Visualizing Diffusion Tensor Imaging Data with Merging Ellipsoids
|
|
|
- Luke Johnston
- 5 years ago
- Views:
Transcription
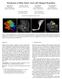
1 Visualizing Diffusion Tensor Imaging Data with Merging Ellipsoids Wei Chen Song Zhang Stephen Correia David F. Tate State Key Lab of CAD&CG, ZJU, China Mississippi State University Brown University Brigham and Women s Hospital Abstract Diffusion tensor fields reveal the underlying anatomical structures in biological tissues such as neural fibers in the brain. Most current methods for visualizing the diffusion tensor field can be categorized into two classes: integral curves and glyphs. Integral curves are continuous and represent the underlying fiber structures, but are prone to integration error and loss of local information. Glyphs are useful for representing local tensor information, but do not convey the connectivity in the anatomical structures well. We introduce a simple yet effective visualization technique that extends the streamball method in flow visualization to tensor ellipsoids. Each tensor ellipsoid represents a local tensor, and either blends with neighboring tensors or breaks away from them depending on their orientations and anisotropies. The resulting visualization shows the connectivity information in the underlying anatomy while characterizing the local tenors in detail. By interactively changing an iso-value parameter, users can examine the diffusion tensor field in the entire spectrum between the continuous integral curves and the discrete glyphs. Expert evaluation indicates that this method conveys very useful visual information about local anisotropy in white matter fibers. Such information was previously unavailable in tractography models. Our method provides a visual tool for assessing variability in DTI fiber tract integrity and its relation to function. Index Terms: Diffusion Tensor Imaging; Visualization; Volume Rendering 1 Introduction Diffusion tensor imaging (DTI) data contains information about water diffusion in biological tissues. The underlying anatomical structures can be retrieved from the diffusion information. Visualizing the diffusion tensor remains challenging due to the complexities of the tensor field and the underlying tensor field. A majority of the previous work on DTI visualization models the diffusion tensor field with either integral curves [2, 21] or glyphs [16, 12, 10, 11]. Integral curves explicitly show the connectivity in the anatomy by integrating over the major eigenvector field of the diffusion tensor field (see Figure 1 ). Integral curve methods, however, have certain limitations. Specifically, considerable information is lost in integral curve methods because the tensor is reduced to a vector. Moreover, integration errors accumulates along the curves. Glyphs show the entire local diffusion tensor information at each glyph location and represent the data faithfully without introducing error. However, impor chenwei@cad.zju.edu.cn szhang@cse.msstate.edu stephen correia@brown.edu dftatephd@mac.com IEEE Pacific Visualization Symposium 2009 April 20-23, Beijing, China /09/$ IEEE tant connectivity information is either lost or hard to interpret from the glyphs. Moreover, the placement of the glyphs can be problematic: if too many glyphs are packed into the visualization volume, visual clutter might prevent users from understanding the inside structures; if too few glyphs are visualized, they will not represent the underlying continuous structures well. Figure 1 shows the ellipsoid glyphs at the sample grid in a diffusion tensor field. Even without the tensors outside the brain, the visualization becomes cluttered by the ellipsoids. Another drawback for glyphs is that they do not represent the physical appearance of the neural fibers and thus require explicit interpretations from users. Figure 1: DTI integral curves methods show the continuous structures such as the white matter fiber tracts well. However, there are integration error hidden in the integral curves and the tensor details are partially lost. Glyphs methods show individual tensors well. However, they do not present the continuous structures and connectivity information well. And they suffer from cluttering if placed on a regular sample grid. We propose a merging ellipsoid method for DTI visualization. We use ellipsoids as the glyphs for diffusion tensors and allow these glyphs to merge or disconnect based on the relative positions of the tensors. Instead of placing the ellipsoids randomly or on a regular grid, we place these ellipsoids on the paths of the integral curves. We choose the integral curves as the sample paths for the ellipsoids because the continuous structures represented by these curves are important and useful for the expert users. Also, the methods used to cull redundant integral curves can be readily transferred to the merging ellipsoids to avoid visual clutters. The downside of the integral curves, namely the lack of detailed tensor information and the integration error, can be compensated by the detailed depiction of the tensors by the merging ellipsoids. The resulting visualization allows users to grasp the connectivity information in the underlying structures as well as examine the local diffusion tensor information in detail. We provide an iso-value parameter for users to interactively change the sizes of the ellipsoids. When ellipsoids merge with neighbors on the integral curves more, a stronger sense of connectivity information is conveyed without sacrificing tensor details. The smaller ellipsoids are more isolated and 145
2 preserve the individual tensor information better while losing some of the connectivity information. By changing the iso-value parameter from the very large to the very small, users can examine both the shapes of the integral curves and the tensor information along the curves with a single view, therefore better understand the nature of these integral curves. For example, an uncertain connectivity in a fiber might be warned by the presence of one or more tensors with rounder shapes (low anisotropy values). We discuss related work in section 2 and describe the method in section 3. We then show the results in section 4. Two neuropsychologists with extensive DTI research experience evaluated our method. Their feedback is documented in section 5. Finally we list our conclusions and future work in section 6. 2 Related Work Numerous methods have been developed in the last decade for DTI visualization [9, 7, 13, 20, 17]. Among them, two classes of methods have relative success in elucidating the underlying anatomical structures. The integral curves method first appeared in Xue et al. [21]. This method reduces the diffusion tensor field to a vector field and traces the streamlines in the vector field. The method is sometimes called fiber tracking [14] or tractography [2]. A main problem of this method is the integration error in the integral curves, especially those running through ambiguous DTI regions (i.e., low linear anisotropy regions). The error will accumulate along the integral curves. Pierpaoli et al. [15] showed a tractography method can produce anatomically incorrect fiber trajectories. Despite these concerns, integral curve methods were useful for showing structures in heart muscle and the cerebral white matter. The resulting fiber structures were shown to be consistent with the known anatomy as validated by histological data [18, 8, 5]. Delmarcelle et al. [6] extend the integral curves method to hyperstreamlines that incorporate more information than a vector from each tensor. However, the hyperstreamlines are sparse in the image space due to their sizes. To place the integral curves appropriately, seeding strategies [22, 19] were designed to prevent visual clutter and generate representative curves. The glyphs method was first applied to DTI data by Pierpaoli et al. [16] using ellipsoids. Laidlaw et al. [12] normalized the ellipsoids to show more in a single image. Kindlmann et al. [10] applied the superquadratics [1] to DTI data to better distinguish between different types of tensors. While many types of glyphs can convey the complete information of a diffusion tensor, the connectivity information in the tensor field is often lost or hard to interpret. Another problem with the glyph method concerns their placement in 3D. Since glyphs need to be at a certain size to be visible, placing a glyph at every data point can block the structures inside while placing them too sparsely only exasperate the loss of connectivity information. Kindlmann et al. [11] densely packed the glyphs into the field using potential energy profiles from the the local tensor values and avoided using the regular grid. Our new method places ellipsoids along the integral curves. It also applies the ideas from streamballs by Brill et al. [4] to the ellipsoids and allows the ellipsoids to merge or disconnect depending on the relationship between the two neighboring tensors. Thus, the merging ellipsoids method preserves both the connectivity information and local tensor details. Moreover, it allows users to judge the uncertainty of the DTI fibers in ambiguous or injured regions by interactively examining the merging ellipsoids. 3 Methods 3.1 Diffusion tensor The diffusion-weighted imaging data was collected from an MRI machine and then fit to a diffusion tensor field. A diffusion tensor is a 3 3 positive definite symmetric matrix: Dxx Dxy Dxz D = D xy D yy D yz (1) D xz D yz D zz The matrix D describes how water molecules propagate assuming free diffusion in a uniform anisotropic medium, such as the coherent fibrous structures in white matter or muscles. D can be decomposed as: D = rλr 1,where e 1, e 2,and e 3 are the three eigenvectors of D, r = ( e 1, e 2, e 3) is a unit rotational matrix, Λ = λ λ 2 0 is the eigenvalue 0 0 λ 3 matrix, with λ 1, λ 2,andλ 3 as the three descending eigenvalues. A diffusion tensor can be represented geometrically by an ellipsoid with their eigenvalues and corresponding eigenvectors mapped to the three axes of the ellipsoid. Thus the diffusion ellipsoid is a natural glyph for the diffusion tensor and not surprisingly one of the first method applied to DTI visualization [16]. 3.2 Ellipsoid placement We place the merging ellipsoids along a set of precomputed integral curves. There are two reasons for choosing this method over other seeding methods such as random seeding, regular grid seeding, and dense packing. First, by placing these ellipsoids along the integral curves, we are able to visualize the inherently continuous structures in the anatomy. Second, we can adjust the density of the ellipsoids by adjusting the distance between the integral curves, taking advantage of the culling process applied in integral curves methods such as [22] or [19]. The set of integral curves were precomputed by integrating in the major eigenvector field of the diffusion tensor field: p(t) = t 0 e 1(p(s))ds (2) where p(t) is the generated curve and e 1 is the major vector field. p(0) is set to the initial point of the integral curve. A dense set of integral curves were initially seeded from every grid point with jittering. These integral curves were thenculledsothatnotwocurveshaveadistancelowerthan a threshold. The distance between the integral curves is defined in [22] as: D t = s1 s 0 s1 s 0 max(dist(s) T t, 0)ds max( dist(s) T t, 0)ds (3) dist(s) T t where s parameterizes the arc length of the shorter curve, s 0 and s 1 are the starting and end points of s, and dist(s) is the shortest distance from location s on the shorter curve to the longer curve. T t ensures that we label two trajectories as different if they differ significantly over any portion of the arc length. The distance between the ellipsoids on the integral curves is set to be the same as the length of the voxel edges, which is 0.85mm for the DTI data used in section
3 3.3 Merging ellipsoids For the sake of simplicity, we adopt the similar notations of [4]. The discrete streamball denotes an isosurface extracted from a scalar influence field F (S, x),whichisgiven as the sum at a given point x of weighted influence functions I i( x) generated by a set of blobby objects: F (S, x) =Σ iω ii i( x) (4) where ω i denotes the strength of the ith influence function. A typical spherical metaball function applies the following polynomial approximation: radius R and the placement of the ellipsoids. The distances among ellipsoids are determined by their placements in associated integral curves, and greatly influence the merging or separation extent. The optimal truncation radius R is computed as the minimum of two distances: R =min(d t,d l 0.7) (7) where D t is the threshold for the ones introduced in Equation 3, and D l denotes the maximum distance between two consecutive tensors in all integral curves. I i( x) = {a f i( x) 6 + b f i( x) 4 + c f i( x) 2 +1, R 6 R 4 R 2 f i( x) R; 0, f i( x) >R. (5) where R is the truncation radius, f i( x) = x s i, and s i is the center of the ith influence function. In our implementation, we use the parameters introduced in [3]: a = 4.0/9.0,b= 17.0/9.0,c= 22.0/9.0. Rather than using the spherical kernel in Equation 5, we represent each diffusion tensor with an elliptical function, yielding: ( e1 ( x si))2 ( e2 ( x si))2 ( e3 ( x si))2 f i( x) = + + (λ 1) 2 (λ 2) 2 (λ 3) 2 (6) Given a list of integral curves, we construct an elliptical function f i( x) for every diffusion tensor on each curve. The threshold on the distance among integral curves introduced in Equation 3 ensures that the influence from one diffusion tensor will be imposed only on its neighboring tensors in its associated integral curve. The sum of all weighted influence functions I i( x) at x, which loops over a regular three-dimensional grid, generates a scalar field F (S, x). By computing an isosurface F (S, x) = C, a global surface representation for the entire diffusion field is constructed. This surface not only depicts the shape and orientation of individual diffusion tensors locally, but also illustrates their connectivity and transition in a smooth and visually plausible fashion. We call this technique merging ellipsoid, as the ellipsoids can blend to others in highly anisotropic regions or disconnect from others in isotropic or isolated regions. Figure 2 compares the effects of the ellipsoid glyph method, the standard streamball method and the merging ellipsoids method for eight diffusion tensors. (e) (f) Figure 3: Visualizing eight diffusion tensors with different iso-values: 0.01, 0.25, 0.51, 0.75, (e) 0.85, (f) The truncation radius R is 1.0. In practice, three eigenvectors of diffusion tensors need to be scaled to the normalized space from the real physical space. This global scaling S t is computed as: S t =2.0 R/r, r =max{r i =min(r c,λ i 1,λ i 2,λ i 3),i=0, 1, 2,...} (8) where λ i 1,λ i 2,λ i 3 are three eigenvectors of the ith diffusion tensor, r c is an adjustable threshold to suppress over-sized tensors. A list of results with different configurations of the truncation radius are compared in Figure 4. A typical choice for R in our experiments ranges from 1.0 to 2.0. Figure 4: The results with different truncation radii: 0.3, 0.5, 1.0. In all cases, the iso-value is 0.5. Figure 2: Visualizing eight diffusion tensors along an integral curve with glyphs, streamballs, and merging ellipsoids (R=1.0). The degree of merging or separation depends on three factors. The first one is the iso-value C that can be adjusted interactively. Its continuous change can reveal the merging process or its reverse. Figure 3 shows a sequence of results with the change of the iso-value. The second and third factors are a pair of complimentary constants: the truncation To summarize, the merging ellipsoids method has the following interesting properties for visualizing a diffusion tensor field: A diffusion tensor produces one elliptical surface, When two diffusion tensors are close, their ellipsoids tend to merge smoothly, If they coincide, a larger ellipsoid is generated, In theory, the entire merging ellipsoid representation is C -continuous, 147
4 If the three eigenvectors are set as identical, our method becomes standard streamball approach [4], If a sequence of ellipsoids are continuously distributed along an integral curve, the hyperstreamline representation [6] is yielded, x) can be extended An individual elliptical function fi ( into other superquadratic functions, yielding the glyph based DTI visualization techniques [10]. 4 Figure 5 shows the merging ellipsoids visualization in a box region of 34mm 34mm 34mm in the corpus callosum area. The fiber paths are generated with the distance between the paths larger than 1.7mm. From the pictures, the anatomical structures such as the corpus callosum and the cingulum bundles are clearly visible. In addition, the individual tensor details and the relationship between the tensors along a path are also revealed by the merging ellipsoids. Results We applied the merging ellipsoids method to the DTI data collected from a normal subject. 12 gradient directions and two b-values 0 and 1000 were used to collect the diffusionweighted images (DWIs) with a 1.5T MRI machine. Diffusion tensor field was then calculated from the DWIs with the in-house software. All experimental results were collected at a laptop equipped with P4 2.2G HZ CPU and 2G host memory. With a given configuration, the scalar field F (S, x) can be precomputed, stored and reloaded for visualization. Its running time depends on the grid resolution and the number of diffusion tensors. In the experiments presented in this paper, the constructions cost range from 15 minutes to 150 minutes with the volume dimension of The visualization of the merging ellipsoids, however, can be performed in real-time. Even when users change the iso-value, the reconstruction of the isosurface can be done in about 0.5 second by means of an unoptimized software implementation. Figure 5: The merging ellipsoids method was applied to the DTI data of a normal subject. The red box indicates the horizontal view of the region-of-interest on the fractional anisotropy map of the brain. The merging ellipsoids capture the connectivity information as well as the tensor details. Sagittal view of the merging ellipsoids. Coronal view of the merging ellipsoids. Figure 6: Comparison of the integral curves method, the simple ellipsoids glyph method, and the merging ellipsoids method in a corpus callosum region of the brain. The red box indicates the horizontal view of the region-of-interest in the brain. The dimension of the region is 17mm 17mm 17mm. The integral curves show the connectivity information but lack the tensor details. These curves also accumulate integration error. Ellipsoid glyphs placed on the integral curves reveal more details in the individual tensors but reduce the connectivity information. The merging ellipsoids capture the connectivity information as well as the tensor details. To illustrate the advantage of the method, Figure 6 shows a smaller box region of 17mm 17mm 17mm and sparser fiber paths visualized by the integral curves, the ellipsoids, and the merging ellipsoids. The distance between the fiber paths is set to be more than 3.4mm in this figure. The integral curves show the connectivity well. But the local tensor details important for users to discern the integrity of a fiber are lost. On the contrary, the ellipsoids method shows the tensor details but does not convey the connectivity information well. The merging ellipsoids combine the advantage of both. This becomes even clearer in Figure 7. Figure 7 illustrates the merging ellipsoids in the same region as Figure 6 with various iso-values. Figure 7,,, and contain merging ellipsoids with decreasing isovalues. The different models visualize the same diffusion tensor field with different emphases on local diffusion tensor information and the global connectivity information. 148
5 Figure 7: The iso-value parameter was adjusted interactively by users to produce a continuous set of merging ellipsoids models that provide information with different emphases on the connectivity information and the tensor details.,,, and were produced by the iso-values set to 0.90, 0.80, 0.60, and The region-of-interest is the same as in Figure 6. Figure 8: The comparison of the three visualization methods in another region-of-interest close to the forceps major. The regionof-interest. The integral curves method. The simple ellipsoid glyph method. The merging ellipsoids method. Figure 8 applies the method for Figure 6 to another region-of-interest closer to the forceps major. Note that the fibers appear to have more isotropic tensors. This may be because the corpus callosum region contains more coherent neural fibers than the forceps major region. The change from high anisotropy region to low anisotropy region on a single fiber can be visualized with the merging ellipsoids better than the other two methods, especially when users change the iso-value interactively. Figure 9 shows that the difference between tensors on a single integral curve can be easily identified with the merging ellipsoids.the difference in the anisotropies is revealed by the shapes of and the blending among the ellipsoids. This helps users to identify the problematic integral curves and the problematic segments on an integral curve. 5 Expert evaluation Two neuropsychologists with extensive experience evaluating white matter DTI tractography metrics examined the results seperately. After examining the merging ellipsoids and working with the visualization tool, they provided the following feedback: This method of visualization is novel yet elegantly simple. The merging ellipsoids provide considerable additional visual information. They permit the user to identify small regions within a single tract that might have areas of low anisotropy. Areas along an integral curve or group of integral curves with low anisotropy could reflect normal anatomy (i.e., crossing fibers) or white matter fibers that have been injured or are at risk of injury. Such visual variability was previously unavailable when selecting tracts of interest in tractography models. This merged presentation of integral curves with diffusion ellipsoids might be most useful as an adjunct to more conventional DTI tract selection. For example, initial tract selection might be performed using the streamtube model [22] and traditional voxel-of-interest or or other selection methods. Once a tract-of-interest is selected, turning on the merging ellipsoid visualization would permit users to refine the selection to target areas of low or high anisotropy. The merging ellipsoids method advances the clinical utility of DTI by improving researcher s ability to discern disease and injury specific tract variability. It has potential to reveal associations that might have otherwise be masked in conventional tract selection methods because of the inclusion of both normal and abnormal fibers. In traditional tractography models, a minimal threshold for linear or fractional anisotropy is set for generating fibers. Fibers will be generated as long as this threshold is met. This approach, however, potentially masks variability in the integrity of the fibers that are rendered, such that in two individuals, a fiber bundle (e.g., cingulum) might appear similar in terms of length and number of fibers even though its underlying structural integrity might actually be poorer in one compared to the other individual. The merging ellipsoids method also allows researchers to identify a small number of fibers at risk (assuming low anisotropy is not due to fiber crossing) within what appears to be an otherwise coherent bundle and test whether or not these at risk fibers have a disproportionate impact on cognition or behavior an association that would otherwise be visually masked by inclusion of the larger set of normal fibers in the tract. 149
6 References Figure 9: This picture shows the difference in tensors along an integral curve. The blue arrow points to the region where the tensors are more anisotropic and well aligned while the red arrow points to the region with more isotropy and uncertainty. 6 Conclusions and future work We propose a new method for visualizing DTI data that merges tensor ellipsoids with integral curves. We use ellipsoids as the glyphs for diffusion tensors and allow these glyphs to merge or disconnect based on the underlying data and a user-controlled visualization parameter. Instead of placing the ellipsoids on a regular grid or randomly, we place these ellipsoids on the paths of the integral curves. User evaluation showed that this method has the advantage over the integral curves methods in that it shows the local tensor details as well as the connectivity information. Our method is especially helpful for representing the ambiguity in the regions where linear anisotropy is lower. By controlling the iso-value parameter, users can see the entire spectrum of visual models from the discrete glyphs to continuous curves. The added interactivity allows the user to gather a more complete understanding of the information in the DTI data. This method has the potential to bridge the two main categories of the DTI visualization methods into a single visualization framework and help elucidate the complicated structures and uncertainty in the DTI data. In future work, we will employ adaptive volume grid to accelerate the computation of the scalar field. Recent progress in programmable graphics hardware has enabled the fast implementation of dynamic branching in GPU. Another interesting goal is to extend the proposed scheme to streamsurface representation for DTI data. We plan to apply this technique to a white matter disease such as the vascular cognitive impairment and further evaluate the effectiveness of this method in discerning the details in the fiber integrity. 7 Acknowledgement This work is partially supported by NSF of China (No ), the Research Initiation Program at Mississippi State University. [1] A. Barr. Superquadrics and angle-preserving transformations. IEEE Computer Graphics and Applications, 1(1):11 23, [2] P. J. Basser, S. Pajevic, C. Pierpaoli, J. Duda, and A. Aldroubi. In vivo fiber tractography using DT-MRI data. Magnetic Resonance in Medicine, 44: , [3] J. Blinn. A generalization of algebraic surface drawing. ACM Transactions on Graphics, 1(3): , [4] M. Brill, H. Hagen, H.-C. Rodrian, W. Djatschin, and S. V. Klimenko. Streamball techniques for flow visualization. In VIS 94: Proceedings of the conference on Visualization 94, pages , Los Alamitos, CA, USA, IEEE Computer Society Press. [5] J. Dauguet, S. Peled, V. Berezovskii, T. Delzescaux, S. K. Warfield, R. Born, and C.-F. Westin. Comparison of fiber tracts derived from in-vivo DTI tractography with 3d histological neural tract tracer reconstruction on a macaque brain. NeuroImage, 37: , [6] T. Delmarcelle and L. Hesselink. Visualizing second-order tensor fields with hyperstream lines. IEEE Computer Graphics and Applications, 13(4):25 33, jul [7] F. Enders, N. Sauber, D. Merhof, P. Hastreiter, C. Nimsky, and M. Stamminger. Visualization of white matter tracts with wrapped streamlines. In Proceedings of IEEE Visualization 2005, pages 51 58, [8] E. Hsu, A. Muzikant, S. Matulevicius, R. Penland, and C. Henriquez. Magnetic resonance myocardial fiberorientation mapping with direct histological correlation. American Journal of Physiology, 274: , [9] C. R. Johnson and C. D. Hansen. Visualization Handbook. Academic Press, June [10] G. Kindlmann. Superquadric tensor glyphs. In Proceeding of The Joint Eurographics - IEEE TCVG Symposium on Visualization, pages , May [11] G. Kindlmann and C.-F. Westin. Diffusion tensor visualization with glyph packing. IEEE Transactions on Visualization and Computer Graphics, 12(5): , [12] D. H. Laidlaw, E. T. Ahrens, davidkremers, M. J. Avalos, C. Readhead, and R. E. Jacobs. Visualizing diffusion tensor images of the mouse spinal cord. In Proceedings of IEEE Visualization 1998, pages IEEE Computer Society Press, October [13] B. Moberts, A. Vilanova, and J. J. van Wijk. Evaluation of fiber clustering methods for diffusion tensor imaging. In Proceedings of IEEE Visualization, pages 65 72, [14] S. Mori and P. van Zijl. Fiber tracking: principles and strategies a technical review. NMR in Biomedicine, 15(7-8): , [15] C. Pierpaoli, A. Barnett, S. Pajevic, A. Virta, and P. Basser. Validation of DT-MRI tractography in the descending motor pathways of human subjects. In International Society for Magnetic Resonance in Medicine (ISMRM), page 501, Glasgow, [16] C. Pierpaoli and P. Basser. Toward a quantitative assessment of diffusion anisotropy. Magnetic Resonance in Medicine, 36(6): , December [17] B. Preim and D. Bartz. Visualization in Medicine. Morgan Kaufmann, [18] D. Scollan, A. Holmes, R. Winslow, and J. Forder. Histological validation of myocardial microstructure obtained from diffusion tensor magnetic resonance imaging. American Journal of Physiology, 275: , [19] A. Vilanova, G. Berenschot, and C. van Pul. DTI visualization with streamsurfaces and evenly-spaced volume seeding. In VisSym 04 Joint Eurographics - I.E.E.E. T.C.V.G. Symposium on Visualization, Conference Proceedings, pages , [20] J. Weickert and H. Hagen. Visualization and Processing of 150
7 Tensor Fields. Springer, [21] R. Xue, P. C. van Zijl, B. J. Crain, M. Solaiyappan, and S. Mori. In vivo three-dimensional reconstruction of rat brain axonal projections by diffusion tensor imaging. Magnetic Resonance in Medicine, 42: , [22] S. Zhang, C. Demiralp, and D. H. Laidlaw. Visualizing diffusion tensor MR images using streamtubes and streamsurfaces. IEEE Transactions on Visualization and Computer Graphics, 9(4): ,
Diffusion Imaging Visualization
 Diffusion Imaging Visualization Thomas Schultz URL: http://cg.cs.uni-bonn.de/schultz/ E-Mail: schultz@cs.uni-bonn.de 1 Outline Introduction to Diffusion Imaging Basic techniques Glyph-based Visualization
Diffusion Imaging Visualization Thomas Schultz URL: http://cg.cs.uni-bonn.de/schultz/ E-Mail: schultz@cs.uni-bonn.de 1 Outline Introduction to Diffusion Imaging Basic techniques Glyph-based Visualization
Evaluation of Local Filter Approaches for Diffusion Tensor based Fiber Tracking
 Evaluation of Local Filter Approaches for Diffusion Tensor based Fiber Tracking D. Merhof 1, M. Buchfelder 2, C. Nimsky 3 1 Visual Computing, University of Konstanz, Konstanz 2 Department of Neurosurgery,
Evaluation of Local Filter Approaches for Diffusion Tensor based Fiber Tracking D. Merhof 1, M. Buchfelder 2, C. Nimsky 3 1 Visual Computing, University of Konstanz, Konstanz 2 Department of Neurosurgery,
Fiber Selection from Diffusion Tensor Data based on Boolean Operators
 Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
The Application of GPU Particle Tracing to Diffusion Tensor Field Visualization
 The Application of GPU Particle Tracing to Diffusion Tensor Field Visualization Polina Kondratieva, Jens Krüger, Rüdiger Westermann Computer Graphics and Visualization Group Technische Universität München
The Application of GPU Particle Tracing to Diffusion Tensor Field Visualization Polina Kondratieva, Jens Krüger, Rüdiger Westermann Computer Graphics and Visualization Group Technische Universität München
HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
Fiber Selection from Diffusion Tensor Data based on Boolean Operators
 Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhofl, G. Greiner 2, M. Buchfelder 3, C. Nimsky4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhofl, G. Greiner 2, M. Buchfelder 3, C. Nimsky4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
8. Tensor Field Visualization
 8. Tensor Field Visualization Tensor: extension of concept of scalar and vector Tensor data for a tensor of level k is given by t i1,i2,,ik (x 1,,x n ) Second-order tensor often represented by matrix Examples:
8. Tensor Field Visualization Tensor: extension of concept of scalar and vector Tensor data for a tensor of level k is given by t i1,i2,,ik (x 1,,x n ) Second-order tensor often represented by matrix Examples:
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes
 Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
Complex Fiber Visualization
 Annales Mathematicae et Informaticae 34 (2007) pp. 103 109 http://www.ektf.hu/tanszek/matematika/ami Complex Fiber Visualization Henrietta Tomán a, Róbert Tornai b, Marianna Zichar c a Department of Computer
Annales Mathematicae et Informaticae 34 (2007) pp. 103 109 http://www.ektf.hu/tanszek/matematika/ami Complex Fiber Visualization Henrietta Tomán a, Róbert Tornai b, Marianna Zichar c a Department of Computer
Technische Universiteit Eindhoven Department of Mathematics and Computer Science. Master s Thesis HIERARCHICAL VISUALIZATION USING FIBER CLUSTERING
 Technische Universiteit Eindhoven Department of Mathematics and Computer Science Master s Thesis HIERARCHICAL VISUALIZATION USING FIBER CLUSTERING by Ing. B. Moberts Supervisors: Dr. A. Vilanova Prof.dr.ir.
Technische Universiteit Eindhoven Department of Mathematics and Computer Science Master s Thesis HIERARCHICAL VISUALIZATION USING FIBER CLUSTERING by Ing. B. Moberts Supervisors: Dr. A. Vilanova Prof.dr.ir.
Automatic Quantification of DTI Parameters along Fiber Bundles
 Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein 1, Simon Hermann 1, Olaf Konrad 1, Horst K. Hahn 1, and Heinz-Otto Peitgen 1 1 MeVis Research, 28359 Bremen Email: klein@mevis.de
Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein 1, Simon Hermann 1, Olaf Konrad 1, Horst K. Hahn 1, and Heinz-Otto Peitgen 1 1 MeVis Research, 28359 Bremen Email: klein@mevis.de
NEURO M203 & BIOMED M263 WINTER 2014
 NEURO M203 & BIOMED M263 WINTER 2014 MRI Lab 2: Neuroimaging Connectivity Lab In today s lab we will work with sample diffusion imaging data and the group averaged fmri data collected during your scanning
NEURO M203 & BIOMED M263 WINTER 2014 MRI Lab 2: Neuroimaging Connectivity Lab In today s lab we will work with sample diffusion imaging data and the group averaged fmri data collected during your scanning
Automatic Quantification of DTI Parameters along Fiber Bundles
 Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein, Simon Hermann, Olaf Konrad, Horst K. Hahn, Heinz-Otto Peitgen MeVis Research, 28359 Bremen Email: klein@mevis.de Abstract. We introduce
Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein, Simon Hermann, Olaf Konrad, Horst K. Hahn, Heinz-Otto Peitgen MeVis Research, 28359 Bremen Email: klein@mevis.de Abstract. We introduce
A Ray-based Approach for Boundary Estimation of Fiber Bundles Derived from Diffusion Tensor Imaging
 A Ray-based Approach for Boundary Estimation of Fiber Bundles Derived from Diffusion Tensor Imaging M. H. A. Bauer 1,3, S. Barbieri 2, J. Klein 2, J. Egger 1,3, D. Kuhnt 1, B. Freisleben 3, H.-K. Hahn
A Ray-based Approach for Boundary Estimation of Fiber Bundles Derived from Diffusion Tensor Imaging M. H. A. Bauer 1,3, S. Barbieri 2, J. Klein 2, J. Egger 1,3, D. Kuhnt 1, B. Freisleben 3, H.-K. Hahn
Diffusion model fitting and tractography: A primer
 Diffusion model fitting and tractography: A primer Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 03/18/10 Why n how Diffusion model fitting and tractography 0/18 Why
Diffusion model fitting and tractography: A primer Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 03/18/10 Why n how Diffusion model fitting and tractography 0/18 Why
An Immersive Virtual Environment for DT-MRI Volume Visualization Applications: a Case Study
 An Immersive Virtual Environment for DT-MRI Volume Visualization Applications: a Case Study S. Zhang, Ç. Demiralp,D.F.Keefe M. DaSilva, D. H. Laidlaw, B. D. Greenberg Brown University P.J. Basser C. Pierpaoli
An Immersive Virtual Environment for DT-MRI Volume Visualization Applications: a Case Study S. Zhang, Ç. Demiralp,D.F.Keefe M. DaSilva, D. H. Laidlaw, B. D. Greenberg Brown University P.J. Basser C. Pierpaoli
Glyph-based Visualization Applications. David H. S. Chung Swansea University
 Glyph-based Applications David H. S. Chung Swansea University Outline Glyph Design Application Flow Event Multi-field Uncertainty Geo-spatial Tensor Medical Outline Glyph Design Application Novel shape
Glyph-based Applications David H. S. Chung Swansea University Outline Glyph Design Application Flow Event Multi-field Uncertainty Geo-spatial Tensor Medical Outline Glyph Design Application Novel shape
An Introduction to Visualization of Diffusion Tensor Imaging and its Applications
 An Introduction to Visualization of Diffusion Tensor Imaging and its Applications A. Vilanova 1, S. Zhang 2, G. Kindlmann 3, and D. Laidlaw 2 1 Department of Biomedical Engineering Eindhoven University
An Introduction to Visualization of Diffusion Tensor Imaging and its Applications A. Vilanova 1, S. Zhang 2, G. Kindlmann 3, and D. Laidlaw 2 1 Department of Biomedical Engineering Eindhoven University
Identifying white-matter fiber bundles in DTI data using an automated proximity-based fiber-clustering method
 1 Identifying white-matter fiber bundles in DTI data using an automated proximity-based fiber-clustering method Song Zhang 1, Stephen Correia 2, and David H. Laidlaw 3 1 Department of Computer Science
1 Identifying white-matter fiber bundles in DTI data using an automated proximity-based fiber-clustering method Song Zhang 1, Stephen Correia 2, and David H. Laidlaw 3 1 Department of Computer Science
Neural Network-Assisted Fiber Tracking of Synthetic and White Matter DT-MR Images
 Neural Network-Assisted Fiber Tracking of Synthetic and White Matter DT-MR Images L.M. San-José-Revuelta, M. Martín-Fernández and C. Alberola-López Abstract In this paper, a recently developed fiber tracking
Neural Network-Assisted Fiber Tracking of Synthetic and White Matter DT-MR Images L.M. San-José-Revuelta, M. Martín-Fernández and C. Alberola-López Abstract In this paper, a recently developed fiber tracking
Saturn User Manual. Rubén Cárdenes. 29th January 2010 Image Processing Laboratory, University of Valladolid. Abstract
 Saturn User Manual Rubén Cárdenes 29th January 2010 Image Processing Laboratory, University of Valladolid Abstract Saturn is a software package for DTI processing and visualization, provided with a graphic
Saturn User Manual Rubén Cárdenes 29th January 2010 Image Processing Laboratory, University of Valladolid Abstract Saturn is a software package for DTI processing and visualization, provided with a graphic
Blood Particle Trajectories in Phase-Contrast-MRI as Minimal Paths Computed with Anisotropic Fast Marching
 Blood Particle Trajectories in Phase-Contrast-MRI as Minimal Paths Computed with Anisotropic Fast Marching Michael Schwenke 1, Anja Hennemuth 1, Bernd Fischer 2, Ola Friman 1 1 Fraunhofer MEVIS, Institute
Blood Particle Trajectories in Phase-Contrast-MRI as Minimal Paths Computed with Anisotropic Fast Marching Michael Schwenke 1, Anja Hennemuth 1, Bernd Fischer 2, Ola Friman 1 1 Fraunhofer MEVIS, Institute
Coloring of DT-MRI Fiber Traces using Laplacian Eigenmaps
 Coloring of DT-MRI Fiber Traces using Laplacian Eigenmaps Anders Brun 1, Hae-Jeong Park 2, Hans Knutsson 3, and Carl-Fredrik Westin 1 1 Laboratory of Mathematics in Imaging, Brigham and Women s Hospital,
Coloring of DT-MRI Fiber Traces using Laplacian Eigenmaps Anders Brun 1, Hae-Jeong Park 2, Hans Knutsson 3, and Carl-Fredrik Westin 1 1 Laboratory of Mathematics in Imaging, Brigham and Women s Hospital,
Multimodal Visualization of DTI and fmri Data using Illustrative Methods
 Multimodal Visualization of DTI and fmri Data using Illustrative Methods Silvia Born 1, Werner Jainek 1, Mario Hlawitschka 2, Gerik Scheuermann 3, Christos Trantakis 4, Jürgen Meixensberger 4, Dirk Bartz
Multimodal Visualization of DTI and fmri Data using Illustrative Methods Silvia Born 1, Werner Jainek 1, Mario Hlawitschka 2, Gerik Scheuermann 3, Christos Trantakis 4, Jürgen Meixensberger 4, Dirk Bartz
Constrained Reconstruction of Sparse Cardiac MR DTI Data
 Constrained Reconstruction of Sparse Cardiac MR DTI Data Ganesh Adluru 1,3, Edward Hsu, and Edward V.R. DiBella,3 1 Electrical and Computer Engineering department, 50 S. Central Campus Dr., MEB, University
Constrained Reconstruction of Sparse Cardiac MR DTI Data Ganesh Adluru 1,3, Edward Hsu, and Edward V.R. DiBella,3 1 Electrical and Computer Engineering department, 50 S. Central Campus Dr., MEB, University
Quantitative MRI of the Brain: Investigation of Cerebral Gray and White Matter Diseases
 Quantities Measured by MR - Quantitative MRI of the Brain: Investigation of Cerebral Gray and White Matter Diseases Static parameters (influenced by molecular environment): T, T* (transverse relaxation)
Quantities Measured by MR - Quantitative MRI of the Brain: Investigation of Cerebral Gray and White Matter Diseases Static parameters (influenced by molecular environment): T, T* (transverse relaxation)
A Novel Interface for Interactive Exploration of DTI Fibers
 A Novel Interface for Interactive Exploration of DTI Fibers Wei Chen, Member, IEEE, Zi ang Ding, Song Zhang, Member, IEEE, Anna MacKay-Brandt, Stephen Correia, Huamin Qu, Member, IEEE, John Allen Crow,
A Novel Interface for Interactive Exploration of DTI Fibers Wei Chen, Member, IEEE, Zi ang Ding, Song Zhang, Member, IEEE, Anna MacKay-Brandt, Stephen Correia, Huamin Qu, Member, IEEE, John Allen Crow,
FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING. Francesca Pizzorni Ferrarese
 FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING Francesca Pizzorni Ferrarese Pipeline overview WM and GM Segmentation Registration Data reconstruction Tractography
FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING Francesca Pizzorni Ferrarese Pipeline overview WM and GM Segmentation Registration Data reconstruction Tractography
n o r d i c B r a i n E x Tutorial DTI Module
 m a k i n g f u n c t i o n a l M R I e a s y n o r d i c B r a i n E x Tutorial DTI Module Please note that this tutorial is for the latest released nordicbrainex. If you are using an older version please
m a k i n g f u n c t i o n a l M R I e a s y n o r d i c B r a i n E x Tutorial DTI Module Please note that this tutorial is for the latest released nordicbrainex. If you are using an older version please
Similarity Coloring of DTI Fiber Tracts
 Similarity Coloring of DTI Fiber Tracts Çağatay Demiralp and David H. Laidlaw Department of Computer Science, Brown University, USA. Abstract. We present a coloring method that conveys spatial relations
Similarity Coloring of DTI Fiber Tracts Çağatay Demiralp and David H. Laidlaw Department of Computer Science, Brown University, USA. Abstract. We present a coloring method that conveys spatial relations
ECE1778 Final Report MRI Visualizer
 ECE1778 Final Report MRI Visualizer David Qixiang Chen Alex Rodionov Word Count: 2408 Introduction We aim to develop a mobile phone/tablet based neurosurgical MRI visualization application with the goal
ECE1778 Final Report MRI Visualizer David Qixiang Chen Alex Rodionov Word Count: 2408 Introduction We aim to develop a mobile phone/tablet based neurosurgical MRI visualization application with the goal
Direct Registration of White Matter Tractographies with Application to Atlas Construction
 Direct Registration of White Matter Tractographies with Application to Atlas Construction Arnaldo Mayer, Hayit Greenspan Medical image processing lab, Tel-Aviv University, Ramat-Aviv, Israel arnakdom@post.tau.ac.il,
Direct Registration of White Matter Tractographies with Application to Atlas Construction Arnaldo Mayer, Hayit Greenspan Medical image processing lab, Tel-Aviv University, Ramat-Aviv, Israel arnakdom@post.tau.ac.il,
HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
Fiber Tract Clustering on Manifolds With Dual Rooted-Graphs
 Fiber Tract Clustering on Manifolds With Dual Rooted-Graphs Andy Tsai 1,2 Carl-Fredrik Westin 1 Alfred O. Hero III 3 Alan S. Willsky 2 1 Dept. of Radiology 2 Dept. of Electrical Eng. 3 Dept. of Electrical
Fiber Tract Clustering on Manifolds With Dual Rooted-Graphs Andy Tsai 1,2 Carl-Fredrik Westin 1 Alfred O. Hero III 3 Alan S. Willsky 2 1 Dept. of Radiology 2 Dept. of Electrical Eng. 3 Dept. of Electrical
Reconstruction of Fiber Trajectories via Population-Based Estimation of Local Orientations
 IDEA Reconstruction of Fiber Trajectories via Population-Based Estimation of Local Orientations Pew-Thian Yap, John H. Gilmore, Weili Lin, Dinggang Shen Email: ptyap@med.unc.edu 2011-09-21 Poster: P2-46-
IDEA Reconstruction of Fiber Trajectories via Population-Based Estimation of Local Orientations Pew-Thian Yap, John H. Gilmore, Weili Lin, Dinggang Shen Email: ptyap@med.unc.edu 2011-09-21 Poster: P2-46-
A GA-based Approach for Parameter Estimation in DT-MRI Tracking Algorithms
 A GA-based Approach for Parameter Estimation in DT-MRI Tracking Algorithms L.M. San-José-Revuelta, M. Martín-Fernández and C. Alberola-López Abstract This paper expands upon previous work of the authors
A GA-based Approach for Parameter Estimation in DT-MRI Tracking Algorithms L.M. San-José-Revuelta, M. Martín-Fernández and C. Alberola-López Abstract This paper expands upon previous work of the authors
Generating Fiber Crossing Phantoms Out of Experimental DWIs
 Generating Fiber Crossing Phantoms Out of Experimental DWIs Matthan Caan 1,2, Anne Willem de Vries 2, Ganesh Khedoe 2,ErikAkkerman 1, Lucas van Vliet 2, Kees Grimbergen 1, and Frans Vos 1,2 1 Department
Generating Fiber Crossing Phantoms Out of Experimental DWIs Matthan Caan 1,2, Anne Willem de Vries 2, Ganesh Khedoe 2,ErikAkkerman 1, Lucas van Vliet 2, Kees Grimbergen 1, and Frans Vos 1,2 1 Department
DIFFUSION TENSOR IMAGING ANALYSIS. Using Analyze
 DIFFUSION TENSOR IMAGING ANALYSIS Using Analyze 2 Table of Contents 1. Introduction page 3 2. Loading DTI Data page 4 3. Computing DTI Maps page 5 4. Defining ROIs for Fiber Tracking page 6 5. Visualizing
DIFFUSION TENSOR IMAGING ANALYSIS Using Analyze 2 Table of Contents 1. Introduction page 3 2. Loading DTI Data page 4 3. Computing DTI Maps page 5 4. Defining ROIs for Fiber Tracking page 6 5. Visualizing
Visualization of DTI fibers using hair-rendering techniques Peeters, T.H.J.M.; Vilanova Bartroli, A.; ter Haar Romenij, B.M.
 Visualization of DTI fibers using hair-rendering techniques Peeters, T.H.J.M.; Vilanova Bartroli, A.; ter Haar Romenij, B.M. Published in: Proceedings of the twelfth annual conference of the Advanced School
Visualization of DTI fibers using hair-rendering techniques Peeters, T.H.J.M.; Vilanova Bartroli, A.; ter Haar Romenij, B.M. Published in: Proceedings of the twelfth annual conference of the Advanced School
Flow Visualisation - Background. CITS4241 Visualisation Lectures 20 and 21
 CITS4241 Visualisation Lectures 20 and 21 Flow Visualisation Flow visualisation is important in both science and engineering From a "theoretical" study of o turbulence or o a fusion reactor plasma, to
CITS4241 Visualisation Lectures 20 and 21 Flow Visualisation Flow visualisation is important in both science and engineering From a "theoretical" study of o turbulence or o a fusion reactor plasma, to
syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets
 syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets Julien Gervais; Lisa Chuah Siemens Healthcare, Magnetic
syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets Julien Gervais; Lisa Chuah Siemens Healthcare, Magnetic
Modeling and Illustration of Muscle from Diffusion Tensor Images
 Online Submission ID: 88 Modeling and Illustration of Muscle from Diffusion Tensor Images Category: Research Abstract Medical illustration has demonstrated its effectiveness to depict salient anatomical
Online Submission ID: 88 Modeling and Illustration of Muscle from Diffusion Tensor Images Category: Research Abstract Medical illustration has demonstrated its effectiveness to depict salient anatomical
Interactive Volume Rendering of Thin Thread Structures within Multivalued Scientific Datasets
 SUBMITTED TO IEEE TVCG, APRIL, 2003 100 Interactive Volume Rendering of Thin Thread Structures within Multivalued Scientific Datasets Andreas Wenger, Daniel F. Keefe, Song Zhang, David H. Laidlaw Abstract
SUBMITTED TO IEEE TVCG, APRIL, 2003 100 Interactive Volume Rendering of Thin Thread Structures within Multivalued Scientific Datasets Andreas Wenger, Daniel F. Keefe, Song Zhang, David H. Laidlaw Abstract
Atelier 2 : Calcul Haute Performance et Sciences du Vivant Forum er juillet, Paris, France
 From Diffusion MR Image Analysis to Whole Brain Connectivity Simulation Jean-Philippe Thiran EPFL Lausanne, Switzerland EPFL - Lausanne HPC in life sciences at EPFL The Blue Brain project: create a biologically
From Diffusion MR Image Analysis to Whole Brain Connectivity Simulation Jean-Philippe Thiran EPFL Lausanne, Switzerland EPFL - Lausanne HPC in life sciences at EPFL The Blue Brain project: create a biologically
Advanced MRI Techniques (and Applications)
 Advanced MRI Techniques (and Applications) Jeffry R. Alger, PhD Department of Neurology Ahmanson-Lovelace Brain Mapping Center Brain Research Institute Jonsson Comprehensive Cancer Center University of
Advanced MRI Techniques (and Applications) Jeffry R. Alger, PhD Department of Neurology Ahmanson-Lovelace Brain Mapping Center Brain Research Institute Jonsson Comprehensive Cancer Center University of
A method of visualization of a brain neural pathway by using critical points and target regions
 Modelling in Medicine and Biology VII 309 A method of visualization of a brain neural pathway by using critical points and target regions A. Doi 1, H. Fujimura 2, M. Nagano 1, T. Inoue 3 & A. Ogawa 3 1
Modelling in Medicine and Biology VII 309 A method of visualization of a brain neural pathway by using critical points and target regions A. Doi 1, H. Fujimura 2, M. Nagano 1, T. Inoue 3 & A. Ogawa 3 1
Dense Glyph Sampling for Visualization
 Dense Glyph Sampling for Visualization Louis Feng 1, Ingrid Hotz 2, Bernd Hamann 1, and Kenneth Joy 1 1 Institute for Data Analysis and Visualization (IDAV), Department of Computer Science, University
Dense Glyph Sampling for Visualization Louis Feng 1, Ingrid Hotz 2, Bernd Hamann 1, and Kenneth Joy 1 1 Institute for Data Analysis and Visualization (IDAV), Department of Computer Science, University
ISMI: A classification index for High Angular Resolution Diffusion Imaging
 ISMI: A classification index for High Angular Resolution Diffusion Imaging D. Röttger, D. Dudai D. Merhof and S. Müller Institute for Computational Visualistics, University of Koblenz-Landau, Germany Visual
ISMI: A classification index for High Angular Resolution Diffusion Imaging D. Röttger, D. Dudai D. Merhof and S. Müller Institute for Computational Visualistics, University of Koblenz-Landau, Germany Visual
A DT-MRI Validation Framework Using Fluoro Data
 A DT-MRI Validation Framework Using Fluoro Data Seniha Esen Yuksel December 14, 2006 Abstract Most of the previous efforts on enhancing the DT-MRI estimation/smoothing have been based on what is assumed
A DT-MRI Validation Framework Using Fluoro Data Seniha Esen Yuksel December 14, 2006 Abstract Most of the previous efforts on enhancing the DT-MRI estimation/smoothing have been based on what is assumed
Advanced Visual Medicine: Techniques for Visual Exploration & Analysis
 Advanced Visual Medicine: Techniques for Visual Exploration & Analysis Interactive Visualization of Multimodal Volume Data for Neurosurgical Planning Felix Ritter, MeVis Research Bremen Multimodal Neurosurgical
Advanced Visual Medicine: Techniques for Visual Exploration & Analysis Interactive Visualization of Multimodal Volume Data for Neurosurgical Planning Felix Ritter, MeVis Research Bremen Multimodal Neurosurgical
Computational Medical Imaging Analysis Chapter 4: Image Visualization
 Computational Medical Imaging Analysis Chapter 4: Image Visualization Jun Zhang Laboratory for Computational Medical Imaging & Data Analysis Department of Computer Science University of Kentucky Lexington,
Computational Medical Imaging Analysis Chapter 4: Image Visualization Jun Zhang Laboratory for Computational Medical Imaging & Data Analysis Department of Computer Science University of Kentucky Lexington,
Diffusion Tensor Imaging and Reading Development
 Diffusion Tensor Imaging and Reading Development Bob Dougherty Stanford Institute for Reading and Learning Reading and Anatomy Every brain is different... Not all brains optimized for highly proficient
Diffusion Tensor Imaging and Reading Development Bob Dougherty Stanford Institute for Reading and Learning Reading and Anatomy Every brain is different... Not all brains optimized for highly proficient
A Stochastic Tractography System and Applications. Tri M. Ngo
 A Stochastic Tractography System and Applications by Tri M. Ngo Submitted to the Department of Electrical Engineering and Computer Science in partial fulfillment of the requirements for the degree of Master
A Stochastic Tractography System and Applications by Tri M. Ngo Submitted to the Department of Electrical Engineering and Computer Science in partial fulfillment of the requirements for the degree of Master
On Classifying Disease-Induced Patterns in the Brain Using Diffusion Tensor Images
 On Classifying Disease-Induced Patterns in the Brain Using Diffusion Tensor Images Peng Wang 1,2 and Ragini Verma 1 1 Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania,
On Classifying Disease-Induced Patterns in the Brain Using Diffusion Tensor Images Peng Wang 1,2 and Ragini Verma 1 1 Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania,
Chapter 4. Clustering Core Atoms by Location
 Chapter 4. Clustering Core Atoms by Location In this chapter, a process for sampling core atoms in space is developed, so that the analytic techniques in section 3C can be applied to local collections
Chapter 4. Clustering Core Atoms by Location In this chapter, a process for sampling core atoms in space is developed, so that the analytic techniques in section 3C can be applied to local collections
MITK-DI. A new Diffusion Imaging Component for MITK. Klaus Fritzsche, Hans-Peter Meinzer
 MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
Diffusion Imaging Models 1: from DTI to HARDI models
 Diffusion Imaging Models 1: from DTI to HARDI models Flavio Dell Acqua, PhD. www.natbrainlab.com flavio.dellacqua@kcl.ac.uk @flaviodellacqua Diffusion Tensor Imaging (DTI) z λ 1 λ 2 The profile of the
Diffusion Imaging Models 1: from DTI to HARDI models Flavio Dell Acqua, PhD. www.natbrainlab.com flavio.dellacqua@kcl.ac.uk @flaviodellacqua Diffusion Tensor Imaging (DTI) z λ 1 λ 2 The profile of the
The organization of the human cerebral cortex estimated by intrinsic functional connectivity
 1 The organization of the human cerebral cortex estimated by intrinsic functional connectivity Journal: Journal of Neurophysiology Author: B. T. Thomas Yeo, et al Link: https://www.ncbi.nlm.nih.gov/pubmed/21653723
1 The organization of the human cerebral cortex estimated by intrinsic functional connectivity Journal: Journal of Neurophysiology Author: B. T. Thomas Yeo, et al Link: https://www.ncbi.nlm.nih.gov/pubmed/21653723
TRACULA: Troubleshooting, visualization, and group analysis
 TRACULA: Troubleshooting, visualization, and group analysis Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 18/11/13 TRACULA: troubleshooting, visualization, group analysis
TRACULA: Troubleshooting, visualization, and group analysis Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 18/11/13 TRACULA: troubleshooting, visualization, group analysis
Fairing Scalar Fields by Variational Modeling of Contours
 Fairing Scalar Fields by Variational Modeling of Contours Martin Bertram University of Kaiserslautern, Germany Abstract Volume rendering and isosurface extraction from three-dimensional scalar fields are
Fairing Scalar Fields by Variational Modeling of Contours Martin Bertram University of Kaiserslautern, Germany Abstract Volume rendering and isosurface extraction from three-dimensional scalar fields are
A Method for Registering Diffusion Weighted Magnetic Resonance Images
 A Method for Registering Diffusion Weighted Magnetic Resonance Images Xiaodong Tao and James V. Miller GE Research, Niskayuna, New York, USA Abstract. Diffusion weighted magnetic resonance (DWMR or DW)
A Method for Registering Diffusion Weighted Magnetic Resonance Images Xiaodong Tao and James V. Miller GE Research, Niskayuna, New York, USA Abstract. Diffusion weighted magnetic resonance (DWMR or DW)
Visualization of White Matter Tracts with Wrapped Streamlines
 Visualization of White Matter Tracts with Wrapped Streamlines Frank Enders Natascha Sauber Dorit Merhof Peter Hastreiter Neurocenter, Dept. of Neurosurgery Neurocenter, Dept. of Neurosurgery Neurocenter,
Visualization of White Matter Tracts with Wrapped Streamlines Frank Enders Natascha Sauber Dorit Merhof Peter Hastreiter Neurocenter, Dept. of Neurosurgery Neurocenter, Dept. of Neurosurgery Neurocenter,
A diffusion tensor imaging software comparison and between control subjects and subjects with known anatomical diagnosis
 UNLV Theses, Dissertations, Professional Papers, and Capstones 12-2011 A diffusion tensor imaging software comparison and between control subjects and subjects with known anatomical diagnosis Michael C.
UNLV Theses, Dissertations, Professional Papers, and Capstones 12-2011 A diffusion tensor imaging software comparison and between control subjects and subjects with known anatomical diagnosis Michael C.
Network connectivity via inference over curvature-regularizing line graphs
 Network connectivity via inference over curvature-regularizing line graphs Asian Conference on Computer Vision Maxwell D. Collins 1,2, Vikas Singh 2,1, Andrew L. Alexander 3 1 Department of Computer Sciences
Network connectivity via inference over curvature-regularizing line graphs Asian Conference on Computer Vision Maxwell D. Collins 1,2, Vikas Singh 2,1, Andrew L. Alexander 3 1 Department of Computer Sciences
Diffusion Tensor Visualization. and the Teem Software that Makes it Go
 Diffusion Tensor Visualization and the Teem Software that Makes it Go Gordon Kindlmann gk@bwh.harvard.edu Laboratory of Mathematics in Imaging Department of Radiology Brigham & Women s Hospital Harvard
Diffusion Tensor Visualization and the Teem Software that Makes it Go Gordon Kindlmann gk@bwh.harvard.edu Laboratory of Mathematics in Imaging Department of Radiology Brigham & Women s Hospital Harvard
Free-Form Fibers: A Whole Brain Fiber-to-DTI Registration Method
 Free-Form Fibers: A Whole Brain Fiber-to-DTI Registration Method Chao Li 1,2, Xiaotian He 2,3, Vincent Mok 4, Winnie Chu 5, Jing Yuan 5, Ying Sun 1, and Xiaogang Wang 2,3 1 Department of Electrical and
Free-Form Fibers: A Whole Brain Fiber-to-DTI Registration Method Chao Li 1,2, Xiaotian He 2,3, Vincent Mok 4, Winnie Chu 5, Jing Yuan 5, Ying Sun 1, and Xiaogang Wang 2,3 1 Department of Electrical and
Parameter Sensitivity Visualization in DTI Fiber Tracking
 Parameter Sensitivity Visualization in DTI Fiber Tracking Ralph Brecheisen, Bram Platel, Anna Vilanova, and Bart ter Haar Romeny Abstract Fiber tracking of Diffusion Tensor Imaging (DTI) data offers a
Parameter Sensitivity Visualization in DTI Fiber Tracking Ralph Brecheisen, Bram Platel, Anna Vilanova, and Bart ter Haar Romeny Abstract Fiber tracking of Diffusion Tensor Imaging (DTI) data offers a
Vector Visualization
 Vector Visualization Vector Visulization Divergence and Vorticity Vector Glyphs Vector Color Coding Displacement Plots Stream Objects Texture-Based Vector Visualization Simplified Representation of Vector
Vector Visualization Vector Visulization Divergence and Vorticity Vector Glyphs Vector Color Coding Displacement Plots Stream Objects Texture-Based Vector Visualization Simplified Representation of Vector
Strategies for Direct Volume Rendering of Diffusion Tensor Fields
 Strategies for Direct Volume Rendering of Diffusion Tensor Fields Gordon Kindlmann, David Weinstein, David Hart Scientific Computing and Imaging Department of Computer Science, University of Utah {gk dmw
Strategies for Direct Volume Rendering of Diffusion Tensor Fields Gordon Kindlmann, David Weinstein, David Hart Scientific Computing and Imaging Department of Computer Science, University of Utah {gk dmw
A Multi-Resolution watershed-based approach for the segmentation of Diffusion Tensor Images
 A Multi-Resolution watershed-based approach for the segmentation of Diffusion Tensor Images P.R. Rodrigues 1, A. Jalba 2, P. Fillard 3, A. Vilanova 1, and B.M. ter Haar Romeny 1 1 Department of Biomedical
A Multi-Resolution watershed-based approach for the segmentation of Diffusion Tensor Images P.R. Rodrigues 1, A. Jalba 2, P. Fillard 3, A. Vilanova 1, and B.M. ter Haar Romeny 1 1 Department of Biomedical
Direct and Comparative Visualization Techniques for HARDI Data
 Direct and Comparative Visualization Techniques for HARDI Data Rik Sonderkamp c 2010 Rik Sonderkamp Delft University of Technology Final Thesis Direct and Comparative Visualization Techniques for HARDI
Direct and Comparative Visualization Techniques for HARDI Data Rik Sonderkamp c 2010 Rik Sonderkamp Delft University of Technology Final Thesis Direct and Comparative Visualization Techniques for HARDI
Similarity Measures for Matching Diffusion Tensor Images
 Similarity Measures for Matching Diffusion Tensor Images Daniel Alexander, James Gee, Ruzena Bajcsy GRASP Lab. and Dept. Radiology U. of Penn. 40 Walnut St., Philadelphia, PA 904, USA daniel2@grip.cis.upenn.edu
Similarity Measures for Matching Diffusion Tensor Images Daniel Alexander, James Gee, Ruzena Bajcsy GRASP Lab. and Dept. Radiology U. of Penn. 40 Walnut St., Philadelphia, PA 904, USA daniel2@grip.cis.upenn.edu
NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2013 May 04.
 NIH Public Access Author Manuscript Published in final edited form as: Med Image Comput Comput Assist Interv. 2005 ; 8(0 1): 180 187. A Hamilton-Jacobi-Bellman approach to high angular resolution diffusion
NIH Public Access Author Manuscript Published in final edited form as: Med Image Comput Comput Assist Interv. 2005 ; 8(0 1): 180 187. A Hamilton-Jacobi-Bellman approach to high angular resolution diffusion
A Generic Lie Group Model for Computer Vision
 A Generic Lie Group Model for Computer Vision Within this research track we follow a generic Lie group approach to computer vision based on recent physiological research on how the primary visual cortex
A Generic Lie Group Model for Computer Vision Within this research track we follow a generic Lie group approach to computer vision based on recent physiological research on how the primary visual cortex
Visualization Computer Graphics I Lecture 20
 15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] November 20, 2003 Doug James Carnegie Mellon University http://www.cs.cmu.edu/~djames/15-462/fall03
15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] November 20, 2003 Doug James Carnegie Mellon University http://www.cs.cmu.edu/~djames/15-462/fall03
Integrated Visualization of Diffusion Tensor and Functional MRI
 University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange Masters Theses Graduate School 8-2003 Integrated Visualization of Diffusion Tensor and Functional MRI Nathaniel Richard
University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange Masters Theses Graduate School 8-2003 Integrated Visualization of Diffusion Tensor and Functional MRI Nathaniel Richard
Classification of Abdominal Tissues by k-means Clustering for 3D Acoustic and Shear-Wave Modeling
 1 Classification of Abdominal Tissues by k-means Clustering for 3D Acoustic and Shear-Wave Modeling Kevin T. Looby klooby@stanford.edu I. ABSTRACT Clutter is an effect that degrades the quality of medical
1 Classification of Abdominal Tissues by k-means Clustering for 3D Acoustic and Shear-Wave Modeling Kevin T. Looby klooby@stanford.edu I. ABSTRACT Clutter is an effect that degrades the quality of medical
Chapter 6 Visualization Techniques for Vector Fields
 Chapter 6 Visualization Techniques for Vector Fields 6.1 Introduction 6.2 Vector Glyphs 6.3 Particle Advection 6.4 Streamlines 6.5 Line Integral Convolution 6.6 Vector Topology 6.7 References 2006 Burkhard
Chapter 6 Visualization Techniques for Vector Fields 6.1 Introduction 6.2 Vector Glyphs 6.3 Particle Advection 6.4 Streamlines 6.5 Line Integral Convolution 6.6 Vector Topology 6.7 References 2006 Burkhard
Analysis of White Matter Fiber Tracts via Fiber Clustering and Parametrization
 Analysis of White Matter Fiber Tracts via Fiber Clustering and Parametrization Sylvain Gouttard 2,4, Isabelle Corouge 1,2, Weili Lin 3, John Gilmore 2, and Guido Gerig 1,2 1 Departments of Computer Science,
Analysis of White Matter Fiber Tracts via Fiber Clustering and Parametrization Sylvain Gouttard 2,4, Isabelle Corouge 1,2, Weili Lin 3, John Gilmore 2, and Guido Gerig 1,2 1 Departments of Computer Science,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy
 The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
Qualitative and quantitative analysis of probabilistic and deterministic fiber tracking
 Qualitative and quantitative analysis of probabilistic and deterministic fiber tracking Jan Klein a, Adrian Grötsch a, Daniel Betz a, Sebastiano Barbieri a, Ola Friman a, Bram Stieltjes b, Helmut Hildebrandt
Qualitative and quantitative analysis of probabilistic and deterministic fiber tracking Jan Klein a, Adrian Grötsch a, Daniel Betz a, Sebastiano Barbieri a, Ola Friman a, Bram Stieltjes b, Helmut Hildebrandt
Visualization Computer Graphics I Lecture 20
 15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 15, 2003 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 15, 2003 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
Motion Tracking and Event Understanding in Video Sequences
 Motion Tracking and Event Understanding in Video Sequences Isaac Cohen Elaine Kang, Jinman Kang Institute for Robotics and Intelligent Systems University of Southern California Los Angeles, CA Objectives!
Motion Tracking and Event Understanding in Video Sequences Isaac Cohen Elaine Kang, Jinman Kang Institute for Robotics and Intelligent Systems University of Southern California Los Angeles, CA Objectives!
THE brain is a massively interconnected organ. Individual
 IEEE TRANSACTIONS ON VISUALIZATION AND COMPUTER GRAPHICS 1 Exploring Connectivity of the Brain s White Matter with Dynamic Queries Anthony Sherbondy, David Akers, Rachel Mackenzie, Robert Dougherty, and
IEEE TRANSACTIONS ON VISUALIZATION AND COMPUTER GRAPHICS 1 Exploring Connectivity of the Brain s White Matter with Dynamic Queries Anthony Sherbondy, David Akers, Rachel Mackenzie, Robert Dougherty, and
Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University
![Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University](/thumbs/90/102611276.jpg) 15-462 Computer Graphics I Lecture 21 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
15-462 Computer Graphics I Lecture 21 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
High-Order Diffusion Tensor Connectivity Mapping on the GPU
 High-Order Diffusion Tensor Connectivity Mapping on the GPU Tim McGraw and Donald Herring Purdue University Abstract. We present an efficient approach to computing white matter fiber connectivity on the
High-Order Diffusion Tensor Connectivity Mapping on the GPU Tim McGraw and Donald Herring Purdue University Abstract. We present an efficient approach to computing white matter fiber connectivity on the
Geometric Representations. Stelian Coros
 Geometric Representations Stelian Coros Geometric Representations Languages for describing shape Boundary representations Polygonal meshes Subdivision surfaces Implicit surfaces Volumetric models Parametric
Geometric Representations Stelian Coros Geometric Representations Languages for describing shape Boundary representations Polygonal meshes Subdivision surfaces Implicit surfaces Volumetric models Parametric
11/1/13. Visualization. Scientific Visualization. Types of Data. Height Field. Contour Curves. Meshes
 CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 2.11] Jernej Barbic University of Southern California Scientific Visualization
CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 2.11] Jernej Barbic University of Southern California Scientific Visualization
Visualization. CSCI 420 Computer Graphics Lecture 26
 CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 11] Jernej Barbic University of Southern California 1 Scientific Visualization
CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 11] Jernej Barbic University of Southern California 1 Scientific Visualization
A SOM-view of oilfield data: A novel vector field visualization for Self-Organizing Maps and its applications in the petroleum industry
 A SOM-view of oilfield data: A novel vector field visualization for Self-Organizing Maps and its applications in the petroleum industry Georg Pölzlbauer, Andreas Rauber (Department of Software Technology
A SOM-view of oilfield data: A novel vector field visualization for Self-Organizing Maps and its applications in the petroleum industry Georg Pölzlbauer, Andreas Rauber (Department of Software Technology
Scaling the Topology of Symmetric, Second-Order Planar Tensor Fields
 Scaling the Topology of Symmetric, Second-Order Planar Tensor Fields Xavier Tricoche, Gerik Scheuermann, and Hans Hagen University of Kaiserslautern, P.O. Box 3049, 67653 Kaiserslautern, Germany E-mail:
Scaling the Topology of Symmetric, Second-Order Planar Tensor Fields Xavier Tricoche, Gerik Scheuermann, and Hans Hagen University of Kaiserslautern, P.O. Box 3049, 67653 Kaiserslautern, Germany E-mail:
MITK-DI. A new Diffusion Imaging Component for MITK. Klaus Fritzsche, Hans-Peter Meinzer
 MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
CS/NEUR125 Brains, Minds, and Machines. Due: Wednesday, April 5
 CS/NEUR125 Brains, Minds, and Machines Lab 8: Using fmri to Discover Language Areas in the Brain Due: Wednesday, April 5 In this lab, you will analyze fmri data from an experiment that was designed to
CS/NEUR125 Brains, Minds, and Machines Lab 8: Using fmri to Discover Language Areas in the Brain Due: Wednesday, April 5 In this lab, you will analyze fmri data from an experiment that was designed to
Mirrored LH Histograms for the Visualization of Material Boundaries
 Mirrored LH Histograms for the Visualization of Material Boundaries Petr Šereda 1, Anna Vilanova 1 and Frans A. Gerritsen 1,2 1 Department of Biomedical Engineering, Technische Universiteit Eindhoven,
Mirrored LH Histograms for the Visualization of Material Boundaries Petr Šereda 1, Anna Vilanova 1 and Frans A. Gerritsen 1,2 1 Department of Biomedical Engineering, Technische Universiteit Eindhoven,
Figure 1. a: Slice of a tensor volume where every \element" of the image matrix corresponds to one component of the tensor D. axes oriented along the
 Level Set Modeling and Segmentation of DT-MRI Brain Data Leonid Zhukov, Ken Museth, David Breen, Ross Whitakery, Alan H. Barr Department of Computer Science,California Institute of Technology Mail Code
Level Set Modeling and Segmentation of DT-MRI Brain Data Leonid Zhukov, Ken Museth, David Breen, Ross Whitakery, Alan H. Barr Department of Computer Science,California Institute of Technology Mail Code
Vector Visualization. CSC 7443: Scientific Information Visualization
 Vector Visualization Vector data A vector is an object with direction and length v = (v x,v y,v z ) A vector field is a field which associates a vector with each point in space The vector data is 3D representation
Vector Visualization Vector data A vector is an object with direction and length v = (v x,v y,v z ) A vector field is a field which associates a vector with each point in space The vector data is 3D representation
GE Healthcare CLINICAL GALLERY. Discovery * MR750w 3.0T. This brochure is intended for European healthcare professionals.
 GE Healthcare CLINICAL GALLERY Discovery * MR750w 3.0T This brochure is intended for European healthcare professionals. NEURO PROPELLER delivers high resolution, motion insensitive imaging in all planes.
GE Healthcare CLINICAL GALLERY Discovery * MR750w 3.0T This brochure is intended for European healthcare professionals. NEURO PROPELLER delivers high resolution, motion insensitive imaging in all planes.
ELEC Dr Reji Mathew Electrical Engineering UNSW
 ELEC 4622 Dr Reji Mathew Electrical Engineering UNSW Review of Motion Modelling and Estimation Introduction to Motion Modelling & Estimation Forward Motion Backward Motion Block Motion Estimation Motion
ELEC 4622 Dr Reji Mathew Electrical Engineering UNSW Review of Motion Modelling and Estimation Introduction to Motion Modelling & Estimation Forward Motion Backward Motion Block Motion Estimation Motion
Multiresolution analysis: theory and applications. Analisi multirisoluzione: teoria e applicazioni
 Multiresolution analysis: theory and applications Analisi multirisoluzione: teoria e applicazioni Course overview Course structure The course is about wavelets and multiresolution Exam Theory: 4 hours
Multiresolution analysis: theory and applications Analisi multirisoluzione: teoria e applicazioni Course overview Course structure The course is about wavelets and multiresolution Exam Theory: 4 hours
Algebraic Methods for Direct and Feature Based Registration of Diffusion Tensor Images
 Algebraic Methods for Direct and Feature Based Registration of Diffusion Tensor Images Alvina Goh and René Vidal Center for Imaging Science, Department of BME, Johns Hopkins University 308B Clark Hall,
Algebraic Methods for Direct and Feature Based Registration of Diffusion Tensor Images Alvina Goh and René Vidal Center for Imaging Science, Department of BME, Johns Hopkins University 308B Clark Hall,
