The Application of GPU Particle Tracing to Diffusion Tensor Field Visualization
|
|
|
- Denis Wilcox
- 5 years ago
- Views:
Transcription
1 The Application of GPU Particle Tracing to Diffusion Tensor Field Visualization Polina Kondratieva, Jens Krüger, Rüdiger Westermann Computer Graphics and Visualization Group Technische Universität München ABSTRACT In this paper we introduce GPU particle tracing for the visualization of 3D diffusion tensor fields. For about half a million particles, reconstruction of diffusion directions from the tensor field, time integration and rendering can be done at interactive rates. Different visualization options like oriented particles of diffusion-dependent shape, stream lines or stream tubes facilitate the use of particle tracing for diffusion tensor visualization. The proposed methods provide efficient and intuitive means to show the dynamics in diffusion tensor fields, and they accommodate the exploration of the diffusion properties of biological tissue. CR Categories: I.3.8 [Computing Methodologies]: Computer Graphics Applications ; J.3 [Computing Application]: Life and Medical Sciences ; I.3.7 [Computer Graphics]: Methodology and Techniques Interaction Techniques; Keywords: Diffusion Tensors, Dynamic Visualization, GPU Particle Tracing and Streamlines, Medical Visualization 1 INTRODUCTION The diffusion properties of biological tissue can be measured using diffusion tensor magnetic resonance imaging (DT-MRI) [1]. The imaging process reveals the diffusion of water molecules depending on the shape and orientation of tissue cells, i.e. within fibrous material the diffusion is anisotropic while there is an equal diffusion probability in real matter of other types. The diffusion probability is characterized by a second order tensor, which describes the deflection of a molecular pathway as a function of spatial position. Approaches to visualize the diffusion in real tissue can be classified into two major categories: glyph-based techniques reveal local variations in diffusion tensor fields by mapping tensor properties like deflection or diffusion probability to the shape or appearance of graphical primitives, i.e. ellipsoids [10], composite shapes [15], or superquadrics [8]. In contrast, directional tracking of massless particles along the most probable diffusion directions in tensor field data [4] allows for the classification of anatomical structures, e.g. the white matter fiber tracks. Different geometric representations like stream lines [14, 12] and stream tubes [17, 3], or stream surfaces [17, 13] have been employed to visualize these structures. To improve the quality and stability of such tracking techniques, regularization and filtering approaches [2, 18, 5] along with heuristics to determine the most probable directions [14, 18] have been proposed. Dedicated color and opacity mapping schemes to visually enhance particular features in diffusion tensor data have been presented in [14, 7, 15, 8]. While tracking based techniques can effectively visualize local features in tensor fields, glyph-based imaging techniques for visu- kondrati@in.tum.de,kruegeje@in.tum.de,westermann@in.tum.de alizing 3D fields can successfully illustrate the global behavior of such fields. However, it is difficult when using such methods to effectively control glyph density, shape and appearance in a way that depicts both the direction structure of the diffusion and the diffusion magnitude. Neither of these techniques allows for interactive exploration of large tensor fields, and they typically fail to visualize and to monitor the diffusion dynamics in real tissue. In this paper, we propose an interactive technique based on GPU particle tracing for tensor field visualization. This method can display the dynamics of large particle sets in flow fields, and it can thus be used to monitor the diffusion in biological tissue in real time. A number of visualization options like oriented texture splats, stream lines and stream tubes provide the user with an effective means for the visual analysis of 3D tensor fields given on a Cartesian grid. 2 DIFFUSION TENSORS The second order diffusion tensor can be expressed mathematically as a 3 3 symmetric semi-positive matrix: D = D xx D xy D xz D yx D yy D yz (1) D zx D zy D zz From a physical point of view, the tensor D describes the probability density for the deflection of molecular pathways in diffusive tissue. Following the common classification, diffusion tensors can be represented as ellipsoids with main, medium, and minor axes corresponding to the eigenvectors e 1, e 2, e 3 with respective eigenvalues λ 1 λ 2 λ 3 of the tensor. The relative differences between the eigenvalues are related to the anisotropy of the diffusion. Three basic types of anisotropy are usually considered in the literature (see Table 1). A thorough discussion of diffusion tensors and other derived quantities can be found in [8, 16]. Table 1: Classification of diffusion tensor anisotropy. Name Coefficient linear c l = λ 1 λ 2 λ 1 + λ 2 + λ 3 planar c p = 2(λ 2 λ 3 ) λ 1 + λ 2 + λ 3 spherical 3λ c s = 3 λ 1 + λ 2 + λ 3 Besides the mapping of local anisotropy to the shape of geometrical icons, a number of different color encoding schemes have been proposed: RGB = (µ 1, µ 2, A 3 ) (2)
2 RGB = (J 4, FA, A 3 ) (3) RGB = (c l, c p, c s ) (4) RGB = FA( e 1x, e 1y, e 1z ) (5) RGB = RA( e 1x, e 1y, e 1z ) (6) Here, the coefficients µ 1, µ 2, A 3, J 4, FA, RA are computed as follows: µ 1 = 3 1 λ i, 1 st central moment of eigenvalues, µ 2 = 3 1 (λ i µ 1 ) 2, 2 nd central moment of eigenvalues, A 3 = (λ i µ 1 ) 3 3 µ 2 µ2, skewness of eigenvalues, J 4 = λi 2, invariant of tensor D, 3 FA = 2 µ2 J 4, RA = fractional anisotropy, µ2 2µ1, relative anisotropy Figure 1: The work cycle of a general particle engine. The advection subsystem takes an initial position, and a vector field to generate a new position which is used for rendering. In the next time-step the initial position is replaced by the advected position and the whole process is repeated from the beginning. 3 GPU PARTICLE ENGINE To interactively explore the dynamics in 3D diffusion tensor fields, we employ a particle system for visualizing steady 3D flow fields on Cartesian grids [9]. We exploit features of recent graphics accelerators to advect particles on the GPU, saving particle positions in graphics memory, and then sending these positions through the GPU again to obtain images in the frame buffer. By using this functionality, particle tracing in Cartesian grids can be entirely performed on the GPU without any read back to application memory. This approach allows for interactive streaming and rendering of millions of particles, and it enables virtual exploration of high resolution fields. In the current scenario, the ability to display the dynamics of large particle sets using visualization options like oriented texture splats, stream lines, and stream tubes provides an intuitive means for the visual analysis of 3D tensor fields that is far beyond existing solutions. Particle advection is performed in the fragment units of programmable GPUs. During particle tracing, particle positions are subsequently read from the current particle container and the result of the advection step is written to an additional container. Containers are internally stored as 2D texture maps. The vector field data is stored in the RGB color components of a 3D texture. To display particle primitives, a vertex array containing as many entries as there are particles is rendered. This array resides in local GPU memory. In the GPU s vertex units, respective particle positions are fetched from the current particle container, and vertex positions are displaced accordingly. This is accomplished using functionality in Shader Model 3.0 [11]. The particle engine provides different visualization options. Oriented 3D icons can be simulated by rendering textured point sprites in combination with a texture atlas. For every icon this atlas contains a set of pre-computed views, of which the one most similar to the actual view is rendered. In addition, the traces of particles can be stored and used to render stream lines, stream balls or stream tubes. 4 TENSOR VISUALIZATION USING PARTICLE TRACING To use the GPU particle engine (see Figure 1) for tensor field visualization, a number of data-specific extensions have been integrated. In particular, the direction vector along which a particle is traced first has to be reconstructed from the tensor field. The basic differences between particle tracing in flow fields and in tensor fields are illustrated in Figure 2. In the following, the specific extensions we have integrated to accommodate the use of particle tracing for tensor field visualization are described: Figure 2: Differences between particle tracing in a vector field and in a tensor field. Note that the advection of a particle in a tensor field as proposed in [14] requires both the current and the last particle direction. The six distinct entries of the diffusion tensor are stored in two 3D RGB texture maps. Because hardware accelerated trilinear interpolation of 32 bit floating point textures is not supported on recent GPUs, we use 16 bit floating point textures in the current implementation. If this precision is not sufficient, hand coded interpolation of 32 bit floating point values can be performed in a fragment shader. As shown in [9], this only results in a slight decrease in performance. At every particle position, the tensor is tri-linearly interpolated from the two 3D texture maps. To derive a vector field for particle tracing, at every particle position the eigen-decomposition of the resampled tensor is computed on the fly 1. Therefore, on the GPU we have implemented a non-iterative analytical algorithm proposed by Hasan et al. [6]. If the tensor is not positive-definite, or if its eigenvalues are degenerate (equal to each other), the propagation process is terminated. For the rendering of textured particle sprites, two different texture atlases for tensor visualization have been designed. The first atlas extends the one proposed in [9] about an additional scaling factor used to emphasize the diffusion 1 The interpolation of precomputed eigenvectors and eigenvalues does not allow for a consistent computation of diffusion direction [8, 18].
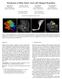
3 anisotropy c a = c l + c p. The second one contains precomputed views of short stream tubes at different orientation and size. By texturing particle sprites with the image of the respective view scaled according to the tensor attributes the appearance of closed stream tubes can be simulated. Because particle tracking in 3D tensor fields along the largest eigenvector of the tensor can lead to ambiguous results (opposite eigenvector directions are both valid), the outgoing particle direction is computed as a linear combination of the deflected incoming direction and the principal eigenvector [14]. A number of different criteria to stop particle propagation have been integrated [8]. In particular, if the fractional anisotropy is less than a given threshold, a particle trace is stopped. In addition, if the fractional anisotropy is above a threshold, it is used to modulate the particle transparency. In this way, particles can be faded out continuously in nearly isotropic regions. The user can interactively select and change tensor-specific parameters, such as the color mapping scheme, the threshold of the fractional anisotropy used to fade out particles, and the propagation algorithm (along the largest eigenvector or along the deflected direction). 5 RESULTS All of our experiments have been done on a NVIDIA Quadro FX 4400 graphics card equipped with 512MB video memory. Rendering was performed into a viewport of pixels. In this section we show screenshots of the dynamic 3D tensor field visualization including various visualization options. Although the images already show the functionality of the particle engine, we should note here that the real benefit of the presented approach is a lot more apparent in the accompanying video. Table 2 gives timings for the advection and rendering of particles. Timings in the second column include all operations that are carried out until updated particle positions are available in the current particle container, i.e. tensor interpolation, eigendecomposition and particle advection using Euler integration. The third and the fourth columns include timings for the rendering of diffusion-aligned point sprites. To demonstrate the dependency of performance from the number of generated fragments, differently sized sprites are used. The last column shows the time that is required to reconstruct different amounts of stream lines of length 100. In all these tests, a tensor data set (canine heart) was visualized. As can be seen, even for large particle sets the GPU implementation still allows for the interactive exploration of tensor fields. It is interesting to note, that with increasing fragment size the rendering stage quickly becomes fragment bound. Moreover, the number of generated fragments strongly depends on viewing parameters, such as field of view, the distance of particles to the camera. As it is rather difficult to provide precise and meaningful timing statistics for different amounts of particles in combination with differently sized point sprites, we give specific timings for all the generated images shown in the following. Also, since for fibers the number of advection steps is proportional to their length, the approximate time for tracking and rendering a fiber of an arbitrary length can be computed as the time needed for one segment multiplied by the number of segments. In contrast to previous approaches, where only the final rendering of precomputed entities can be done interactively, the proposed method provides an even more intuitive means to explore high resolution diffusion tensor fields. In particular, the user can interactively select the seed density of particles as well as parameters specific to particles advection only oriented splats lines length pixels 7 7 pixels Table 2: Application performance in frames per second, for different amounts of particles and for stream tubes each consisting of 100 segments. the propagation process. By using these options, pathways can be visualized at almost arbitrary resolution. The advection of oriented particles allows for the simultaneous exploration of both local and global diffusion tensor properties. Fiber structures can be observed without that particle positions have to be connected. Even in still images the fibrous structures in the investigated tissue can clearly be seen. Figures 3 to 9 show a number of examples that were generated using the presented particle engine. Two data sets were used to demonstrate the effectiveness of particle tracking for tensor field visualization: a human brain data set of resolution and a canine heart data set of resolution Both data sets consist of six diffusion tensor components and an additional confidence value per voxel. Different visualization options and color mappings were selected to emphasize particular diffusion properties and anatomical structures in these data sets. In all examples, visual exploration was performed in real-time, thus allowing for an effective and intuitive analysis of biological tissue. In Figure 3 colored point sprites of size 5 5 pixels were rendered to visualize the human brain data set. The FA-based color mapping scheme (equation 5) was used. By fading out particles in regions showing low anisotropy, highly anisotropic brain structures, such as corpus callosum, corona radiata and pyramid, are emphasized and can clearly be distinguished in the generated image. Figure 3: Visualization of the diffusion tensor field measured in a human brain. Below a given anisotropy threshold the transparency of rendered particle primitives is inversely proportional to the measured anisotropy. In this way, particles in regions of low anisotropy are continuously faded out. 64K particles of size 5 5 pixels were rendered at roughly 40 frames per second. In Figure 4, diffusion-dependent oriented sprites were used to
4 render a close up view of the corona radiata. According to the FAbased color mapping scheme, an optic tract can be clearly distinguished by green color. To generate the image, 64K point sprites of size pixels were rendered at 6.1 frames per second. The left image in Figure 5 shows a visualization of the corona radiata using stream tubes. The seed box from which particle traces were initially released is shown as wire frame in red. Overall, 512 traces of a maximum length of 600 were traced. Textured point sprites of size pixels were rendered using the texture atlas described above. Reconstruction and rendering was performed at 2.4 frames per second. As can be seen, this visualization option helps to track the bunches of fibers with biologically similar properties, and it allows the expert to follow the paths of single fibers. On the right of Figure 5 the superior longitudinal fasciculus is rendered by means of colored stream lines. The axial slice along the fasciculus was chosen as seed region. 512 lines of a maximum length of 600 were computed and displayed at 10 frames per second. Both images show nicely the property of stream tubes and stream lines to effectively reveal spatial relationships between different tracks. The sagittal, coronal, and axial slices through the human brain are visualized in Figure 6 using diffusion-dependent sprites. In combination, shape and color of the rendered ellipsoids give a good impression of the degree of anisotropy and the main diffusion direction within the corresponding brain structures. The brain structures themselves, such as pyramidal tracts, U-shaped fibers, superior longitudinal fasciculus and others, are clearly visible in all slices. The axial and sagittal slices were rendered using 64K particles of size pixels at 10.8 frame per second. The coronal slice was produced using 256K particles of size pixels at 4.5 frames per second. Visualizations of the canine heart data set are shown in Figures 7, 8, 9. Figure 7 shows the longitudinal and latitudinal heart slices, rendered with textured point sprites. The orientation of the sprites corresponds to the helical construction of the heart muscle, and the spatial positions depict the heart structure consisting of four chambers and several valves in between. The animation of 64K particles of size pixels runs at roughly 8 frames per second. Figure 8 shows images of the heart data set rendered from different view points using aligned diffusion-dependent point sprites. In the interactive animation, the clockwise and counter-clockwise motion of particles along the muscle structure can be clearly tracked. Both images were rendered using 64K particles at and pixels per point sprite, respectively. Accordingly, the application performance dropped from 9.4 frames per second to 7.4 frames per second. Stream tubes were used in Figure 9 to visualize the heart data set. On the right, tubes were colored according to equation 3. Both images were generated using 1K stream tubes of maximum length of 1K in combination with point sprites of size 5 5 pixels to render the texture-based tube segments. Reconstruction and rendering was done at 1.5 frames per second. 6 CONCLUSION We have presented an efficient and effective visualization system for 3D diffusion tensor fields. This system allows for the visual exploration of such fields at interactive rates and at arbitrary level of detail. The user can interactively select regions of interest by positioning a particle probe of adjustable position and size. A number of different visualization options can be selected to visualize local as well as global features in the diffusion process. By using this system, local variations in diffusion tensor fields as well as anatomical structures can be visualized at interactive rates. Due to the possibility to simulate the dynamic behavior of massless particles in the derived diffusion field, a very intuitive approach to understanding diffusion tensor fields has been presented. In contrast to previous methods, real-time advection and rendering of large particle sets produces animations that closely and intuitively mimic the underlying dynamic diffusion process. ACKNOWLEDGMENTS We would like to thank Prof. Edward W. Hsu, Department of Bioengineering, Duke University, Leonid Zhukov and Gordon Kindlmann for providing the tensor field data sets. REFERENCES [1] P.J. Basser, J. Mattiello, and D. LeBihan. MR diffusion tensor spectroscopy and imaging. Biophysical Journal, pages , [2] O. Coulon, D. Alexander, and S.R. Arridge. A regularization scheme for diffusion tensor magnetic resonance images. Lecture Notes in Computer Science, 2082:92 105, [3] Marco DaSilva, Song Zhang, Cagatay Demiralp, and David H. Laidlaw. Visualizing diffusion tensor volume differences. In Visualization 01 Work in Progress Proceedings, pages 16 17, October [4] Thierry Delmarcelle and Lambertus Hesselink. Visualization of second order tensor fields and matrix data. In Proceedings IEEE Visualization, pages , [5] C. Gossl, L. Fahrmeir, B. Putz, L.M. Auer, and D.P. Auer. Fiber tracking from DTI using linear state space models: Detectability of the pyramidal tract. In NeuroImage, volume 16, pages , [6] K. M. Hasan. Analytical computation of the eigenvalues and eigenvectors in DT MRI. IEEE Computer Graphics Applications, pages 36 46, [7] G. Kindlmann and D. Weinstein. Hue balls and lit tensors for direct volume rendering of diffusion tensor field. In Proceedings IEEE Visualization, pages , [8] Gordon Kindlmann. DTI Visualization and Analysis of Diffusion Tensor Fields. PhD thesis, University of Utah, [9] J. Krueger, P. Kipfer, P. Kondratieva, and R. Wetsermann. A particle system for interactive visualization of 3d flows. IEEE Transactions on Visualization and Computer Graphics, to appear. [10] David H. Laidlaw, Eric T. Ahrens, David Kremers, Matthew J. Avalos, Carol Readhead, and Russell E. Jacobs. Visualizing diffusion tensor images of the mouse spinal cord. In Proceedings IEEE Visualization, pages , [11] Microsoft. Shader model 3.0 specification [12] S. Mori, B.J. Crain, V.P. Chacko, and P.C. van Zijl. Three dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol, 45(2): , [13] A. Vilanova, G.Berenschot, and C. van Pul. DTI visualization with streamsurfaces and evenly spaced volume seeding. In VisSym 04 Joint EG IEEE TCVG Symposium on Visualization, pages , [14] David M. Weinstein, Gordon L. Kindlmann, and Eric C. Lundberg. Tensorlines: Advection-diffusion based propagation through diffusion tensor fields. In IEEE Visualization 99, pages , [15] C.-F. Westin, S.E. Maier, H. Mamata, A. Nabavi, F.A. Jolesz, and R. Kikinis. Processing and visualization for diffusion tensor MRI. Medical Image Analysis, 6:93 108, [16] Masutani Y., Aoki S., Abe O., Hayashi N., and Otomo K. MR diffusion tensor imaging: recent advance and new techniques for diffusion tensor visualization. European Journal of Radiology, 1(46):53 66, [17] Song Zhang, Charles T. Curry, Daniel S. Morris, and David H. Laidlaw. Streamtubes and streamsurfaces for visualizing diffusion tensor MRI volume images. In Visualization 00 Work in Progress, October [18] L. Zhukov and A. Barr. Oriented tensor reconstruction: tracing neural pathways from diffusion tensor MRI. In Proceedings of the IEEE Visualization 02, pages , 2002.
5 Figure 4: Brain structures: By using the color mapping scheme described in equation 5, the optic tract (green) and corona radiata (blue) fibers can be clearly distinguished. At 6.1 frames per second, 64K particles can be advected, and rendered using textured sprites of size pixels. Figure 5: Brain structures: On the left, corona radiata is visualized using stream tubes. On the right, the axial slice along the longitudinal fasciculus is rendered by means of simple stream lines. The FA-based color mapping scheme (equation 5) is applied. 512 fibers with a maximum length of 600 can be generated and rendered at 2.4 and 10 frames per second, respectively. Figure 6: Brain structures: Axial (left), coronal (middle) and sagittal (right) slices through the brain are shown. The coronal and sagittal slices are color coded using equation 5, and equation 4 is used to color the axial slice. The axial and sagittal slices were rendered using 64K particles of size pixels at 10.8 frames per second. The image of the coronal slice was generated at 4.5 frames per second using 256K particles of size pixels.
6 Figure 7: Longitudinal (top) and latitudinal (bottom) slices of the heart muscle are reconstructed and rendered at 8 frames per second. The helical orientation of the heart muscle fiber becomes apparent from the latitudinal slice. Both slices also depict the four-chambered structure of the heart. The FA-based color mapping scheme was chosen. Figure 8: Heart muscle: Multiple views of the data set are shown. The helical structure of the heart muscle can be easily tracked on both images. The FA-based color mapping scheme (equation 5) was applied. 64K particles of size pixels (left) and pixels (right) were animated at 9.4 and 7.4 frames per second, respectively. Figure 9: Heart muscle: Multiple views of the data set are shown. The color mapping schemes described in equation 5 (left) and equation 3 (right) were used. 1K stream tubes of maximum length of 1K were reconstructed and rendered with point sprites of size 5 5 pixels. Both images were generated at 1.5 frames per second.
8. Tensor Field Visualization
 8. Tensor Field Visualization Tensor: extension of concept of scalar and vector Tensor data for a tensor of level k is given by t i1,i2,,ik (x 1,,x n ) Second-order tensor often represented by matrix Examples:
8. Tensor Field Visualization Tensor: extension of concept of scalar and vector Tensor data for a tensor of level k is given by t i1,i2,,ik (x 1,,x n ) Second-order tensor often represented by matrix Examples:
Evaluation of Local Filter Approaches for Diffusion Tensor based Fiber Tracking
 Evaluation of Local Filter Approaches for Diffusion Tensor based Fiber Tracking D. Merhof 1, M. Buchfelder 2, C. Nimsky 3 1 Visual Computing, University of Konstanz, Konstanz 2 Department of Neurosurgery,
Evaluation of Local Filter Approaches for Diffusion Tensor based Fiber Tracking D. Merhof 1, M. Buchfelder 2, C. Nimsky 3 1 Visual Computing, University of Konstanz, Konstanz 2 Department of Neurosurgery,
HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
Fiber Selection from Diffusion Tensor Data based on Boolean Operators
 Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Complex Fiber Visualization
 Annales Mathematicae et Informaticae 34 (2007) pp. 103 109 http://www.ektf.hu/tanszek/matematika/ami Complex Fiber Visualization Henrietta Tomán a, Róbert Tornai b, Marianna Zichar c a Department of Computer
Annales Mathematicae et Informaticae 34 (2007) pp. 103 109 http://www.ektf.hu/tanszek/matematika/ami Complex Fiber Visualization Henrietta Tomán a, Róbert Tornai b, Marianna Zichar c a Department of Computer
Diffusion Imaging Visualization
 Diffusion Imaging Visualization Thomas Schultz URL: http://cg.cs.uni-bonn.de/schultz/ E-Mail: schultz@cs.uni-bonn.de 1 Outline Introduction to Diffusion Imaging Basic techniques Glyph-based Visualization
Diffusion Imaging Visualization Thomas Schultz URL: http://cg.cs.uni-bonn.de/schultz/ E-Mail: schultz@cs.uni-bonn.de 1 Outline Introduction to Diffusion Imaging Basic techniques Glyph-based Visualization
Saturn User Manual. Rubén Cárdenes. 29th January 2010 Image Processing Laboratory, University of Valladolid. Abstract
 Saturn User Manual Rubén Cárdenes 29th January 2010 Image Processing Laboratory, University of Valladolid Abstract Saturn is a software package for DTI processing and visualization, provided with a graphic
Saturn User Manual Rubén Cárdenes 29th January 2010 Image Processing Laboratory, University of Valladolid Abstract Saturn is a software package for DTI processing and visualization, provided with a graphic
Technische Universiteit Eindhoven Department of Mathematics and Computer Science. Master s Thesis HIERARCHICAL VISUALIZATION USING FIBER CLUSTERING
 Technische Universiteit Eindhoven Department of Mathematics and Computer Science Master s Thesis HIERARCHICAL VISUALIZATION USING FIBER CLUSTERING by Ing. B. Moberts Supervisors: Dr. A. Vilanova Prof.dr.ir.
Technische Universiteit Eindhoven Department of Mathematics and Computer Science Master s Thesis HIERARCHICAL VISUALIZATION USING FIBER CLUSTERING by Ing. B. Moberts Supervisors: Dr. A. Vilanova Prof.dr.ir.
Fiber Selection from Diffusion Tensor Data based on Boolean Operators
 Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhofl, G. Greiner 2, M. Buchfelder 3, C. Nimsky4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhofl, G. Greiner 2, M. Buchfelder 3, C. Nimsky4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Automatic Quantification of DTI Parameters along Fiber Bundles
 Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein 1, Simon Hermann 1, Olaf Konrad 1, Horst K. Hahn 1, and Heinz-Otto Peitgen 1 1 MeVis Research, 28359 Bremen Email: klein@mevis.de
Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein 1, Simon Hermann 1, Olaf Konrad 1, Horst K. Hahn 1, and Heinz-Otto Peitgen 1 1 MeVis Research, 28359 Bremen Email: klein@mevis.de
Automatic Quantification of DTI Parameters along Fiber Bundles
 Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein, Simon Hermann, Olaf Konrad, Horst K. Hahn, Heinz-Otto Peitgen MeVis Research, 28359 Bremen Email: klein@mevis.de Abstract. We introduce
Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein, Simon Hermann, Olaf Konrad, Horst K. Hahn, Heinz-Otto Peitgen MeVis Research, 28359 Bremen Email: klein@mevis.de Abstract. We introduce
Visualizing Diffusion Tensor Imaging Data with Merging Ellipsoids
 Visualizing Diffusion Tensor Imaging Data with Merging Ellipsoids Wei Chen Song Zhang Stephen Correia David F. Tate State Key Lab of CAD&CG, ZJU, China Mississippi State University Brown University Brigham
Visualizing Diffusion Tensor Imaging Data with Merging Ellipsoids Wei Chen Song Zhang Stephen Correia David F. Tate State Key Lab of CAD&CG, ZJU, China Mississippi State University Brown University Brigham
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes
 Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
On Classifying Disease-Induced Patterns in the Brain Using Diffusion Tensor Images
 On Classifying Disease-Induced Patterns in the Brain Using Diffusion Tensor Images Peng Wang 1,2 and Ragini Verma 1 1 Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania,
On Classifying Disease-Induced Patterns in the Brain Using Diffusion Tensor Images Peng Wang 1,2 and Ragini Verma 1 1 Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania,
DIFFUSION TENSOR IMAGING ANALYSIS. Using Analyze
 DIFFUSION TENSOR IMAGING ANALYSIS Using Analyze 2 Table of Contents 1. Introduction page 3 2. Loading DTI Data page 4 3. Computing DTI Maps page 5 4. Defining ROIs for Fiber Tracking page 6 5. Visualizing
DIFFUSION TENSOR IMAGING ANALYSIS Using Analyze 2 Table of Contents 1. Introduction page 3 2. Loading DTI Data page 4 3. Computing DTI Maps page 5 4. Defining ROIs for Fiber Tracking page 6 5. Visualizing
NEURO M203 & BIOMED M263 WINTER 2014
 NEURO M203 & BIOMED M263 WINTER 2014 MRI Lab 2: Neuroimaging Connectivity Lab In today s lab we will work with sample diffusion imaging data and the group averaged fmri data collected during your scanning
NEURO M203 & BIOMED M263 WINTER 2014 MRI Lab 2: Neuroimaging Connectivity Lab In today s lab we will work with sample diffusion imaging data and the group averaged fmri data collected during your scanning
Neural Network-Assisted Fiber Tracking of Synthetic and White Matter DT-MR Images
 Neural Network-Assisted Fiber Tracking of Synthetic and White Matter DT-MR Images L.M. San-José-Revuelta, M. Martín-Fernández and C. Alberola-López Abstract In this paper, a recently developed fiber tracking
Neural Network-Assisted Fiber Tracking of Synthetic and White Matter DT-MR Images L.M. San-José-Revuelta, M. Martín-Fernández and C. Alberola-López Abstract In this paper, a recently developed fiber tracking
UberFlow: A GPU-Based Particle Engine
 UberFlow: A GPU-Based Particle Engine Peter Kipfer Mark Segal Rüdiger Westermann Technische Universität München ATI Research Technische Universität München Motivation Want to create, modify and render
UberFlow: A GPU-Based Particle Engine Peter Kipfer Mark Segal Rüdiger Westermann Technische Universität München ATI Research Technische Universität München Motivation Want to create, modify and render
Flow Visualisation - Background. CITS4241 Visualisation Lectures 20 and 21
 CITS4241 Visualisation Lectures 20 and 21 Flow Visualisation Flow visualisation is important in both science and engineering From a "theoretical" study of o turbulence or o a fusion reactor plasma, to
CITS4241 Visualisation Lectures 20 and 21 Flow Visualisation Flow visualisation is important in both science and engineering From a "theoretical" study of o turbulence or o a fusion reactor plasma, to
n o r d i c B r a i n E x Tutorial DTI Module
 m a k i n g f u n c t i o n a l M R I e a s y n o r d i c B r a i n E x Tutorial DTI Module Please note that this tutorial is for the latest released nordicbrainex. If you are using an older version please
m a k i n g f u n c t i o n a l M R I e a s y n o r d i c B r a i n E x Tutorial DTI Module Please note that this tutorial is for the latest released nordicbrainex. If you are using an older version please
FAST AND MEMORY EFFICIENT GPU-BASED RENDERING OF TENSOR DATA
 FAST AND MEMORY EFFICIENT GPU-BASED RENDERING OF TENSOR DATA Mario Hlawitschka*, Sebastian Eichelbaum #, and Gerik Scheuermann* University of Leipzig PF 100920, 04009 Leipzig *{hlawitschka,scheuermann}@informatik.uni-leipzig.de
FAST AND MEMORY EFFICIENT GPU-BASED RENDERING OF TENSOR DATA Mario Hlawitschka*, Sebastian Eichelbaum #, and Gerik Scheuermann* University of Leipzig PF 100920, 04009 Leipzig *{hlawitschka,scheuermann}@informatik.uni-leipzig.de
Diffusion Tensor Imaging and Reading Development
 Diffusion Tensor Imaging and Reading Development Bob Dougherty Stanford Institute for Reading and Learning Reading and Anatomy Every brain is different... Not all brains optimized for highly proficient
Diffusion Tensor Imaging and Reading Development Bob Dougherty Stanford Institute for Reading and Learning Reading and Anatomy Every brain is different... Not all brains optimized for highly proficient
Glyph-based Visualization Applications. David H. S. Chung Swansea University
 Glyph-based Applications David H. S. Chung Swansea University Outline Glyph Design Application Flow Event Multi-field Uncertainty Geo-spatial Tensor Medical Outline Glyph Design Application Novel shape
Glyph-based Applications David H. S. Chung Swansea University Outline Glyph Design Application Flow Event Multi-field Uncertainty Geo-spatial Tensor Medical Outline Glyph Design Application Novel shape
Diffusion model fitting and tractography: A primer
 Diffusion model fitting and tractography: A primer Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 03/18/10 Why n how Diffusion model fitting and tractography 0/18 Why
Diffusion model fitting and tractography: A primer Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 03/18/10 Why n how Diffusion model fitting and tractography 0/18 Why
Volume Visualization. Part 1 (out of 3) Volume Data. Where do the data come from? 3D Data Space How are volume data organized?
 Volume Data Volume Visualization Part 1 (out of 3) Where do the data come from? Medical Application Computed Tomographie (CT) Magnetic Resonance Imaging (MR) Materials testing Industrial-CT Simulation
Volume Data Volume Visualization Part 1 (out of 3) Where do the data come from? Medical Application Computed Tomographie (CT) Magnetic Resonance Imaging (MR) Materials testing Industrial-CT Simulation
A Method for Registering Diffusion Weighted Magnetic Resonance Images
 A Method for Registering Diffusion Weighted Magnetic Resonance Images Xiaodong Tao and James V. Miller GE Research, Niskayuna, New York, USA Abstract. Diffusion weighted magnetic resonance (DWMR or DW)
A Method for Registering Diffusion Weighted Magnetic Resonance Images Xiaodong Tao and James V. Miller GE Research, Niskayuna, New York, USA Abstract. Diffusion weighted magnetic resonance (DWMR or DW)
Chapter 6 Visualization Techniques for Vector Fields
 Chapter 6 Visualization Techniques for Vector Fields 6.1 Introduction 6.2 Vector Glyphs 6.3 Particle Advection 6.4 Streamlines 6.5 Line Integral Convolution 6.6 Vector Topology 6.7 References 2006 Burkhard
Chapter 6 Visualization Techniques for Vector Fields 6.1 Introduction 6.2 Vector Glyphs 6.3 Particle Advection 6.4 Streamlines 6.5 Line Integral Convolution 6.6 Vector Topology 6.7 References 2006 Burkhard
An Immersive Virtual Environment for DT-MRI Volume Visualization Applications: a Case Study
 An Immersive Virtual Environment for DT-MRI Volume Visualization Applications: a Case Study S. Zhang, Ç. Demiralp,D.F.Keefe M. DaSilva, D. H. Laidlaw, B. D. Greenberg Brown University P.J. Basser C. Pierpaoli
An Immersive Virtual Environment for DT-MRI Volume Visualization Applications: a Case Study S. Zhang, Ç. Demiralp,D.F.Keefe M. DaSilva, D. H. Laidlaw, B. D. Greenberg Brown University P.J. Basser C. Pierpaoli
Volume Visualization
 Volume Visualization Part 1 (out of 3) Overview: Volume Visualization Introduction to volume visualization On volume data Surface vs. volume rendering Overview: Techniques Simple methods Slicing, cuberille
Volume Visualization Part 1 (out of 3) Overview: Volume Visualization Introduction to volume visualization On volume data Surface vs. volume rendering Overview: Techniques Simple methods Slicing, cuberille
An Introduction to Visualization of Diffusion Tensor Imaging and its Applications
 An Introduction to Visualization of Diffusion Tensor Imaging and its Applications A. Vilanova 1, S. Zhang 2, G. Kindlmann 3, and D. Laidlaw 2 1 Department of Biomedical Engineering Eindhoven University
An Introduction to Visualization of Diffusion Tensor Imaging and its Applications A. Vilanova 1, S. Zhang 2, G. Kindlmann 3, and D. Laidlaw 2 1 Department of Biomedical Engineering Eindhoven University
Multimodal Visualization of DTI and fmri Data using Illustrative Methods
 Multimodal Visualization of DTI and fmri Data using Illustrative Methods Silvia Born 1, Werner Jainek 1, Mario Hlawitschka 2, Gerik Scheuermann 3, Christos Trantakis 4, Jürgen Meixensberger 4, Dirk Bartz
Multimodal Visualization of DTI and fmri Data using Illustrative Methods Silvia Born 1, Werner Jainek 1, Mario Hlawitschka 2, Gerik Scheuermann 3, Christos Trantakis 4, Jürgen Meixensberger 4, Dirk Bartz
Strategies for Direct Volume Rendering of Diffusion Tensor Fields
 Strategies for Direct Volume Rendering of Diffusion Tensor Fields Gordon Kindlmann, David Weinstein, David Hart Scientific Computing and Imaging Department of Computer Science, University of Utah {gk dmw
Strategies for Direct Volume Rendering of Diffusion Tensor Fields Gordon Kindlmann, David Weinstein, David Hart Scientific Computing and Imaging Department of Computer Science, University of Utah {gk dmw
Mirrored LH Histograms for the Visualization of Material Boundaries
 Mirrored LH Histograms for the Visualization of Material Boundaries Petr Šereda 1, Anna Vilanova 1 and Frans A. Gerritsen 1,2 1 Department of Biomedical Engineering, Technische Universiteit Eindhoven,
Mirrored LH Histograms for the Visualization of Material Boundaries Petr Šereda 1, Anna Vilanova 1 and Frans A. Gerritsen 1,2 1 Department of Biomedical Engineering, Technische Universiteit Eindhoven,
Volume Illumination & Vector Field Visualisation
 Volume Illumination & Vector Field Visualisation Visualisation Lecture 11 Institute for Perception, Action & Behaviour School of Informatics Volume Illumination & Vector Vis. 1 Previously : Volume Rendering
Volume Illumination & Vector Field Visualisation Visualisation Lecture 11 Institute for Perception, Action & Behaviour School of Informatics Volume Illumination & Vector Vis. 1 Previously : Volume Rendering
CS GPU and GPGPU Programming Lecture 2: Introduction; GPU Architecture 1. Markus Hadwiger, KAUST
 CS 380 - GPU and GPGPU Programming Lecture 2: Introduction; GPU Architecture 1 Markus Hadwiger, KAUST Reading Assignment #2 (until Feb. 17) Read (required): GLSL book, chapter 4 (The OpenGL Programmable
CS 380 - GPU and GPGPU Programming Lecture 2: Introduction; GPU Architecture 1 Markus Hadwiger, KAUST Reading Assignment #2 (until Feb. 17) Read (required): GLSL book, chapter 4 (The OpenGL Programmable
ECE1778 Final Report MRI Visualizer
 ECE1778 Final Report MRI Visualizer David Qixiang Chen Alex Rodionov Word Count: 2408 Introduction We aim to develop a mobile phone/tablet based neurosurgical MRI visualization application with the goal
ECE1778 Final Report MRI Visualizer David Qixiang Chen Alex Rodionov Word Count: 2408 Introduction We aim to develop a mobile phone/tablet based neurosurgical MRI visualization application with the goal
A Ray-based Approach for Boundary Estimation of Fiber Bundles Derived from Diffusion Tensor Imaging
 A Ray-based Approach for Boundary Estimation of Fiber Bundles Derived from Diffusion Tensor Imaging M. H. A. Bauer 1,3, S. Barbieri 2, J. Klein 2, J. Egger 1,3, D. Kuhnt 1, B. Freisleben 3, H.-K. Hahn
A Ray-based Approach for Boundary Estimation of Fiber Bundles Derived from Diffusion Tensor Imaging M. H. A. Bauer 1,3, S. Barbieri 2, J. Klein 2, J. Egger 1,3, D. Kuhnt 1, B. Freisleben 3, H.-K. Hahn
Course Review. Computer Animation and Visualisation. Taku Komura
 Course Review Computer Animation and Visualisation Taku Komura Characters include Human models Virtual characters Animal models Representation of postures The body has a hierarchical structure Many types
Course Review Computer Animation and Visualisation Taku Komura Characters include Human models Virtual characters Animal models Representation of postures The body has a hierarchical structure Many types
Visualization Computer Graphics I Lecture 20
 15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 15, 2003 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 15, 2003 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University
![Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University](/thumbs/90/102611276.jpg) 15-462 Computer Graphics I Lecture 21 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
15-462 Computer Graphics I Lecture 21 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
Visualization. Images are used to aid in understanding of data. Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26]
![Visualization. Images are used to aid in understanding of data. Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26] Visualization. Images are used to aid in understanding of data. Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26]](/thumbs/74/70771954.jpg) Visualization Images are used to aid in understanding of data Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26] Tumor SCI, Utah Scientific Visualization Visualize large
Visualization Images are used to aid in understanding of data Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26] Tumor SCI, Utah Scientific Visualization Visualize large
11/1/13. Visualization. Scientific Visualization. Types of Data. Height Field. Contour Curves. Meshes
 CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 2.11] Jernej Barbic University of Southern California Scientific Visualization
CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 2.11] Jernej Barbic University of Southern California Scientific Visualization
Visualization. CSCI 420 Computer Graphics Lecture 26
 CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 11] Jernej Barbic University of Southern California 1 Scientific Visualization
CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 11] Jernej Barbic University of Southern California 1 Scientific Visualization
A Study of Medical Image Analysis System
 Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
Data Representation in Visualisation
 Data Representation in Visualisation Visualisation Lecture 4 Taku Komura Institute for Perception, Action & Behaviour School of Informatics Taku Komura Data Representation 1 Data Representation We have
Data Representation in Visualisation Visualisation Lecture 4 Taku Komura Institute for Perception, Action & Behaviour School of Informatics Taku Komura Data Representation 1 Data Representation We have
Lecture overview. Visualisatie BMT. Vector algorithms. Vector algorithms. Time animation. Time animation
 Visualisatie BMT Lecture overview Vector algorithms Tensor algorithms Modeling algorithms Algorithms - 2 Arjan Kok a.j.f.kok@tue.nl 1 2 Vector algorithms Vector 2 or 3 dimensional representation of direction
Visualisatie BMT Lecture overview Vector algorithms Tensor algorithms Modeling algorithms Algorithms - 2 Arjan Kok a.j.f.kok@tue.nl 1 2 Vector algorithms Vector 2 or 3 dimensional representation of direction
Visualization and Analysis of White Matter Structural Asymmetry in Diffusion Tensor MRI Data
 Magnetic Resonance in Medicine 51:140 147 (2004) Visualization and Analysis of White Matter Structural Asymmetry in Diffusion Tensor MRI Data Song Zhang, 1 Mark E. Bastin, 2 * David H. Laidlaw, 1 Saurabh
Magnetic Resonance in Medicine 51:140 147 (2004) Visualization and Analysis of White Matter Structural Asymmetry in Diffusion Tensor MRI Data Song Zhang, 1 Mark E. Bastin, 2 * David H. Laidlaw, 1 Saurabh
Interactive Volume Rendering of Thin Thread Structures within Multivalued Scientific Datasets
 SUBMITTED TO IEEE TVCG, APRIL, 2003 100 Interactive Volume Rendering of Thin Thread Structures within Multivalued Scientific Datasets Andreas Wenger, Daniel F. Keefe, Song Zhang, David H. Laidlaw Abstract
SUBMITTED TO IEEE TVCG, APRIL, 2003 100 Interactive Volume Rendering of Thin Thread Structures within Multivalued Scientific Datasets Andreas Wenger, Daniel F. Keefe, Song Zhang, David H. Laidlaw Abstract
Volume visualization. Volume visualization. Volume visualization methods. Sources of volume visualization. Sources of volume visualization
 Volume visualization Volume visualization Volumes are special cases of scalar data: regular 3D grids of scalars, typically interpreted as density values. Each data value is assumed to describe a cubic
Volume visualization Volume visualization Volumes are special cases of scalar data: regular 3D grids of scalars, typically interpreted as density values. Each data value is assumed to describe a cubic
Visualization of DTI fibers using hair-rendering techniques Peeters, T.H.J.M.; Vilanova Bartroli, A.; ter Haar Romenij, B.M.
 Visualization of DTI fibers using hair-rendering techniques Peeters, T.H.J.M.; Vilanova Bartroli, A.; ter Haar Romenij, B.M. Published in: Proceedings of the twelfth annual conference of the Advanced School
Visualization of DTI fibers using hair-rendering techniques Peeters, T.H.J.M.; Vilanova Bartroli, A.; ter Haar Romenij, B.M. Published in: Proceedings of the twelfth annual conference of the Advanced School
Coloring of DT-MRI Fiber Traces using Laplacian Eigenmaps
 Coloring of DT-MRI Fiber Traces using Laplacian Eigenmaps Anders Brun 1, Hae-Jeong Park 2, Hans Knutsson 3, and Carl-Fredrik Westin 1 1 Laboratory of Mathematics in Imaging, Brigham and Women s Hospital,
Coloring of DT-MRI Fiber Traces using Laplacian Eigenmaps Anders Brun 1, Hae-Jeong Park 2, Hans Knutsson 3, and Carl-Fredrik Westin 1 1 Laboratory of Mathematics in Imaging, Brigham and Women s Hospital,
CIS 467/602-01: Data Visualization
 CIS 467/60-01: Data Visualization Isosurfacing and Volume Rendering Dr. David Koop Fields and Grids Fields: values come from a continuous domain, infinitely many values - Sampled at certain positions to
CIS 467/60-01: Data Visualization Isosurfacing and Volume Rendering Dr. David Koop Fields and Grids Fields: values come from a continuous domain, infinitely many values - Sampled at certain positions to
Volume Rendering. Computer Animation and Visualisation Lecture 9. Taku Komura. Institute for Perception, Action & Behaviour School of Informatics
 Volume Rendering Computer Animation and Visualisation Lecture 9 Taku Komura Institute for Perception, Action & Behaviour School of Informatics Volume Rendering 1 Volume Data Usually, a data uniformly distributed
Volume Rendering Computer Animation and Visualisation Lecture 9 Taku Komura Institute for Perception, Action & Behaviour School of Informatics Volume Rendering 1 Volume Data Usually, a data uniformly distributed
Vector Visualization
 Vector Visualization Vector Visulization Divergence and Vorticity Vector Glyphs Vector Color Coding Displacement Plots Stream Objects Texture-Based Vector Visualization Simplified Representation of Vector
Vector Visualization Vector Visulization Divergence and Vorticity Vector Glyphs Vector Color Coding Displacement Plots Stream Objects Texture-Based Vector Visualization Simplified Representation of Vector
Splatting Illuminated Ellipsoids with Depth Correction
 Splatting Illuminated Ellipsoids with Depth Correction Stefan Gumhold University of Tübingen, WSI/GRIS Sand 14, 72076 Tübingen, Germany Email: gumhold@gris.uni-tuebingen.de Abstract Ellipsoids are important
Splatting Illuminated Ellipsoids with Depth Correction Stefan Gumhold University of Tübingen, WSI/GRIS Sand 14, 72076 Tübingen, Germany Email: gumhold@gris.uni-tuebingen.de Abstract Ellipsoids are important
A Generic Lie Group Model for Computer Vision
 A Generic Lie Group Model for Computer Vision Within this research track we follow a generic Lie group approach to computer vision based on recent physiological research on how the primary visual cortex
A Generic Lie Group Model for Computer Vision Within this research track we follow a generic Lie group approach to computer vision based on recent physiological research on how the primary visual cortex
A method of visualization of a brain neural pathway by using critical points and target regions
 Modelling in Medicine and Biology VII 309 A method of visualization of a brain neural pathway by using critical points and target regions A. Doi 1, H. Fujimura 2, M. Nagano 1, T. Inoue 3 & A. Ogawa 3 1
Modelling in Medicine and Biology VII 309 A method of visualization of a brain neural pathway by using critical points and target regions A. Doi 1, H. Fujimura 2, M. Nagano 1, T. Inoue 3 & A. Ogawa 3 1
Visualization of White Matter Tracts with Wrapped Streamlines
 Visualization of White Matter Tracts with Wrapped Streamlines Frank Enders Natascha Sauber Dorit Merhof Peter Hastreiter Neurocenter, Dept. of Neurosurgery Neurocenter, Dept. of Neurosurgery Neurocenter,
Visualization of White Matter Tracts with Wrapped Streamlines Frank Enders Natascha Sauber Dorit Merhof Peter Hastreiter Neurocenter, Dept. of Neurosurgery Neurocenter, Dept. of Neurosurgery Neurocenter,
Generating Fiber Crossing Phantoms Out of Experimental DWIs
 Generating Fiber Crossing Phantoms Out of Experimental DWIs Matthan Caan 1,2, Anne Willem de Vries 2, Ganesh Khedoe 2,ErikAkkerman 1, Lucas van Vliet 2, Kees Grimbergen 1, and Frans Vos 1,2 1 Department
Generating Fiber Crossing Phantoms Out of Experimental DWIs Matthan Caan 1,2, Anne Willem de Vries 2, Ganesh Khedoe 2,ErikAkkerman 1, Lucas van Vliet 2, Kees Grimbergen 1, and Frans Vos 1,2 1 Department
syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets
 syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets Julien Gervais; Lisa Chuah Siemens Healthcare, Magnetic
syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets Julien Gervais; Lisa Chuah Siemens Healthcare, Magnetic
Analysis of White Matter Fiber Tracts via Fiber Clustering and Parametrization
 Analysis of White Matter Fiber Tracts via Fiber Clustering and Parametrization Sylvain Gouttard 2,4, Isabelle Corouge 1,2, Weili Lin 3, John Gilmore 2, and Guido Gerig 1,2 1 Departments of Computer Science,
Analysis of White Matter Fiber Tracts via Fiber Clustering and Parametrization Sylvain Gouttard 2,4, Isabelle Corouge 1,2, Weili Lin 3, John Gilmore 2, and Guido Gerig 1,2 1 Departments of Computer Science,
HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
Applications of Explicit Early-Z Culling
 Applications of Explicit Early-Z Culling Jason L. Mitchell ATI Research Pedro V. Sander ATI Research Introduction In past years, in the SIGGRAPH Real-Time Shading course, we have covered the details of
Applications of Explicit Early-Z Culling Jason L. Mitchell ATI Research Pedro V. Sander ATI Research Introduction In past years, in the SIGGRAPH Real-Time Shading course, we have covered the details of
Triangle Rasterization
 Triangle Rasterization Computer Graphics COMP 770 (236) Spring 2007 Instructor: Brandon Lloyd 2/07/07 1 From last time Lines and planes Culling View frustum culling Back-face culling Occlusion culling
Triangle Rasterization Computer Graphics COMP 770 (236) Spring 2007 Instructor: Brandon Lloyd 2/07/07 1 From last time Lines and planes Culling View frustum culling Back-face culling Occlusion culling
An Efficient Approach for Emphasizing Regions of Interest in Ray-Casting based Volume Rendering
 An Efficient Approach for Emphasizing Regions of Interest in Ray-Casting based Volume Rendering T. Ropinski, F. Steinicke, K. Hinrichs Institut für Informatik, Westfälische Wilhelms-Universität Münster
An Efficient Approach for Emphasizing Regions of Interest in Ray-Casting based Volume Rendering T. Ropinski, F. Steinicke, K. Hinrichs Institut für Informatik, Westfälische Wilhelms-Universität Münster
MITK-DI. A new Diffusion Imaging Component for MITK. Klaus Fritzsche, Hans-Peter Meinzer
 MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
Network connectivity via inference over curvature-regularizing line graphs
 Network connectivity via inference over curvature-regularizing line graphs Asian Conference on Computer Vision Maxwell D. Collins 1,2, Vikas Singh 2,1, Andrew L. Alexander 3 1 Department of Computer Sciences
Network connectivity via inference over curvature-regularizing line graphs Asian Conference on Computer Vision Maxwell D. Collins 1,2, Vikas Singh 2,1, Andrew L. Alexander 3 1 Department of Computer Sciences
Fundamental Algorithms
 Fundamental Algorithms Fundamental Algorithms 3-1 Overview This chapter introduces some basic techniques for visualizing different types of scientific data sets. We will categorize visualization methods
Fundamental Algorithms Fundamental Algorithms 3-1 Overview This chapter introduces some basic techniques for visualizing different types of scientific data sets. We will categorize visualization methods
Scalar Data. Visualization Torsten Möller. Weiskopf/Machiraju/Möller
 Scalar Data Visualization Torsten Möller Weiskopf/Machiraju/Möller Overview Basic strategies Function plots and height fields Isolines Color coding Volume visualization (overview) Classification Segmentation
Scalar Data Visualization Torsten Möller Weiskopf/Machiraju/Möller Overview Basic strategies Function plots and height fields Isolines Color coding Volume visualization (overview) Classification Segmentation
Direct and Comparative Visualization Techniques for HARDI Data
 Direct and Comparative Visualization Techniques for HARDI Data Rik Sonderkamp c 2010 Rik Sonderkamp Delft University of Technology Final Thesis Direct and Comparative Visualization Techniques for HARDI
Direct and Comparative Visualization Techniques for HARDI Data Rik Sonderkamp c 2010 Rik Sonderkamp Delft University of Technology Final Thesis Direct and Comparative Visualization Techniques for HARDI
Volume Graphics Introduction
 High-Quality Volume Graphics on Consumer PC Hardware Volume Graphics Introduction Joe Kniss Gordon Kindlmann Markus Hadwiger Christof Rezk-Salama Rüdiger Westermann Motivation (1) Motivation (2) Scientific
High-Quality Volume Graphics on Consumer PC Hardware Volume Graphics Introduction Joe Kniss Gordon Kindlmann Markus Hadwiger Christof Rezk-Salama Rüdiger Westermann Motivation (1) Motivation (2) Scientific
FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING. Francesca Pizzorni Ferrarese
 FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING Francesca Pizzorni Ferrarese Pipeline overview WM and GM Segmentation Registration Data reconstruction Tractography
FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING Francesca Pizzorni Ferrarese Pipeline overview WM and GM Segmentation Registration Data reconstruction Tractography
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy
 The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
Vector Visualization. CSC 7443: Scientific Information Visualization
 Vector Visualization Vector data A vector is an object with direction and length v = (v x,v y,v z ) A vector field is a field which associates a vector with each point in space The vector data is 3D representation
Vector Visualization Vector data A vector is an object with direction and length v = (v x,v y,v z ) A vector field is a field which associates a vector with each point in space The vector data is 3D representation
MITK-DI. A new Diffusion Imaging Component for MITK. Klaus Fritzsche, Hans-Peter Meinzer
 MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
Clipping. CSC 7443: Scientific Information Visualization
 Clipping Clipping to See Inside Obscuring critical information contained in a volume data Contour displays show only exterior visible surfaces Isosurfaces can hide other isosurfaces Other displays can
Clipping Clipping to See Inside Obscuring critical information contained in a volume data Contour displays show only exterior visible surfaces Isosurfaces can hide other isosurfaces Other displays can
Motion Tracking and Event Understanding in Video Sequences
 Motion Tracking and Event Understanding in Video Sequences Isaac Cohen Elaine Kang, Jinman Kang Institute for Robotics and Intelligent Systems University of Southern California Los Angeles, CA Objectives!
Motion Tracking and Event Understanding in Video Sequences Isaac Cohen Elaine Kang, Jinman Kang Institute for Robotics and Intelligent Systems University of Southern California Los Angeles, CA Objectives!
Visualization Computer Graphics I Lecture 20
 15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] November 20, 2003 Doug James Carnegie Mellon University http://www.cs.cmu.edu/~djames/15-462/fall03
15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] November 20, 2003 Doug James Carnegie Mellon University http://www.cs.cmu.edu/~djames/15-462/fall03
Data Visualization (DSC 530/CIS )
 Data Visualization (DSC 530/CIS 60-0) Isosurfaces & Volume Rendering Dr. David Koop Fields & Grids Fields: - Values come from a continuous domain, infinitely many values - Sampled at certain positions
Data Visualization (DSC 530/CIS 60-0) Isosurfaces & Volume Rendering Dr. David Koop Fields & Grids Fields: - Values come from a continuous domain, infinitely many values - Sampled at certain positions
Integrated Visualization of Diffusion Tensor and Functional MRI
 University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange Masters Theses Graduate School 8-2003 Integrated Visualization of Diffusion Tensor and Functional MRI Nathaniel Richard
University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange Masters Theses Graduate School 8-2003 Integrated Visualization of Diffusion Tensor and Functional MRI Nathaniel Richard
Part I: Theoretical Background and Integration-Based Methods
 Large Vector Field Visualization: Theory and Practice Part I: Theoretical Background and Integration-Based Methods Christoph Garth Overview Foundations Time-Varying Vector Fields Numerical Integration
Large Vector Field Visualization: Theory and Practice Part I: Theoretical Background and Integration-Based Methods Christoph Garth Overview Foundations Time-Varying Vector Fields Numerical Integration
Blood Particle Trajectories in Phase-Contrast-MRI as Minimal Paths Computed with Anisotropic Fast Marching
 Blood Particle Trajectories in Phase-Contrast-MRI as Minimal Paths Computed with Anisotropic Fast Marching Michael Schwenke 1, Anja Hennemuth 1, Bernd Fischer 2, Ola Friman 1 1 Fraunhofer MEVIS, Institute
Blood Particle Trajectories in Phase-Contrast-MRI as Minimal Paths Computed with Anisotropic Fast Marching Michael Schwenke 1, Anja Hennemuth 1, Bernd Fischer 2, Ola Friman 1 1 Fraunhofer MEVIS, Institute
Vector Field Visualisation
 Vector Field Visualisation Computer Animation and Visualization Lecture 14 Institute for Perception, Action & Behaviour School of Informatics Visualising Vectors Examples of vector data: meteorological
Vector Field Visualisation Computer Animation and Visualization Lecture 14 Institute for Perception, Action & Behaviour School of Informatics Visualising Vectors Examples of vector data: meteorological
A Novel Contrast for DTI Visualization for Thalamus Delineation
 A Novel Contrast for DTI Visualization for Thalamus Delineation Xian Fan a, Meredith Thompson a,b, John A. Bogovic a, Pierre-Louis Bazin c, Jerry L. Prince a,c a Johns Hopkins University, Baltimore, MD,
A Novel Contrast for DTI Visualization for Thalamus Delineation Xian Fan a, Meredith Thompson a,b, John A. Bogovic a, Pierre-Louis Bazin c, Jerry L. Prince a,c a Johns Hopkins University, Baltimore, MD,
45 µm polystyrene bead embedded in scattering tissue phantom. (a,b) raw images under oblique
 Phase gradient microscopy in thick tissue with oblique back-illumination Tim N Ford, Kengyeh K Chu & Jerome Mertz Supplementary Figure 1: Comparison of added versus subtracted raw OBM images 45 µm polystyrene
Phase gradient microscopy in thick tissue with oblique back-illumination Tim N Ford, Kengyeh K Chu & Jerome Mertz Supplementary Figure 1: Comparison of added versus subtracted raw OBM images 45 µm polystyrene
3D vector fields. Contents. Introduction 3D vector field topology Representation of particle lines. 3D LIC Combining different techniques
 3D vector fields Scientific Visualization (Part 9) PD Dr.-Ing. Peter Hastreiter Contents Introduction 3D vector field topology Representation of particle lines Path lines Ribbons Balls Tubes Stream tetrahedra
3D vector fields Scientific Visualization (Part 9) PD Dr.-Ing. Peter Hastreiter Contents Introduction 3D vector field topology Representation of particle lines Path lines Ribbons Balls Tubes Stream tetrahedra
9. Three Dimensional Object Representations
 9. Three Dimensional Object Representations Methods: Polygon and Quadric surfaces: For simple Euclidean objects Spline surfaces and construction: For curved surfaces Procedural methods: Eg. Fractals, Particle
9. Three Dimensional Object Representations Methods: Polygon and Quadric surfaces: For simple Euclidean objects Spline surfaces and construction: For curved surfaces Procedural methods: Eg. Fractals, Particle
Direct Registration of White Matter Tractographies with Application to Atlas Construction
 Direct Registration of White Matter Tractographies with Application to Atlas Construction Arnaldo Mayer, Hayit Greenspan Medical image processing lab, Tel-Aviv University, Ramat-Aviv, Israel arnakdom@post.tau.ac.il,
Direct Registration of White Matter Tractographies with Application to Atlas Construction Arnaldo Mayer, Hayit Greenspan Medical image processing lab, Tel-Aviv University, Ramat-Aviv, Israel arnakdom@post.tau.ac.il,
Scalar Data. CMPT 467/767 Visualization Torsten Möller. Weiskopf/Machiraju/Möller
 Scalar Data CMPT 467/767 Visualization Torsten Möller Weiskopf/Machiraju/Möller Overview Basic strategies Function plots and height fields Isolines Color coding Volume visualization (overview) Classification
Scalar Data CMPT 467/767 Visualization Torsten Möller Weiskopf/Machiraju/Möller Overview Basic strategies Function plots and height fields Isolines Color coding Volume visualization (overview) Classification
THE brain is a massively interconnected organ. Individual
 IEEE TRANSACTIONS ON VISUALIZATION AND COMPUTER GRAPHICS 1 Exploring Connectivity of the Brain s White Matter with Dynamic Queries Anthony Sherbondy, David Akers, Rachel Mackenzie, Robert Dougherty, and
IEEE TRANSACTIONS ON VISUALIZATION AND COMPUTER GRAPHICS 1 Exploring Connectivity of the Brain s White Matter with Dynamic Queries Anthony Sherbondy, David Akers, Rachel Mackenzie, Robert Dougherty, and
Adaptive Point Cloud Rendering
 1 Adaptive Point Cloud Rendering Project Plan Final Group: May13-11 Christopher Jeffers Eric Jensen Joel Rausch Client: Siemens PLM Software Client Contact: Michael Carter Adviser: Simanta Mitra 4/29/13
1 Adaptive Point Cloud Rendering Project Plan Final Group: May13-11 Christopher Jeffers Eric Jensen Joel Rausch Client: Siemens PLM Software Client Contact: Michael Carter Adviser: Simanta Mitra 4/29/13
Constrained Reconstruction of Sparse Cardiac MR DTI Data
 Constrained Reconstruction of Sparse Cardiac MR DTI Data Ganesh Adluru 1,3, Edward Hsu, and Edward V.R. DiBella,3 1 Electrical and Computer Engineering department, 50 S. Central Campus Dr., MEB, University
Constrained Reconstruction of Sparse Cardiac MR DTI Data Ganesh Adluru 1,3, Edward Hsu, and Edward V.R. DiBella,3 1 Electrical and Computer Engineering department, 50 S. Central Campus Dr., MEB, University
NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2013 May 04.
 NIH Public Access Author Manuscript Published in final edited form as: Med Image Comput Comput Assist Interv. 2005 ; 8(0 1): 180 187. A Hamilton-Jacobi-Bellman approach to high angular resolution diffusion
NIH Public Access Author Manuscript Published in final edited form as: Med Image Comput Comput Assist Interv. 2005 ; 8(0 1): 180 187. A Hamilton-Jacobi-Bellman approach to high angular resolution diffusion
Unveiling the White Matter Microstructure in 22q11.2 Deletion Syndrome. Julio Villalón
 Unveiling the White Matter Microstructure in 22q11.2 Deletion Syndrome Julio Villalón What is 22q11.2 Deletion Syndrome? Yes, there is a mouse model!! Modified from Meechan et al. 2015 How are neurons
Unveiling the White Matter Microstructure in 22q11.2 Deletion Syndrome Julio Villalón What is 22q11.2 Deletion Syndrome? Yes, there is a mouse model!! Modified from Meechan et al. 2015 How are neurons
ISMI: A classification index for High Angular Resolution Diffusion Imaging
 ISMI: A classification index for High Angular Resolution Diffusion Imaging D. Röttger, D. Dudai D. Merhof and S. Müller Institute for Computational Visualistics, University of Koblenz-Landau, Germany Visual
ISMI: A classification index for High Angular Resolution Diffusion Imaging D. Röttger, D. Dudai D. Merhof and S. Müller Institute for Computational Visualistics, University of Koblenz-Landau, Germany Visual
Quantitative MRI of the Brain: Investigation of Cerebral Gray and White Matter Diseases
 Quantities Measured by MR - Quantitative MRI of the Brain: Investigation of Cerebral Gray and White Matter Diseases Static parameters (influenced by molecular environment): T, T* (transverse relaxation)
Quantities Measured by MR - Quantitative MRI of the Brain: Investigation of Cerebral Gray and White Matter Diseases Static parameters (influenced by molecular environment): T, T* (transverse relaxation)
Scientific Visualization. CSC 7443: Scientific Information Visualization
 Scientific Visualization Scientific Datasets Gaining insight into scientific data by representing the data by computer graphics Scientific data sources Computation Real material simulation/modeling (e.g.,
Scientific Visualization Scientific Datasets Gaining insight into scientific data by representing the data by computer graphics Scientific data sources Computation Real material simulation/modeling (e.g.,
Atelier 2 : Calcul Haute Performance et Sciences du Vivant Forum er juillet, Paris, France
 From Diffusion MR Image Analysis to Whole Brain Connectivity Simulation Jean-Philippe Thiran EPFL Lausanne, Switzerland EPFL - Lausanne HPC in life sciences at EPFL The Blue Brain project: create a biologically
From Diffusion MR Image Analysis to Whole Brain Connectivity Simulation Jean-Philippe Thiran EPFL Lausanne, Switzerland EPFL - Lausanne HPC in life sciences at EPFL The Blue Brain project: create a biologically
Flow Visualisation 1
 Flow Visualisation Visualisation Lecture 13 Institute for Perception, Action & Behaviour School of Informatics Flow Visualisation 1 Flow Visualisation... so far Vector Field Visualisation vector fields
Flow Visualisation Visualisation Lecture 13 Institute for Perception, Action & Behaviour School of Informatics Flow Visualisation 1 Flow Visualisation... so far Vector Field Visualisation vector fields
White Pixel Artifact. Caused by a noise spike during acquisition Spike in K-space <--> sinusoid in image space
 White Pixel Artifact Caused by a noise spike during acquisition Spike in K-space sinusoid in image space Susceptibility Artifacts Off-resonance artifacts caused by adjacent regions with different
White Pixel Artifact Caused by a noise spike during acquisition Spike in K-space sinusoid in image space Susceptibility Artifacts Off-resonance artifacts caused by adjacent regions with different
