularization is necessary. Thirion proposed to apply a Gaussian filter to each of the three components of f. The standard deviation σ of the filter can be used to control the deformability of the model. The larger σ, the less deformable the image. A P 3
4 In our implementation the static image is the patient data set I P and the model image is the (global aligned) atlas data set I A. The matching is first computed on coarse downsampled images then successively on images with a finer spatial resolution. Each final solution at one scale serves to initialize the registration at the next finer scale. This strategy speeds up the computations and the convergence of the algorithm. By this technique macroscopic features are aligned first and afterwards microscopic features. Due to the strong anatomical variations in soft tissues a correct inter-patient matching of the whole pelvis region is nearly impossible. Hence, for the planning of orthopedic interventions the atlas-matching is limited to bones and surrounding voxel. For the coarse levels of the multiresolution strategy segmented atlas and patient data are used. Therefore, an automatic segmentation of the bone structures of the patient data is applied using conventional threshold based methods and morphological operators. The registration of the segmented atlas and patient data leads to a fast and robust matching of the bone surfaces. In order to enable a good adjustment of internal bone structures and to be tolerant in relation to errors of the threshold based segmentation, the original CT-volumes are used for the registration process on the finest resolution. The demons are placed in the static patient data volume. Due to the criterion I 0, for segmented images the demons are located on the object contours. For the registration of the gray value CT data the demons positions are determined by the result of the threshold based segmentation and a following dilatation. By this strategy, only bone structures are aligned and the matching of non-corresponding soft tissue is avoided. Figure shows a CT slice of a patient s data set overlaid with the contours of the atlas bone structures before and after the freeform registration. A good fit of atlas and patient data is obtained, but nevertheless small deviations can occur. 5. Recognition of anatomical structures and surface generation The atlas information is transferred by means of a nearest-neighbor approach to the patient data. Based on the threshold segmentation of the patient s bone structures the label of the nearest structure in the transformed atlas data set is assigned to each segmented voxel. Thus, bony patient voxels, which are not covered by an atlas label, are added to the nearest bone structure. For every labeled bone structure a surface model is constructed applying the marching cubes algorithm to the original CT data [9]. The chosen threshold must be the same as for the automatic segmentation. It can be determined by choosing standard hounsfield units for the corticalis ( ~ 100 HU ). Before the surface is generated, all voxels above the threshold, which are not belonging to the actual label, are set to values below the threshold. By using original CT data for the surface generation rather than segmented data, smooth surfaces with appropriate normals and gradients are generated. A triangle decimation algorithm [10] is used to reduce the number of the generated polygons significantly. The position of the anatomical point landmarks is determined by calculating the nearest point onto the patient s bone surface for every landmark in the transformed atlas data set. Figure 3 shows the surface models generated from the atlas data using the algorithm described above. The position of the anatomical landmarks is indicated by small blue points. The models consist of about triangles due to the high resolution of mm. 5. Automatic determination of orthopedic parameters 4
5 Fig. : CT slice of a patient s data set overlaid with the edges of the atlas bone structures (white) before (left) and after the non-rigid registration (right). Fig. 3: Three dimensional surface model of the female atlas data. The separated structures are presented in different colours. 6. Results and Conclusion For a first evaluation of the presented registration method, we register the two atlas data sets of the woman and the man. The image volumes are resliced to a resolution of mm resulting in voxels. To obtain a good starting position an affine pre-registration is performed. For an automatic labeling of the female bone structures both atlas data sets are binarized using the manual segmentation results. These binary data sets are registered and the labels from the male data set are transferred to the female. In comparison to the manual segmentation results 99.5% of the bony voxels are labeled correctly. Hence, a nearly perfect automatic labeling of the regarded anatomical structures is possible. In practice, for the patient data set only a rough threshold based segmentation is available. Therefore, a matching is performed on gray value data to evaluate the recognition accuracy in the practical application. During the registration process the CT data set of the visible female is treated as a patient data set. The female bone structures are labeled automatically by the transformed male atlas information. For 98.5% of the bony voxels the correct labels are assigned. In figure 4 the distance from the female bone surface to the bone surface of the male after the registration is visualized. The light surface regions correspond to distances near to the voxel resolution. Inaccuracies are introduced by varying anatomical details and intensities (e.g. 5
6 different calcification of bones). Furthermore, the smoothness constrain of the deformation field establishes a compromise between intensity resemblance and uniform local deformations. The presented methods are a central step to provide the surgeon with all conditions needed for the virtual planning without interactive pre-processing steps. Beside the separated surface models orthopedic measures of the patient s hip are necessary for a successful planning. By defining point landmarks and measures in the atlas we have done the first step in this direction. Since an automatic measuring of the patient s hip needs a very precise landmark localization we are developing algorithms for an automatic correction of landmark positions in a post-processing step. Fig. 4: Visualization of the surface fit: For every point located on the surface of the visualized bone the distance to the registered atlas surface is represented by gray scale values. References [1] K. H. Höhne, M. Bomanns, M. Riemer, R. Schubert, U. Tiede, W. Lierse: A 3D Anatomical Atlas Based on a Volume Model. IEEE Comput. Graphics Appl., 1, 4 (199), pp [] R. Kikinis, M. E. Shenton, D. V. Iosifescu, R. W. McCarley, P. Saiviroonporn, H. H. Hokama, A. Robatino, D. Metcal, C. G. Wible, C. M. Portas, R. M. Donnino and F. A. Jolesz: A Digital Brain Atlas for Surgical Planning, Model-Driven Segmentation and Teaching, IEEE Trans. Visualiz. Comput. Graphics, 3 (1996), pp [3] D. L. Collins, T. M. Peters, W. Dai, A. C. Evans: Model Based Segmentation of Individual Brain Structures from MRI Data. In R. A. Robb (ed.), Visualization in Biomedical Computing II, Proc. SPIE 1808, Chapel Hill, 199, pp [4] S. Warfield, J. Dengler, J. Zaers, C. R. G. Guttmann, W. M. Wells, G. J. Ettinger, J. Hiller, R. Kikinis: Automatic Identification of Gray Matter Structures from MRI to Improve the Segmentation of White Matter Lesions. In Proc. of Medical Robotics and Computer Assisted Surgery, 1995, pp [5] M. J. Ackermann, The Visible Human Project: A Resource for Anatomical Visualization. In: B. Cesnik, A.T. McCray, J.-R. Scherrer (ed.), 9 th World Congress on Medical Informatics, MEDINFO 98. IOS Press, Seoul, Korea, 1998, pp [6] H. Handels, J. Ehrhardt, W. Plötz and S.J.Pöppl, Computer-Assisted Planning and Simulation of Hip Operations Using Virtual Three-Dimensional Models. In: P. Kokol, B. Zupan, J. Stare, M. Premik, R. Engelbrecht (ed.), Medical Informatics Europe, MIE 99, IOS Press, Amsterdam, 1999, pp [7] W. A. Barrett and N.E. Mortensen, An Interactive Live-Wire Boundary Extraction, Medical Image Analysis, 1, 1 (1996) [8] J. P. Thirion, Non-Rigid Matching Using Demons, Proc. Int. Conf. Computer Vision and Pattern Recognition (CVPR 96),
7 [9] W.E. Lorensen and H.E. Cline, Marching Cubes: A High Resolution 3-D Surface Construction Algorithm, Computer Graphics, 1,4 (1987) [10] W.J. Schroeder, J.A. Zarge and W.E. Lorensen, Decimation of Triangle Meshes. In: SIGGRAPH9, Chicago, 199, pp
Simon K. Warfield, Michael Kaus, Ferenc A. Jolesz and Ron Kikinis
 Medical Image Analysis (2000) volume 4, number 1, pp 43 55 c Elsevier Science BV Adaptive, Template Moderated, Spatially Varying Statistical Classification Simon K. Warfield, Michael Kaus, Ferenc A. Jolesz
Medical Image Analysis (2000) volume 4, number 1, pp 43 55 c Elsevier Science BV Adaptive, Template Moderated, Spatially Varying Statistical Classification Simon K. Warfield, Michael Kaus, Ferenc A. Jolesz
Calculating the Distance Map for Binary Sampled Data
 MITSUBISHI ELECTRIC RESEARCH LABORATORIES http://www.merl.com Calculating the Distance Map for Binary Sampled Data Sarah F. Frisken Gibson TR99-6 December 999 Abstract High quality rendering and physics-based
MITSUBISHI ELECTRIC RESEARCH LABORATORIES http://www.merl.com Calculating the Distance Map for Binary Sampled Data Sarah F. Frisken Gibson TR99-6 December 999 Abstract High quality rendering and physics-based
Whole Body MRI Intensity Standardization
 Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Image Registration I
 Image Registration I Comp 254 Spring 2002 Guido Gerig Image Registration: Motivation Motivation for Image Registration Combine images from different modalities (multi-modality registration), e.g. CT&MRI,
Image Registration I Comp 254 Spring 2002 Guido Gerig Image Registration: Motivation Motivation for Image Registration Combine images from different modalities (multi-modality registration), e.g. CT&MRI,
Rigid and Deformable Vasculature-to-Image Registration : a Hierarchical Approach
 Rigid and Deformable Vasculature-to-Image Registration : a Hierarchical Approach Julien Jomier and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel
Rigid and Deformable Vasculature-to-Image Registration : a Hierarchical Approach Julien Jomier and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel
2 Michael E. Leventon and Sarah F. F. Gibson a b c d Fig. 1. (a, b) Two MR scans of a person's knee. Both images have high resolution in-plane, but ha
 Model Generation from Multiple Volumes using Constrained Elastic SurfaceNets Michael E. Leventon and Sarah F. F. Gibson 1 MIT Artificial Intelligence Laboratory, Cambridge, MA 02139, USA leventon@ai.mit.edu
Model Generation from Multiple Volumes using Constrained Elastic SurfaceNets Michael E. Leventon and Sarah F. F. Gibson 1 MIT Artificial Intelligence Laboratory, Cambridge, MA 02139, USA leventon@ai.mit.edu
A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images
 A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images Jianhua Yao 1, Russell Taylor 2 1. Diagnostic Radiology Department, Clinical Center,
A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images Jianhua Yao 1, Russell Taylor 2 1. Diagnostic Radiology Department, Clinical Center,
Scene-Based Segmentation of Multiple Muscles from MRI in MITK
 Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
Segmentation of Bony Structures with Ligament Attachment Sites
 Segmentation of Bony Structures with Ligament Attachment Sites Heiko Seim 1, Hans Lamecker 1, Markus Heller 2, Stefan Zachow 1 1 Visualisierung und Datenanalyse, Zuse-Institut Berlin (ZIB), 14195 Berlin
Segmentation of Bony Structures with Ligament Attachment Sites Heiko Seim 1, Hans Lamecker 1, Markus Heller 2, Stefan Zachow 1 1 Visualisierung und Datenanalyse, Zuse-Institut Berlin (ZIB), 14195 Berlin
A Study of Medical Image Analysis System
 Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
Parallel Non-Rigid Registration on a Cluster of Workstations
 Parallel Non-Rigid Registration on a Cluster of Workstations Radu Stefanescu, Xavier Pennec, Nicholas Ayache INRIA Sophia, Epidaure, 004 Rte des Lucioles, F-0690 Sophia-Antipolis Cedex {Radu.Stefanescu,
Parallel Non-Rigid Registration on a Cluster of Workstations Radu Stefanescu, Xavier Pennec, Nicholas Ayache INRIA Sophia, Epidaure, 004 Rte des Lucioles, F-0690 Sophia-Antipolis Cedex {Radu.Stefanescu,
An Automated Registration Algorithm for Measuring MRI Subcortical Brain Structures 1,2
 NEUROIMAGE 6, 13 25 (1997) ARTICLE NO. NI970274 An Automated Registration Algorithm for Measuring MRI Subcortical Brain Structures 1,2 Dan V. Iosifescu,* Martha E. Shenton,* Simon K. Warfield, Ron Kikinis,
NEUROIMAGE 6, 13 25 (1997) ARTICLE NO. NI970274 An Automated Registration Algorithm for Measuring MRI Subcortical Brain Structures 1,2 Dan V. Iosifescu,* Martha E. Shenton,* Simon K. Warfield, Ron Kikinis,
Non linear Registration of Pre and Intraoperative Volume Data Based On Piecewise Linear Transformations
 Non linear Registration of Pre and Intraoperative Volume Data Based On Piecewise Linear Transformations C. Rezk Salama, P. Hastreiter, G. Greiner, T. Ertl University of Erlangen, Computer Graphics Group
Non linear Registration of Pre and Intraoperative Volume Data Based On Piecewise Linear Transformations C. Rezk Salama, P. Hastreiter, G. Greiner, T. Ertl University of Erlangen, Computer Graphics Group
The Insight Toolkit. Image Registration Algorithms & Frameworks
 The Insight Toolkit Image Registration Algorithms & Frameworks Registration in ITK Image Registration Framework Multi Resolution Registration Framework Components PDE Based Registration FEM Based Registration
The Insight Toolkit Image Registration Algorithms & Frameworks Registration in ITK Image Registration Framework Multi Resolution Registration Framework Components PDE Based Registration FEM Based Registration
Fast Interactive Region of Interest Selection for Volume Visualization
 Fast Interactive Region of Interest Selection for Volume Visualization Dominik Sibbing and Leif Kobbelt Lehrstuhl für Informatik 8, RWTH Aachen, 20 Aachen Email: {sibbing,kobbelt}@informatik.rwth-aachen.de
Fast Interactive Region of Interest Selection for Volume Visualization Dominik Sibbing and Leif Kobbelt Lehrstuhl für Informatik 8, RWTH Aachen, 20 Aachen Email: {sibbing,kobbelt}@informatik.rwth-aachen.de
Additional file 1: Online Supplementary Material 1
 Additional file 1: Online Supplementary Material 1 Calyn R Moulton and Michael J House School of Physics, University of Western Australia, Crawley, Western Australia. Victoria Lye, Colin I Tang, Michele
Additional file 1: Online Supplementary Material 1 Calyn R Moulton and Michael J House School of Physics, University of Western Australia, Crawley, Western Australia. Victoria Lye, Colin I Tang, Michele
An Automated Measurement of Subcortical Brain MR Structures in Schizophrenia
 An Automated Measurement of Subcortical Brain MR Structures in Schizophrenia Dan V. Iosifescu 1, Martha E. Shenton 1, Simon K. Warfield 3, Ron Kikinis 2, Joachim Dengler 4, Ferenc A. Jolesz 2, and Robert
An Automated Measurement of Subcortical Brain MR Structures in Schizophrenia Dan V. Iosifescu 1, Martha E. Shenton 1, Simon K. Warfield 3, Ron Kikinis 2, Joachim Dengler 4, Ferenc A. Jolesz 2, and Robert
CT Protocol Clinical Graphics Move Forward 3D motion analysis service
 CT Protocol Clinical Graphics Move Forward 3D motion analysis service Version 1.4 Description of contents This document describes the CT protocol of scans for Clinical Graphics Move Forward 3D Motion Simulation
CT Protocol Clinical Graphics Move Forward 3D motion analysis service Version 1.4 Description of contents This document describes the CT protocol of scans for Clinical Graphics Move Forward 3D Motion Simulation
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes
 Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
Preprocessing II: Between Subjects John Ashburner
 Preprocessing II: Between Subjects John Ashburner Pre-processing Overview Statistics or whatever fmri time-series Anatomical MRI Template Smoothed Estimate Spatial Norm Motion Correct Smooth Coregister
Preprocessing II: Between Subjects John Ashburner Pre-processing Overview Statistics or whatever fmri time-series Anatomical MRI Template Smoothed Estimate Spatial Norm Motion Correct Smooth Coregister
MR IMAGE SEGMENTATION
 MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
Non-Rigid Image Registration III
 Non-Rigid Image Registration III CS6240 Multimedia Analysis Leow Wee Kheng Department of Computer Science School of Computing National University of Singapore Leow Wee Kheng (CS6240) Non-Rigid Image Registration
Non-Rigid Image Registration III CS6240 Multimedia Analysis Leow Wee Kheng Department of Computer Science School of Computing National University of Singapore Leow Wee Kheng (CS6240) Non-Rigid Image Registration
Biomedical Paper AnatomyBrowser: A Novel Approach to Visualization and Integration of Medical Information
 Computer Aided Surgery 4:129 143 (1999) Biomedical Paper AnatomyBrowser: A Novel Approach to Visualization and Integration of Medical Information P. Golland, M.Sc., R. Kikinis, M.D., M. Halle, Ph.D., C.
Computer Aided Surgery 4:129 143 (1999) Biomedical Paper AnatomyBrowser: A Novel Approach to Visualization and Integration of Medical Information P. Golland, M.Sc., R. Kikinis, M.D., M. Halle, Ph.D., C.
Segmentation of 3D CT Volume Images Using a Single 2D Atlas
 Segmentation of 3D CT Volume Images Using a Single 2D Atlas Feng Ding 1, Wee Kheng Leow 1, and Shih-Chang Wang 2 1 Dept. of Computer Science, National University of Singapore, 3 Science Drive 2, Singapore
Segmentation of 3D CT Volume Images Using a Single 2D Atlas Feng Ding 1, Wee Kheng Leow 1, and Shih-Chang Wang 2 1 Dept. of Computer Science, National University of Singapore, 3 Science Drive 2, Singapore
Non-rigid Image Registration
 Overview Non-rigid Image Registration Introduction to image registration - he goal of image registration - Motivation for medical image registration - Classification of image registration - Nonrigid registration
Overview Non-rigid Image Registration Introduction to image registration - he goal of image registration - Motivation for medical image registration - Classification of image registration - Nonrigid registration
Advanced Visual Medicine: Techniques for Visual Exploration & Analysis
 Advanced Visual Medicine: Techniques for Visual Exploration & Analysis Interactive Visualization of Multimodal Volume Data for Neurosurgical Planning Felix Ritter, MeVis Research Bremen Multimodal Neurosurgical
Advanced Visual Medicine: Techniques for Visual Exploration & Analysis Interactive Visualization of Multimodal Volume Data for Neurosurgical Planning Felix Ritter, MeVis Research Bremen Multimodal Neurosurgical
VETOT, Volume Estimation and Tracking Over Time: Framework and Validation
 VETOT, Volume Estimation and Tracking Over Time: Framework and Validation Jean-Philippe Guyon 1, Mark Foskey 1, Jisung Kim 1, Zeynep Firat 2, Barb Davis 3, and Stephen Aylward 1 1 Computer Aided Display
VETOT, Volume Estimation and Tracking Over Time: Framework and Validation Jean-Philippe Guyon 1, Mark Foskey 1, Jisung Kim 1, Zeynep Firat 2, Barb Davis 3, and Stephen Aylward 1 1 Computer Aided Display
Segmentation of 3-D medical image data sets with a combination of region based initial segmentation and active surfaces
 Header for SPIE use Segmentation of 3-D medical image data sets with a combination of region based initial segmentation and active surfaces Regina Pohle, Thomas Behlau, Klaus D. Toennies Otto-von-Guericke
Header for SPIE use Segmentation of 3-D medical image data sets with a combination of region based initial segmentation and active surfaces Regina Pohle, Thomas Behlau, Klaus D. Toennies Otto-von-Guericke
A Virtual Reality Training System for Pediatric Sonography
 1 A Virtual Reality Training System for Pediatric Sonography W. Arkhurst a, A. Pommert a, E. Richter b, H. Frederking a, S.-I. Kim ab, R. Schubert a, and K. H. Höhne a a Institute of Mathematics and Computer
1 A Virtual Reality Training System for Pediatric Sonography W. Arkhurst a, A. Pommert a, E. Richter b, H. Frederking a, S.-I. Kim ab, R. Schubert a, and K. H. Höhne a a Institute of Mathematics and Computer
A Novel Nonrigid Registration Algorithm and Applications
 A Novel Nonrigid Registration Algorithm and Applications J. Rexilius 1, S.K. Warfield 1, C.R.G. Guttmann 1, X. Wei 1, R. Benson 2, L. Wolfson 2, M. Shenton 1, H. Handels 3, and R. Kikinis 1 1 Surgical
A Novel Nonrigid Registration Algorithm and Applications J. Rexilius 1, S.K. Warfield 1, C.R.G. Guttmann 1, X. Wei 1, R. Benson 2, L. Wolfson 2, M. Shenton 1, H. Handels 3, and R. Kikinis 1 1 Surgical
Elastic registration of medical images using finite element meshes
 Elastic registration of medical images using finite element meshes Hartwig Grabowski Institute of Real-Time Computer Systems & Robotics, University of Karlsruhe, D-76128 Karlsruhe, Germany. Email: grabow@ira.uka.de
Elastic registration of medical images using finite element meshes Hartwig Grabowski Institute of Real-Time Computer Systems & Robotics, University of Karlsruhe, D-76128 Karlsruhe, Germany. Email: grabow@ira.uka.de
Edge-Preserving Denoising for Segmentation in CT-Images
 Edge-Preserving Denoising for Segmentation in CT-Images Eva Eibenberger, Anja Borsdorf, Andreas Wimmer, Joachim Hornegger Lehrstuhl für Mustererkennung, Friedrich-Alexander-Universität Erlangen-Nürnberg
Edge-Preserving Denoising for Segmentation in CT-Images Eva Eibenberger, Anja Borsdorf, Andreas Wimmer, Joachim Hornegger Lehrstuhl für Mustererkennung, Friedrich-Alexander-Universität Erlangen-Nürnberg
Problem Solving Assignment 1
 CS6240 Problem Solving Assignment 1 p. 1/20 Problem Solving Assignment 1 CS6240 Multimedia Analysis Daniel Dahlmeier National University of Singapore CS6240 Problem Solving Assignment 1 p. 2/20 Introduction
CS6240 Problem Solving Assignment 1 p. 1/20 Problem Solving Assignment 1 CS6240 Multimedia Analysis Daniel Dahlmeier National University of Singapore CS6240 Problem Solving Assignment 1 p. 2/20 Introduction
Nonlinear Registration and Template Driven. Segmentation
 Nonlinear Registration and Template Driven Segmentation Simon Warfield Andre Robatino Joachim Dengler Ferenc Jolesz Ron Kikinis I Introduction Registration is the process of the alignment of medical image
Nonlinear Registration and Template Driven Segmentation Simon Warfield Andre Robatino Joachim Dengler Ferenc Jolesz Ron Kikinis I Introduction Registration is the process of the alignment of medical image
NIH Public Access Author Manuscript Proc Soc Photo Opt Instrum Eng. Author manuscript; available in PMC 2014 October 07.
 NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
Comparison of Different Metrics for Appearance-model-based 2D/3D-registration with X-ray Images
 Comparison of Different Metrics for Appearance-model-based 2D/3D-registration with X-ray Images Philipp Steininger 1, Karl D. Fritscher 1, Gregor Kofler 1, Benedikt Schuler 1, Markus Hänni 2, Karsten Schwieger
Comparison of Different Metrics for Appearance-model-based 2D/3D-registration with X-ray Images Philipp Steininger 1, Karl D. Fritscher 1, Gregor Kofler 1, Benedikt Schuler 1, Markus Hänni 2, Karsten Schwieger
coding of various parts showing different features, the possibility of rotation or of hiding covering parts of the object's surface to gain an insight
 Three-Dimensional Object Reconstruction from Layered Spatial Data Michael Dangl and Robert Sablatnig Vienna University of Technology, Institute of Computer Aided Automation, Pattern Recognition and Image
Three-Dimensional Object Reconstruction from Layered Spatial Data Michael Dangl and Robert Sablatnig Vienna University of Technology, Institute of Computer Aided Automation, Pattern Recognition and Image
Volumetric Deformable Models for Simulation of Laparoscopic Surgery
 Volumetric Deformable Models for Simulation of Laparoscopic Surgery S. Cotin y, H. Delingette y, J.M. Clément z V. Tassetti z, J. Marescaux z, N. Ayache y y INRIA, Epidaure Project 2004, route des Lucioles,
Volumetric Deformable Models for Simulation of Laparoscopic Surgery S. Cotin y, H. Delingette y, J.M. Clément z V. Tassetti z, J. Marescaux z, N. Ayache y y INRIA, Epidaure Project 2004, route des Lucioles,
Anatomical landmark and region mapping based on a template surface deformation for foot bone morphology
 Anatomical landmark and region mapping based on a template surface deformation for foot bone morphology Jaeil Kim 1, Sang Gyo Seo 2, Dong Yeon Lee 2, Jinah Park 1 1 Department of Computer Science, KAIST,
Anatomical landmark and region mapping based on a template surface deformation for foot bone morphology Jaeil Kim 1, Sang Gyo Seo 2, Dong Yeon Lee 2, Jinah Park 1 1 Department of Computer Science, KAIST,
Where are we now? Structural MRI processing and analysis
 Where are we now? Structural MRI processing and analysis Pierre-Louis Bazin bazin@cbs.mpg.de Leipzig, Germany Structural MRI processing: why bother? Just use the standards? SPM FreeSurfer FSL However:
Where are we now? Structural MRI processing and analysis Pierre-Louis Bazin bazin@cbs.mpg.de Leipzig, Germany Structural MRI processing: why bother? Just use the standards? SPM FreeSurfer FSL However:
Integrated Approaches to Non-Rigid Registration in Medical Images
 Work. on Appl. of Comp. Vision, pg 102-108. 1 Integrated Approaches to Non-Rigid Registration in Medical Images Yongmei Wang and Lawrence H. Staib + Departments of Electrical Engineering and Diagnostic
Work. on Appl. of Comp. Vision, pg 102-108. 1 Integrated Approaches to Non-Rigid Registration in Medical Images Yongmei Wang and Lawrence H. Staib + Departments of Electrical Engineering and Diagnostic
STATISTICAL ATLAS-BASED SUB-VOXEL SEGMENTATION OF 3D BRAIN MRI
 STATISTICA ATAS-BASED SUB-VOXE SEGMENTATION OF 3D BRAIN MRI Marcel Bosc 1,2, Fabrice Heitz 1, Jean-Paul Armspach 2 (1) SIIT UMR-7005 CNRS / Strasbourg I University, 67400 Illkirch, France (2) IPB UMR-7004
STATISTICA ATAS-BASED SUB-VOXE SEGMENTATION OF 3D BRAIN MRI Marcel Bosc 1,2, Fabrice Heitz 1, Jean-Paul Armspach 2 (1) SIIT UMR-7005 CNRS / Strasbourg I University, 67400 Illkirch, France (2) IPB UMR-7004
Landmark-based 3D Elastic Registration of Pre- and Postoperative Liver CT Data
 Landmark-based 3D Elastic Registration of Pre- and Postoperative Liver CT Data An Experimental Comparison Thomas Lange 1, Stefan Wörz 2, Karl Rohr 2, Peter M. Schlag 3 1 Experimental and Clinical Research
Landmark-based 3D Elastic Registration of Pre- and Postoperative Liver CT Data An Experimental Comparison Thomas Lange 1, Stefan Wörz 2, Karl Rohr 2, Peter M. Schlag 3 1 Experimental and Clinical Research
CT IMAGE PROCESSING IN HIP ARTHROPLASTY
 U.P.B. Sci. Bull., Series C, Vol. 75, Iss. 3, 2013 ISSN 2286 3540 CT IMAGE PROCESSING IN HIP ARTHROPLASTY Anca MORAR 1, Florica MOLDOVEANU 2, Alin MOLDOVEANU 3, Victor ASAVEI 4, Alexandru EGNER 5 The use
U.P.B. Sci. Bull., Series C, Vol. 75, Iss. 3, 2013 ISSN 2286 3540 CT IMAGE PROCESSING IN HIP ARTHROPLASTY Anca MORAR 1, Florica MOLDOVEANU 2, Alin MOLDOVEANU 3, Victor ASAVEI 4, Alexandru EGNER 5 The use
Methods for data preprocessing
 Methods for data preprocessing John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. Overview Voxel-Based Morphometry Morphometry in general Volumetrics VBM preprocessing
Methods for data preprocessing John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. Overview Voxel-Based Morphometry Morphometry in general Volumetrics VBM preprocessing
3D VISUALIZATION OF SEGMENTED CRUCIATE LIGAMENTS 1. INTRODUCTION
 JOURNAL OF MEDICAL INFORMATICS & TECHNOLOGIES Vol. 10/006, ISSN 164-6037 Paweł BADURA * cruciate ligament, segmentation, fuzzy connectedness,3d visualization 3D VISUALIZATION OF SEGMENTED CRUCIATE LIGAMENTS
JOURNAL OF MEDICAL INFORMATICS & TECHNOLOGIES Vol. 10/006, ISSN 164-6037 Paweł BADURA * cruciate ligament, segmentation, fuzzy connectedness,3d visualization 3D VISUALIZATION OF SEGMENTED CRUCIATE LIGAMENTS
RIGID IMAGE REGISTRATION
 RIGID IMAGE REGISTRATION Duygu Tosun-Turgut, Ph.D. Center for Imaging of Neurodegenerative Diseases Department of Radiology and Biomedical Imaging duygu.tosun@ucsf.edu What is registration? Image registration
RIGID IMAGE REGISTRATION Duygu Tosun-Turgut, Ph.D. Center for Imaging of Neurodegenerative Diseases Department of Radiology and Biomedical Imaging duygu.tosun@ucsf.edu What is registration? Image registration
Norbert Schuff VA Medical Center and UCSF
 Norbert Schuff Medical Center and UCSF Norbert.schuff@ucsf.edu Medical Imaging Informatics N.Schuff Course # 170.03 Slide 1/67 Objective Learn the principle segmentation techniques Understand the role
Norbert Schuff Medical Center and UCSF Norbert.schuff@ucsf.edu Medical Imaging Informatics N.Schuff Course # 170.03 Slide 1/67 Objective Learn the principle segmentation techniques Understand the role
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION
 ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
ABSTRACT 1. INTRODUCTION 2. METHODS
 Finding Seeds for Segmentation Using Statistical Fusion Fangxu Xing *a, Andrew J. Asman b, Jerry L. Prince a,c, Bennett A. Landman b,c,d a Department of Electrical and Computer Engineering, Johns Hopkins
Finding Seeds for Segmentation Using Statistical Fusion Fangxu Xing *a, Andrew J. Asman b, Jerry L. Prince a,c, Bennett A. Landman b,c,d a Department of Electrical and Computer Engineering, Johns Hopkins
Efficient population registration of 3D data
 Efficient population registration of 3D data Lilla Zöllei 1, Erik Learned-Miller 2, Eric Grimson 1, William Wells 1,3 1 Computer Science and Artificial Intelligence Lab, MIT; 2 Dept. of Computer Science,
Efficient population registration of 3D data Lilla Zöllei 1, Erik Learned-Miller 2, Eric Grimson 1, William Wells 1,3 1 Computer Science and Artificial Intelligence Lab, MIT; 2 Dept. of Computer Science,
Level Set Evolution with Region Competition: Automatic 3-D Segmentation of Brain Tumors
 1 Level Set Evolution with Region Competition: Automatic 3-D Segmentation of Brain Tumors 1 Sean Ho, 2 Elizabeth Bullitt, and 1,3 Guido Gerig 1 Department of Computer Science, 2 Department of Surgery,
1 Level Set Evolution with Region Competition: Automatic 3-D Segmentation of Brain Tumors 1 Sean Ho, 2 Elizabeth Bullitt, and 1,3 Guido Gerig 1 Department of Computer Science, 2 Department of Surgery,
Biomedical Image Processing
 Biomedical Image Processing Jason Thong Gabriel Grant 1 2 Motivation from the Medical Perspective MRI, CT and other biomedical imaging devices were designed to assist doctors in their diagnosis and treatment
Biomedical Image Processing Jason Thong Gabriel Grant 1 2 Motivation from the Medical Perspective MRI, CT and other biomedical imaging devices were designed to assist doctors in their diagnosis and treatment
Registration-Based Segmentation of Medical Images
 School of Computing National University of Singapore Graduate Research Paper Registration-Based Segmentation of Medical Images by Li Hao under guidance of A/Prof. Leow Wee Kheng July, 2006 Abstract Medical
School of Computing National University of Singapore Graduate Research Paper Registration-Based Segmentation of Medical Images by Li Hao under guidance of A/Prof. Leow Wee Kheng July, 2006 Abstract Medical
Comparison of Vessel Segmentations using STAPLE
 Comparison of Vessel Segmentations using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel Hill, Department
Comparison of Vessel Segmentations using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel Hill, Department
Automatic Subthalamic Nucleus Targeting for Deep Brain Stimulation. A Validation Study
 Automatic Subthalamic Nucleus Targeting for Deep Brain Stimulation. A Validation Study F. Javier Sánchez Castro a, Claudio Pollo a,b, Jean-Guy Villemure b, Jean-Philippe Thiran a a École Polytechnique
Automatic Subthalamic Nucleus Targeting for Deep Brain Stimulation. A Validation Study F. Javier Sánchez Castro a, Claudio Pollo a,b, Jean-Guy Villemure b, Jean-Philippe Thiran a a École Polytechnique
2D-3D Registration using Gradient-based MI for Image Guided Surgery Systems
 2D-3D Registration using Gradient-based MI for Image Guided Surgery Systems Yeny Yim 1*, Xuanyi Chen 1, Mike Wakid 1, Steve Bielamowicz 2, James Hahn 1 1 Department of Computer Science, The George Washington
2D-3D Registration using Gradient-based MI for Image Guided Surgery Systems Yeny Yim 1*, Xuanyi Chen 1, Mike Wakid 1, Steve Bielamowicz 2, James Hahn 1 1 Department of Computer Science, The George Washington
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge
 Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
Probabilistic Registration of 3-D Medical Images
 Probabilistic Registration of 3-D Medical Images Mei Chen, Takeo Kanade, Dean Pomerleau, Jeff Schneider CMU-RI-TR-99-16 The Robotics Institute Carnegie Mellon University Pittsburgh, Pennsylvania 1513 July,
Probabilistic Registration of 3-D Medical Images Mei Chen, Takeo Kanade, Dean Pomerleau, Jeff Schneider CMU-RI-TR-99-16 The Robotics Institute Carnegie Mellon University Pittsburgh, Pennsylvania 1513 July,
A Binary Entropy Measure to Assess Nonrigid Registration Algorithms
 A Binary Entropy Measure to Assess Nonrigid Registration Algorithms Simon K. Warfield 1, Jan Rexilius 1, Petra S. Huppi 2, Terrie E. Inder 3, Erik G. Miller 1, William M. Wells III 1, Gary P. Zientara
A Binary Entropy Measure to Assess Nonrigid Registration Algorithms Simon K. Warfield 1, Jan Rexilius 1, Petra S. Huppi 2, Terrie E. Inder 3, Erik G. Miller 1, William M. Wells III 1, Gary P. Zientara
Learning-based Neuroimage Registration
 Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
Norbert Schuff Professor of Radiology VA Medical Center and UCSF
 Norbert Schuff Professor of Radiology Medical Center and UCSF Norbert.schuff@ucsf.edu 2010, N.Schuff Slide 1/67 Overview Definitions Role of Segmentation Segmentation methods Intensity based Shape based
Norbert Schuff Professor of Radiology Medical Center and UCSF Norbert.schuff@ucsf.edu 2010, N.Schuff Slide 1/67 Overview Definitions Role of Segmentation Segmentation methods Intensity based Shape based
Computational Neuroanatomy
 Computational Neuroanatomy John Ashburner john@fil.ion.ucl.ac.uk Smoothing Motion Correction Between Modality Co-registration Spatial Normalisation Segmentation Morphometry Overview fmri time-series kernel
Computational Neuroanatomy John Ashburner john@fil.ion.ucl.ac.uk Smoothing Motion Correction Between Modality Co-registration Spatial Normalisation Segmentation Morphometry Overview fmri time-series kernel
A Non-Linear Image Registration Scheme for Real-Time Liver Ultrasound Tracking using Normalized Gradient Fields
 A Non-Linear Image Registration Scheme for Real-Time Liver Ultrasound Tracking using Normalized Gradient Fields Lars König, Till Kipshagen and Jan Rühaak Fraunhofer MEVIS Project Group Image Registration,
A Non-Linear Image Registration Scheme for Real-Time Liver Ultrasound Tracking using Normalized Gradient Fields Lars König, Till Kipshagen and Jan Rühaak Fraunhofer MEVIS Project Group Image Registration,
An Object-Based Volumetric Deformable Atlas for the Improved Localization of Neuroanatomy in MR Images
 An Object-Based Volumetric Deformable Atlas for the Improved Localization of Neuroanatomy in MR Images Tim McInerney 1,2 and Ron Kikinis 3 1 University of Toronto, Toronto, ON, Canada M5S 3H5 2 Massachusetts
An Object-Based Volumetric Deformable Atlas for the Improved Localization of Neuroanatomy in MR Images Tim McInerney 1,2 and Ron Kikinis 3 1 University of Toronto, Toronto, ON, Canada M5S 3H5 2 Massachusetts
CHAPTER 2. Morphometry on rodent brains. A.E.H. Scheenstra J. Dijkstra L. van der Weerd
 CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
Comparison of Vessel Segmentations Using STAPLE
 Comparison of Vessel Segmentations Using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, The University of North Carolina at Chapel Hill, Department
Comparison of Vessel Segmentations Using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, The University of North Carolina at Chapel Hill, Department
Labeling the Brain Surface Using a Deformable Multiresolution Mesh
 Labeling the Brain Surface Using a Deformable Multiresolution Mesh Sylvain Jaume 1, Benoît Macq 2, and Simon K. Warfield 3 1 Multiresolution Modeling Group, California Institute of Technology, 256-80,
Labeling the Brain Surface Using a Deformable Multiresolution Mesh Sylvain Jaume 1, Benoît Macq 2, and Simon K. Warfield 3 1 Multiresolution Modeling Group, California Institute of Technology, 256-80,
Introduction to Medical Image Processing
 Introduction to Medical Image Processing Δ Essential environments of a medical imaging system Subject Image Analysis Energy Imaging System Images Image Processing Feature Images Image processing may be
Introduction to Medical Image Processing Δ Essential environments of a medical imaging system Subject Image Analysis Energy Imaging System Images Image Processing Feature Images Image processing may be
2D Rigid Registration of MR Scans using the 1d Binary Projections
 2D Rigid Registration of MR Scans using the 1d Binary Projections Panos D. Kotsas Abstract This paper presents the application of a signal intensity independent registration criterion for 2D rigid body
2D Rigid Registration of MR Scans using the 1d Binary Projections Panos D. Kotsas Abstract This paper presents the application of a signal intensity independent registration criterion for 2D rigid body
3D Surface Reconstruction of the Brain based on Level Set Method
 3D Surface Reconstruction of the Brain based on Level Set Method Shijun Tang, Bill P. Buckles, and Kamesh Namuduri Department of Computer Science & Engineering Department of Electrical Engineering University
3D Surface Reconstruction of the Brain based on Level Set Method Shijun Tang, Bill P. Buckles, and Kamesh Namuduri Department of Computer Science & Engineering Department of Electrical Engineering University
Dr. Ulas Bagci
 Lecture 9: Deformable Models and Segmentation CAP-Computer Vision Lecture 9-Deformable Models and Segmentation Dr. Ulas Bagci bagci@ucf.edu Lecture 9: Deformable Models and Segmentation Motivation A limitation
Lecture 9: Deformable Models and Segmentation CAP-Computer Vision Lecture 9-Deformable Models and Segmentation Dr. Ulas Bagci bagci@ucf.edu Lecture 9: Deformable Models and Segmentation Motivation A limitation
1. INTRODUCTION ABSTRACT
 Incorporating Tissue Excision in Deformable Image Registration: A Modified Demons Algorithm for Cone-Beam CT-Guided Surgery S.Nithiananthan, a D. Mirota, b A. Uneri, b S. Schafer, a Y. Otake, b J.W. Stayman,
Incorporating Tissue Excision in Deformable Image Registration: A Modified Demons Algorithm for Cone-Beam CT-Guided Surgery S.Nithiananthan, a D. Mirota, b A. Uneri, b S. Schafer, a Y. Otake, b J.W. Stayman,
Assessing Accuracy Factors in Deformable 2D/3D Medical Image Registration Using a Statistical Pelvis Model
 Assessing Accuracy Factors in Deformable 2D/3D Medical Image Registration Using a Statistical Pelvis Model Jianhua Yao National Institute of Health Bethesda, MD USA jyao@cc.nih.gov Russell Taylor The Johns
Assessing Accuracy Factors in Deformable 2D/3D Medical Image Registration Using a Statistical Pelvis Model Jianhua Yao National Institute of Health Bethesda, MD USA jyao@cc.nih.gov Russell Taylor The Johns
3D Volume Mesh Generation of Human Organs Using Surface Geometries Created from the Visible Human Data Set
 3D Volume Mesh Generation of Human Organs Using Surface Geometries Created from the Visible Human Data Set John M. Sullivan, Jr., Ziji Wu, and Anand Kulkarni Worcester Polytechnic Institute Worcester,
3D Volume Mesh Generation of Human Organs Using Surface Geometries Created from the Visible Human Data Set John M. Sullivan, Jr., Ziji Wu, and Anand Kulkarni Worcester Polytechnic Institute Worcester,
David Wagner, Kaan Divringi, Can Ozcan Ozen Engineering
 Internal Forces of the Femur: An Automated Procedure for Applying Boundary Conditions Obtained From Inverse Dynamic Analysis to Finite Element Simulations David Wagner, Kaan Divringi, Can Ozcan Ozen Engineering
Internal Forces of the Femur: An Automated Procedure for Applying Boundary Conditions Obtained From Inverse Dynamic Analysis to Finite Element Simulations David Wagner, Kaan Divringi, Can Ozcan Ozen Engineering
VOLUMETRIC HARMONIC MAP
 COMMUNICATIONS IN INFORMATION AND SYSTEMS c 2004 International Press Vol. 3, No. 3, pp. 191-202, March 2004 004 VOLUMETRIC HARMONIC MAP YALIN WANG, XIANFENG GU, AND SHING-TUNG YAU Abstract. We develop
COMMUNICATIONS IN INFORMATION AND SYSTEMS c 2004 International Press Vol. 3, No. 3, pp. 191-202, March 2004 004 VOLUMETRIC HARMONIC MAP YALIN WANG, XIANFENG GU, AND SHING-TUNG YAU Abstract. We develop
Scalar Data. Visualization Torsten Möller. Weiskopf/Machiraju/Möller
 Scalar Data Visualization Torsten Möller Weiskopf/Machiraju/Möller Overview Basic strategies Function plots and height fields Isolines Color coding Volume visualization (overview) Classification Segmentation
Scalar Data Visualization Torsten Möller Weiskopf/Machiraju/Möller Overview Basic strategies Function plots and height fields Isolines Color coding Volume visualization (overview) Classification Segmentation
Atlas Based Segmentation of the prostate in MR images
 Atlas Based Segmentation of the prostate in MR images Albert Gubern-Merida and Robert Marti Universitat de Girona, Computer Vision and Robotics Group, Girona, Spain {agubern,marly}@eia.udg.edu Abstract.
Atlas Based Segmentation of the prostate in MR images Albert Gubern-Merida and Robert Marti Universitat de Girona, Computer Vision and Robotics Group, Girona, Spain {agubern,marly}@eia.udg.edu Abstract.
Automatic Extraction of Femur Contours from Hip X-ray Images
 Automatic Extraction of Femur Contours from Hip X-ray Images Ying Chen 1, Xianhe Ee 1, Wee Kheng Leow 1, and Tet Sen Howe 2 1 Dept of Computer Science, National University of Singapore, 3 Science Drive
Automatic Extraction of Femur Contours from Hip X-ray Images Ying Chen 1, Xianhe Ee 1, Wee Kheng Leow 1, and Tet Sen Howe 2 1 Dept of Computer Science, National University of Singapore, 3 Science Drive
Image Registration. Prof. Dr. Lucas Ferrari de Oliveira UFPR Informatics Department
 Image Registration Prof. Dr. Lucas Ferrari de Oliveira UFPR Informatics Department Introduction Visualize objects inside the human body Advances in CS methods to diagnosis, treatment planning and medical
Image Registration Prof. Dr. Lucas Ferrari de Oliveira UFPR Informatics Department Introduction Visualize objects inside the human body Advances in CS methods to diagnosis, treatment planning and medical
Basic principles of MR image analysis. Basic principles of MR image analysis. Basic principles of MR image analysis
 Basic principles of MR image analysis Basic principles of MR image analysis Julien Milles Leiden University Medical Center Terminology of fmri Brain extraction Registration Linear registration Non-linear
Basic principles of MR image analysis Basic principles of MR image analysis Julien Milles Leiden University Medical Center Terminology of fmri Brain extraction Registration Linear registration Non-linear
A Navigation System for Minimally Invasive Abdominal Intervention Surgery Robot
 A Navigation System for Minimally Invasive Abdominal Intervention Surgery Robot Weiming ZHAI, Yannan ZHAO and Peifa JIA State Key Laboratory of Intelligent Technology and Systems Tsinghua National Laboratory
A Navigation System for Minimally Invasive Abdominal Intervention Surgery Robot Weiming ZHAI, Yannan ZHAO and Peifa JIA State Key Laboratory of Intelligent Technology and Systems Tsinghua National Laboratory
City, University of London Institutional Repository
 City Research Online City, University of London Institutional Repository Citation: Doan, H., Slabaugh, G.G., Unal, G.B. & Fang, T. (2006). Semi-Automatic 3-D Segmentation of Anatomical Structures of Brain
City Research Online City, University of London Institutional Repository Citation: Doan, H., Slabaugh, G.G., Unal, G.B. & Fang, T. (2006). Semi-Automatic 3-D Segmentation of Anatomical Structures of Brain
Open Topology: A Toolkit for Brain Isosurface Correction
 Open Topology: A Toolkit for Brain Isosurface Correction Sylvain Jaume 1, Patrice Rondao 2, and Benoît Macq 2 1 National Institute of Research in Computer Science and Control, INRIA, France, sylvain@mit.edu,
Open Topology: A Toolkit for Brain Isosurface Correction Sylvain Jaume 1, Patrice Rondao 2, and Benoît Macq 2 1 National Institute of Research in Computer Science and Control, INRIA, France, sylvain@mit.edu,
Nonrigid Registration with Adaptive, Content-Based Filtering of the Deformation Field
 Nonrigid Registration with Adaptive, Content-Based Filtering of the Deformation Field Marius Staring*, Stefan Klein and Josien P.W. Pluim Image Sciences Institute, University Medical Center Utrecht, P.O.
Nonrigid Registration with Adaptive, Content-Based Filtering of the Deformation Field Marius Staring*, Stefan Klein and Josien P.W. Pluim Image Sciences Institute, University Medical Center Utrecht, P.O.
Volume visualization. Volume visualization. Volume visualization methods. Sources of volume visualization. Sources of volume visualization
 Volume visualization Volume visualization Volumes are special cases of scalar data: regular 3D grids of scalars, typically interpreted as density values. Each data value is assumed to describe a cubic
Volume visualization Volume visualization Volumes are special cases of scalar data: regular 3D grids of scalars, typically interpreted as density values. Each data value is assumed to describe a cubic
Medical Image Registration by Maximization of Mutual Information
 Medical Image Registration by Maximization of Mutual Information EE 591 Introduction to Information Theory Instructor Dr. Donald Adjeroh Submitted by Senthil.P.Ramamurthy Damodaraswamy, Umamaheswari Introduction
Medical Image Registration by Maximization of Mutual Information EE 591 Introduction to Information Theory Instructor Dr. Donald Adjeroh Submitted by Senthil.P.Ramamurthy Damodaraswamy, Umamaheswari Introduction
Knowledge-Based Deformable Matching for Pathology Detection
 Knowledge-Based Deformable Matching for Pathology Detection Thesis Proposal Mei Chen CMU-RI-TR-97-20 The Robotics Institute Carnegie Mellon University Pittsburgh, Pennsylvania 15213 May 1997 c 1997 Carnegie
Knowledge-Based Deformable Matching for Pathology Detection Thesis Proposal Mei Chen CMU-RI-TR-97-20 The Robotics Institute Carnegie Mellon University Pittsburgh, Pennsylvania 15213 May 1997 c 1997 Carnegie
A Haptic VR Milling Surgery Simulator Using High-Resolution CT-Data.
 A Haptic VR Milling Surgery Simulator Using High-Resolution CT-Data. Magnus ERIKSSON a, Mark DIXON b and Jan WIKANDER a. a The Mechatronics Lab/Machine Design, KTH, Stockholm, Sweden b SenseGraphics AB,
A Haptic VR Milling Surgery Simulator Using High-Resolution CT-Data. Magnus ERIKSSON a, Mark DIXON b and Jan WIKANDER a. a The Mechatronics Lab/Machine Design, KTH, Stockholm, Sweden b SenseGraphics AB,
Reconstruction of complete 3D object model from multi-view range images.
 Header for SPIE use Reconstruction of complete 3D object model from multi-view range images. Yi-Ping Hung *, Chu-Song Chen, Ing-Bor Hsieh, Chiou-Shann Fuh Institute of Information Science, Academia Sinica,
Header for SPIE use Reconstruction of complete 3D object model from multi-view range images. Yi-Ping Hung *, Chu-Song Chen, Ing-Bor Hsieh, Chiou-Shann Fuh Institute of Information Science, Academia Sinica,
Curvature guided surface registration using level sets
 Curvature guided surface registration using level sets Marcel Lüthi, Thomas Albrecht, Thomas Vetter Department of Computer Science, University of Basel, Switzerland Abstract. We present a new approach
Curvature guided surface registration using level sets Marcel Lüthi, Thomas Albrecht, Thomas Vetter Department of Computer Science, University of Basel, Switzerland Abstract. We present a new approach
Image Registration with Local Rigidity Constraints
 Image Registration with Local Rigidity Constraints Jan Modersitzki Institute of Mathematics, University of Lübeck, Wallstraße 40, D-23560 Lübeck Email: modersitzki@math.uni-luebeck.de Abstract. Registration
Image Registration with Local Rigidity Constraints Jan Modersitzki Institute of Mathematics, University of Lübeck, Wallstraße 40, D-23560 Lübeck Email: modersitzki@math.uni-luebeck.de Abstract. Registration
Volume Illumination and Segmentation
 Volume Illumination and Segmentation Computer Animation and Visualisation Lecture 13 Institute for Perception, Action & Behaviour School of Informatics Overview Volume illumination Segmentation Volume
Volume Illumination and Segmentation Computer Animation and Visualisation Lecture 13 Institute for Perception, Action & Behaviour School of Informatics Overview Volume illumination Segmentation Volume
Fmri Spatial Processing
 Educational Course: Fmri Spatial Processing Ray Razlighi Jun. 8, 2014 Spatial Processing Spatial Re-alignment Geometric distortion correction Spatial Normalization Smoothing Why, When, How, Which Why is
Educational Course: Fmri Spatial Processing Ray Razlighi Jun. 8, 2014 Spatial Processing Spatial Re-alignment Geometric distortion correction Spatial Normalization Smoothing Why, When, How, Which Why is
A MORPHOLOGY-BASED FILTER STRUCTURE FOR EDGE-ENHANCING SMOOTHING
 Proceedings of the 1994 IEEE International Conference on Image Processing (ICIP-94), pp. 530-534. (Austin, Texas, 13-16 November 1994.) A MORPHOLOGY-BASED FILTER STRUCTURE FOR EDGE-ENHANCING SMOOTHING
Proceedings of the 1994 IEEE International Conference on Image Processing (ICIP-94), pp. 530-534. (Austin, Texas, 13-16 November 1994.) A MORPHOLOGY-BASED FILTER STRUCTURE FOR EDGE-ENHANCING SMOOTHING
Using Probability Maps for Multi organ Automatic Segmentation
 Using Probability Maps for Multi organ Automatic Segmentation Ranveer Joyseeree 1,2, Óscar Jiménez del Toro1, and Henning Müller 1,3 1 University of Applied Sciences Western Switzerland (HES SO), Sierre,
Using Probability Maps for Multi organ Automatic Segmentation Ranveer Joyseeree 1,2, Óscar Jiménez del Toro1, and Henning Müller 1,3 1 University of Applied Sciences Western Switzerland (HES SO), Sierre,
A Method for Registering Diffusion Weighted Magnetic Resonance Images
 A Method for Registering Diffusion Weighted Magnetic Resonance Images Xiaodong Tao and James V. Miller GE Research, Niskayuna, New York, USA Abstract. Diffusion weighted magnetic resonance (DWMR or DW)
A Method for Registering Diffusion Weighted Magnetic Resonance Images Xiaodong Tao and James V. Miller GE Research, Niskayuna, New York, USA Abstract. Diffusion weighted magnetic resonance (DWMR or DW)
Anomaly Detection through Registration
 Anomaly Detection through Registration Mei Chen Takeo Kanade Henry A. Rowley Dean Pomerleau meichen@cs.cmu.edu tk@cs.cmu.edu har@cs.cmu.edu pomerlea@cs.cmu.edu School of Computer Science, Carnegie Mellon
Anomaly Detection through Registration Mei Chen Takeo Kanade Henry A. Rowley Dean Pomerleau meichen@cs.cmu.edu tk@cs.cmu.edu har@cs.cmu.edu pomerlea@cs.cmu.edu School of Computer Science, Carnegie Mellon
Using Distance Maps for Accurate Surface Representation in Sampled Volumes
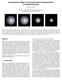 Using Distance Maps for Accurate Surface Representation in Sampled Volumes SarahF.F.Gibson MERL A Mitsubishi Electric Research Laboratory 201 Broadway, Cambridge, MA, 02139 (a) (b) (c) (d) Figure 1: Shaded,
Using Distance Maps for Accurate Surface Representation in Sampled Volumes SarahF.F.Gibson MERL A Mitsubishi Electric Research Laboratory 201 Broadway, Cambridge, MA, 02139 (a) (b) (c) (d) Figure 1: Shaded,
