Quantitative Evaluation of Skull Stripping Techniques on Magnetic Resonance Images
|
|
|
- Maude Scott
- 6 years ago
- Views:
Transcription
1 Proceedings of the World Congress on Electrical Engineering and Computer Systems and Science (EECSS 2015) Barcelona, Spain July 13-14, 2015 Paper No. 310 Quantitative Evaluation of Skull Stripping Techniques on Magnetic Resonance Images Farahnaz Hosseini, Hossein Ebrahimpourkomleh Department of Computer Engineering,University of Kashan 6KM ghotbravandiblvd,kashan, Iran Mehrnaz KhodamHazrati Department of Electrical and Computer Engineering,University of Florida Gainesville, Fl. USA Abstract -Magnetic resonance (MR) brain images are often employed to diagnose diseases or to detect abnormalities such as brain tumors inhuman beings. Whole brain segmentation, also known as skull stripping, is an essential pre-processing step to reduce unwanted information and to remove non-brain backgrounds from these images. Automated and intelligent skull stripping techniques can facilitate and expedite the entire process of extracting accurate diagnostic information from MR images. This paper presents a nonparametric approach to quantitatively evaluate the skull stripping methods such as brain surface extractor (BSE), brain extraction tool (BET) and a new approach based on the wavelet transform. We applied these methods to three datasets obtained from Sina and Beheshti hospitals in Iran for detecting and eliminating the skull region. By calculating the Hausdorff distance between each image and the manually segmented image as a gold standard, we provide a qualitative framework to compare the performance of the three algorithms. Keywords: Human brain images, MRI, Skull stripping, Hausdorff distance, Wavelet transform, BET, BSE. 1. Introduction Magnetic Resonance Imaging (MRI) is an extremely valuable tool for detection of brain abnormalities such as brain tumors. Artefacts and undesired tissues affect the quality of processing and may lead to diagnostic confusion. Thus, one of the important steps in the processing of brain images is skull stripping where the brain tissue is completely segmented from the skull. It is done for the purpose of clearing away non-brain backgrounds and reducing unwanted information from the MR images. Since the manual segmentation is very time consuming and prone to errors, various methods have been developed to automatically remove extra-cerebral tissues without human intervention. In recent years, several semi-automatic and automatic skull stripping techniques were introduced in the literature. Approaches such as brain extraction tool (BET) (Smith, 2012), the brain surface extractor (BSE) (Shattuck et al, 2001), and the hybrid watershed (HWA) (Segonne et al, 2004) have been applied widely for this purpose. In general, the automatic skull stripping can be categorized into three major methods: morphology, deformable model and intensity based methods. There are a few certain metrics that are used to evaluate the performance of the segmentation algorithms. The segmentation accuracy indices such as Tanimoto coefficient or correlation are usually used to evaluate the general performance of a method. A threshold value is mainly applied using the Otsu method (Otsu, 1979) which minimizes the within-class variance between two classes. We conducted a comprehensive literature review on the available methods to study the comparison factor
2 Atkins et al. (1998) proposed a method based on the regionthat integrates anisotropic diffusion filtering, gray level thresholding, binary morphology and active contour models snakes. Their method created a mask based on a threshold that was determined by fitting a single Gaussian curve to the histogram of the image.two images, weight T1 and T2, was tested. The accuracy of their method depends on the threshold value, and with choosing the threshold equal to 0.5 for T1 weighted images, their method worked better than the image intensity alone. Boesen et al. (2003) compared four methods of skull stripping (McStrip, SPM, BET, and BSE). Based on their comparison, BSE and BET were faster than SPM and McStrip. Skull stripping in SPM is performed by masking out the brain region after calculating a threshold based on both gray and white matter of the brain image. When compared to McStrip manual operation, it showed higher accuracy on T1 images. A technique was introduced by Ségonneet al. (2004)for skull stripping based on watershed algorithm and deformable surface. They computed different coefficients to compare the performance of various segmentation techniques. Risk evaluation and Jaccard similarity were used to evaluate their method which is a measure of probability of a miss and probability of false detection. Later, Ségonneet al (2014) provided a very simple but powerful method for skull removal. Their method is based on the combination of watershed algorithm and deformable to determine the boundary of the brain in the T1 images. Forkert et al. (2008) introduced a method which contains three main steps: 1) The first phase contains pre-processing and noise removal, 2) In the second phase, extracted boundary points of the brain are calculated, 3) The third phase is the correction method. They used 18 data sets automatically and the average segmentation accuracy was 99.18%. Somasundaramet al. (2010) came up with a method based on clustering for detecting the brain boundaries inside the skull. A 2D growing scheme was used to connect together the clusters and also remove the skull area.prasad et al (2011) applied deformable organism scheme to successfully stripped skull using images T1 weight. Speier et al (2011) used Robex method for skull stripping. Their results showed this method was better than BSE, BET and HWA. They used T1 weight images taken from Glioblastoma patients. Wang et al (2011) proposed a method, a combination of Atlas, and a deformable method. Galdameset al. (2011) presented a skull stripping based on deformable model and histogram analysis. Their method is applied in two steps: pre segmentation and the deformable method which is based on thresholds and morphological operator. Balanet al. (2012) proposed a method based on histogram analysis and compared the segmentation accuracy between their proposed method and two widely used techniques, namely BSE and BET. Based on this factor, they reported that their proposed method outperforms these methods. Pratibha(2013) compared two methods of skull stripping (BET, BSE) over two datasets containing T1, T2. They showed that BSE outperforms BET in segmentation of brain images. In 2013, Roy et al. presented a simple and effective automatic skull removal method that comprises of both statistical and computational elements. This paper evaluatesa modified version of fully automated parameter free approach proposed by (Roy et al, 2013) and compared it with already established methods using three different datasets. The proposed method is able to detect the boundary of the brain and to separate brain from skull and background areas with a high acceptance rate. The algorithm is based on wavelet transformation and the convex hull algorithm. The aim of this work is to present a quantitative measure to evaluate the available algorithms. Here, we suggest that the Hausdorff distance between each image and a gold standard provides useful information to quantitatively evaluate each method. Finallywe compared the performance of the above algorithm as well as BSE and BET methods in a qualitative way. 2. Datasets Three different datasets were used in this study. These databases contain 21, 20 and 11 slices of brain MR images, respectively. They are standard datasets in the axial imaging which have been provided in Sina and Beheshti hospitals of Iran
3 3. Skull Stripping Algorithms Brain Extraction Tools This paper evaluated and compared three different types of brain extraction tools, namely, brain surface extraction (BSE), brain extraction tool (BET), and a modified version of the recently introduced wavelet-based approach linked to convex hull algorithm Brain-surface Extraction Software (BSE) BSE applies a series of mathematical morphological operators to the edge map found in the edge detection step before detecting the brain region. The first step is called erosion with a 3D cross element, which expands the edges into each voxel that shares a face with an edge voxel. The result is that small connections less than 2 voxels wide will break. For images with resolution of 1mm, this corresponds to a 2mm structure. This is sufficient to erode most cranial nerves and to eliminate most noise related connections between the brain and scalp portions of the MRI. With finer resolution images, it may be necessary to increase the erosion size up to 2 in order to separate the brain. In the next step, BSE segments the eroded edge map into several connected regions that are bounded by the edges. BSE then analyses these regions to select a candidate brain region based on the size of the connected region, the average intensity of the connected region and the position of the centroid of the connected region. Here BrainSuite software is used to extract and parameterize the inner and outer surfaces of the cerebral cortex and to segment and label grey and white matter structures. This software can automatically process magnetic resonance images. This tool can remove none brain particles from the brain (Web-1) Brain Extraction Tool Brain Extraction Tool or BET was introduced by Smith (2002) as an accurate and robust method which utilizes an automatically evolving deformable model to fit the brains surface. The algorithm finds intensity values in images and defines a threshold for separating brain and none brain areas. By calculating the intensity histogram, values with the highest and lowest intensity in images are found and consequently a threshold is defined. In the next step, a triangular tessellation of a sphere s surface is initialized inside the brain, and allowed to slowly deform, one vertex at a time.if this step doesn t clean surface satisfactory, then the algorithm runs again by a higher smoothness constraint. Finally, the outer surface of the skull is removed (Pratibha, 2013).In this study, we used Mango (Multi-image Analysis GUI)which is a famous viewer and analytical tool for segmentation of brain. The BET plug in installed in this program was used to strip of skull in our datasets Wavelet-based Approach We have modified the algorithm introduced in (Huttenlocher et al, 1993),in order to find the best stripped image based on a quantitative measure along with brain extraction tools and brain surface extraction methods. This algorithm contains six steps: Bineralization, wavelet transformation, interpolation, labelling and filtering algorithm which is composed of convex hull. In this study, we compared the effect of different parameters to determine the optimum setting based on a quantitative measure. These steps are shown in Figure 1. A) Binary image In the first step, the original image is binerized in order to remove background and grey levels. Bineralization decreases the size of the image and therefore speeds up the process of identifying the image in the next steps. B) Applying wavelet algorithm In the original algorithm two-dimensional wavelet decomposition was applied using db1 wavelet. We studied a wide range of mother wavelets based on the original brain image and then applied the wavelet algorithm in two levels. It causes the unwanted information to be blurred in the image and also reduces the dimensions of the image
4 C) Image interpolation The image obtained after wavelet transformation has smaller size compared to the original image. In order to retrieve the original image size an interpolation step is required. The next step for identify the parts of the skull is labelling. D) Labelling This section is supposed to remove the additional places in the image. We have labelled the parts of the image so that all the adjacent parts and pieces are numbered. It also finds the largest label and sets it to zero. In other words, the brain is the greatest piece which is kept and the small pieces around the skull are eliminated. E) Convex hull algorithm In the last step, after labelling, the edges of the image that are soft get removed. We applied the convex hull algorithm on the image to perfectly extract the mask. Figure 1 demonstrates the algorithm of the skull stripping algorithm based on (roy et al, 2013). Fig. 1.Scheme of the skull stripping algorithm based on (Roy et al, 2013) 4. Convex Hull Algorithm The smallest convex set which comprises S in the Euclidean space is convex hull of the set Sof points. As a formal situation, the convex hull can be described as cross point all convex sets which consist S or as the convexes combinations of all dots in S. It can be visualized as a rubber stripe pulled around Swhen S is a bounded subset (Berg et al, 2000). According to the second description, convex hulls can be generalized by stretching from Euclidean spaces to true vector spaces (Knuth et al, 1992)
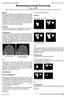
5 5. Hausdorff Distance Hausdorff distance is a metric for gauging the closeness between two subsets in a metric space. Based on this definition, we have compared the distance between cleaned images and a reference image in our datasets. Sample images are shown in the result section. The calculated distance between two images can be interpreted as a similarity index. The mathematical definition is as follows (Agarwal et al, 2010): Consider two sets A and B.A*B R that Distance between components in the sets of A and B are considered. For a A every member of the set B is considered and also for every member of the set B, b B to the set Hausdorff distance is calculated. The Hausdorff distance directional between A and B is defined as: h(a, B) = sup a A d(a, B) And the Hausdorff distance between image A and image B is calculated as follows: H(A, B) = max [h(a, B), h(b, A)] The distance of each component calculates their Hausdorff with members of the other set, which is the minimum Hausdorff distance. One of the weaknesses of Hausdorff distance is being sensitive to outliers in the data. One of the possible methods of root-mean-square is the Hausdorff distance between images A and B which is determined by:. h R (A, B) = (d2 (a, B)da) A. da A 1/2 Where A is the automatically skull stripped region and B is the brain region of the manually stripped image. In order to calculate the maximum distance: h R (A, B) = max{h R (A, B), h R (B, A)} Finally, Hausdorff distance is calculated: h s (A, B) =. (d(a, B)da) A. A da 6. Result and Discussion In this article, the skull region and non-brain background have been removed by applying three selected methods. Skull stripping results are illustrated in Figure 2 for sample images from each dataset. The results of BSE show that some of non-brain tissues are not skull stripped. According to given parameters as can be visualized in Figure 2, BET removed the skull of the brain better than BSE. Table 1 describe metrics used for the following tests. Table 2 shows comparison metrics for BET and BSE and db1-wavelet on Hausdorff distance. Table 1 shows the parameters used for BSE and BET. In the first dataset, the db1 wavelet method has not been able to remove the skull perfectly from all images and also as shown above, it has not done perfectly the removal of the skull in the second dataset which is one of the drawbacks of this method. As shown in Table 2, the average Hausdorff distances using BET method are considerably smaller than the other two methods in dataset 2 which shows this method has successfully removed the skull. When comparing these three methods, it can be seen that BET outperforms BSE and db1 wavelet in segmentation of brain image. Figure 3 shows the comparison of brain extraction tools by the averaged Hausdorff distances over all three datasets. The results show that 310-5
6 the wavelet based method was not able to fully detect and remove the skull in some images, however, the skull in some images was perfectly stripped. Main images BET BSE Wavelet-based Fig. 2. Illustration of skull stripping results on sample images from each dataset. First column shows the input images (I 1,2,3 ) and the second column shows the results of BET method and the third column shows BSE results and finally the last column shows the results of wavelet-based approach. Table.1. Parameters used in the comparative evaluation Parameters Dataset 1 Dataset 2 Dataset 3 Diffuse iterations Diffuse constant Edge constant Erosion size Fractional intensity Threshold Table. 2. Comparison of BET, BSE and Wavelet method based on Hausdorff distance (mean value) Data BET BSE Wavelet Image I Image I Image I
7 BET BSE Wavelet Fig. 3. Comparison of brain extraction tools by the averaged Hausdorff distances over all three datasets 7. Conclusion Brain segmentation is widely used as the main step in analyzing brain images. Fully automated skull stripping helps extract fast and accurate diagnostic information; however, it can only be realized through robust and reliable processing algorithms. This paper evaluated and compared three different types of brain extraction tools, namely, brain surface extraction (BSE), brain extraction tool (BET), and a modified version of the recently introduced wavelet-based approach linked to convex hull algorithm (Roy et al, 2013). Previously, we have studied the effect of using different mother wavelets in the latter method. Ultimately, we have selected a db1 mother wavelet. To evaluate the performance of the segmentation algorithms we used the Hausdorff distance as a measure of similarity between the segmented image and the reference image. In average the waveletbased approach showed the lowest Hausdorff distance in dataset 1 and a comparable value in two other datasets. There are a few segmentation accuracy indices such as Tanimoto coefficient or correlation that are usually used to evaluate the general performance of a method. A threshold value is required in most cases. Our measure is a parameter free index that can be used as a measure of segmentation accuracy on MR segmented images. References Agarwal, P. K., Peled, S. H., Sharir, M., & Wang, Y. (2010). Hausdor_ Distance Under Translation For Pointsand Balls. Journal of the ACM, V(N), Atkins, M.S., & Mackiewich, B.T. (1998). Fully Automatic Segmentation Of The Brain In MRI. IEEE Trans. Med. Imaging, 17, Balan, A.G. R., Marcela, A.J.M.T., Ribeiro, X., Marques, P. M. A., & Traina, C. (2012). Smart Histogram Analysis Applied To The Skull-Stripping Problem In T1-Weighted MRI. Computers in Biology and Medicine, 42, De Berg, M., van Kreveld, M., Overmars, M., & Schwarzkopf, O. (2000). Computational Geometry: Algorithms and Applications. Springer Boesen, K., Rehm, K., Schaper, K., Stoltzner, S., Woods, R., & Rottenberg, D. (2003). Quantitative Comparison of Four Brain Extraction Algorithms. Conf. on Functional Mapping of the Human Brain, 19, (2). Forkert, N. D., Säring, D., Fiehler, J., Illies, T., Färber, M., Möller, D., & Handels, H. (2008). Fully Automatic Skull-Stripping in 3D Time-of-Flight MRI Image Sequences. Eurographics Workshop on Visual Computing for Biomedicine. Galdamesa, F.J., Jaillet, D.F., & Pereza, C.A. (2011). An Accurate Skull Stripping Method Based on Simplex Meshes and Histogram Analysis in Magnetic Resonance Images. Rapport de recherche RR- LIRIS-019. Huttenlocher, D.P., Klanderman, G. A., & Rucklidge, W. J. (1993). Comparing Images Using the Hausdorff Distance. IEEE Transactions on pattern analysis and machine intelligence, 15(9)
8 Kass, M., Witkin, A., & Terzopoulos, D. (1987). Snakes: Active Contour Models. Int. J. Comput. Vision 1, Knuth, D.E., & Hulls, A. (1992). Lecture Notes in Computer Science, 606. Berlin: Springer Verlag. ISBN: Pratibha. (2013). Performance Evaluation of Skull Stripping Methods and Tools. International Journal of Computer Trends and Technology, 4(9). Prasad, G., Joshi, A. A., Thompson, P. M., Toga, A.W., Shattuck, D. W., & Terzopoulos, D. (2011). Skull Stripping Eith Deformable Organisms. IEEE Journal, Roy, S., Nag, S., Maitra, I. K., & Bandyopadhyay, S. K. (2013). Artefact Removal and Skull Elimination from MRI of Brain Image. International Journal of Scientific & Engineering Research, 4(6), ISSN Ségonne, F., Dale, A. M., Busa, E., Glessner, M., Salat, D., Hahn, H. K., & Fischla, B. (2004). A Hybrid Approach To The Skull Stripping Problem In MRI. ELSEVIER NeuroImage, 22, Shattuck, D.W., Sandor-Leahy, S.R., Schaper, K.A., Rottenberg, D.A., & Leahy, R.M. (2001). Magnetic Resonance Image Tissue Classification Using A Partial Volume Model. NeuroImage, 13(5), Smith, S.M. (2002). Fast Robust Automated Brain Extraction. Human Brain Mapping, 17(3), Somasundaram, K., & Siva Shankar, R. (2010). Skull Stripping of MRI Using Clustering and 2D Region Growing Method. Image Processing NCIMP. Speier, W., Iglesias, J. E., El-Kara, L., Tu, Z., & Arnold, C. (2011). Robust Skull Stripping of Clinical Glioblastoma Multiforme Data. G. Fichtinger, A. Martel and T. MICCAI, Part III, LNCS 6893, Wang, Y., Nie, J., Yap, P., Shi, F., Guo, L., & Shen, D. (2011). Robust Deformable-Surface-Based Skull- Stripping for Large-Scale Studies. G. Fichtinger, A. Martel, and T. Peters MICCAI, Part III, LNCS 6893, Web sites: Web-1:
Brain Portion Peeling from T2 Axial MRI Head Scans using Clustering and Morphological Operation
 159 Brain Portion Peeling from T2 Axial MRI Head Scans using Clustering and Morphological Operation K. Somasundaram Image Processing Lab Dept. of Computer Science and Applications Gandhigram Rural Institute
159 Brain Portion Peeling from T2 Axial MRI Head Scans using Clustering and Morphological Operation K. Somasundaram Image Processing Lab Dept. of Computer Science and Applications Gandhigram Rural Institute
AN EFFICIENT SKULL STRIPPING ALGORITHM USING CONNECTED REGIONS AND MORPHOLOGICAL OPERATION
 AN EFFICIENT SKULL STRIPPING ALGORITHM USING CONNECTED REGIONS AND MORPHOLOGICAL OPERATION Shijin Kumar P. S. 1 and Dharun V. S. 2 1 Department of Electronics and Communication Engineering, Noorul Islam
AN EFFICIENT SKULL STRIPPING ALGORITHM USING CONNECTED REGIONS AND MORPHOLOGICAL OPERATION Shijin Kumar P. S. 1 and Dharun V. S. 2 1 Department of Electronics and Communication Engineering, Noorul Islam
City, University of London Institutional Repository
 City Research Online City, University of London Institutional Repository Citation: Doan, H., Slabaugh, G.G., Unal, G.B. & Fang, T. (2006). Semi-Automatic 3-D Segmentation of Anatomical Structures of Brain
City Research Online City, University of London Institutional Repository Citation: Doan, H., Slabaugh, G.G., Unal, G.B. & Fang, T. (2006). Semi-Automatic 3-D Segmentation of Anatomical Structures of Brain
A Method for Filling Holes in Objects of Medical Images Using Region Labeling and Run Length Encoding Schemes
 110 Image Processing (NCIMP 2010) Image Processing (NCIMP 2010) Editor: K. Somasundaram Allied Publishers A Method for Filling Holes in Objects of Medical Images Using Region Labeling and Run Length Encoding
110 Image Processing (NCIMP 2010) Image Processing (NCIMP 2010) Editor: K. Somasundaram Allied Publishers A Method for Filling Holes in Objects of Medical Images Using Region Labeling and Run Length Encoding
Topology Correction for Brain Atlas Segmentation using a Multiscale Algorithm
 Topology Correction for Brain Atlas Segmentation using a Multiscale Algorithm Lin Chen and Gudrun Wagenknecht Central Institute for Electronics, Research Center Jülich, Jülich, Germany Email: l.chen@fz-juelich.de
Topology Correction for Brain Atlas Segmentation using a Multiscale Algorithm Lin Chen and Gudrun Wagenknecht Central Institute for Electronics, Research Center Jülich, Jülich, Germany Email: l.chen@fz-juelich.de
MRI BRAIN NUCLEI SEGMENTATION AND EVALUATION OF SEGMENTED NUCLEI WITH BET AND BSE
 VOL. 10, NO. 11, JUNE 015 ISSN 1819-6608 006-015 Asian Research Publishing Network (ARPN). All rights reserved. MRI BRAIN NUCLEI SEGMENTATION AND EVALUATION OF SEGMENTED NUCLEI WITH BET AND BSE D. Selvaraj
VOL. 10, NO. 11, JUNE 015 ISSN 1819-6608 006-015 Asian Research Publishing Network (ARPN). All rights reserved. MRI BRAIN NUCLEI SEGMENTATION AND EVALUATION OF SEGMENTED NUCLEI WITH BET AND BSE D. Selvaraj
SEGMENTATION OF STROKE REGIONS FROM DWI AND ADC SEQUENCES USING A MODIFIED WATERSHED METHOD
 SEGMENTATION OF STROKE REGIONS FROM DWI AND ADC SEQUENCES USING A MODIFIED WATERSHED METHOD Ravi S. 1, A.M. Khan 2 1 Research Student, Dept. of Electronics, Mangalore University, Mangalagangotri, India
SEGMENTATION OF STROKE REGIONS FROM DWI AND ADC SEQUENCES USING A MODIFIED WATERSHED METHOD Ravi S. 1, A.M. Khan 2 1 Research Student, Dept. of Electronics, Mangalore University, Mangalagangotri, India
Computers in Biology and Medicine
 Computers in Biology and Medicine 42 (212) 59 522 Contents lists available at SciVerse ScienceDirect Computers in Biology and Medicine journal homepage: www.elsevier.com/locate/cbm Smart histogram analysis
Computers in Biology and Medicine 42 (212) 59 522 Contents lists available at SciVerse ScienceDirect Computers in Biology and Medicine journal homepage: www.elsevier.com/locate/cbm Smart histogram analysis
The organization of the human cerebral cortex estimated by intrinsic functional connectivity
 1 The organization of the human cerebral cortex estimated by intrinsic functional connectivity Journal: Journal of Neurophysiology Author: B. T. Thomas Yeo, et al Link: https://www.ncbi.nlm.nih.gov/pubmed/21653723
1 The organization of the human cerebral cortex estimated by intrinsic functional connectivity Journal: Journal of Neurophysiology Author: B. T. Thomas Yeo, et al Link: https://www.ncbi.nlm.nih.gov/pubmed/21653723
SKULL STRIPPING OF MRI USING CLUSTERING AND RESONANCE METHOD
 International Journal of Knowledge Management & e-learning Volume 3 Number 1 January-June 2011 pp. 19-23 SKULL STRIPPING OF MRI USING CLUSTERING AND RESONANCE METHOD K. Somasundaram 1 & R. Siva Shankar
International Journal of Knowledge Management & e-learning Volume 3 Number 1 January-June 2011 pp. 19-23 SKULL STRIPPING OF MRI USING CLUSTERING AND RESONANCE METHOD K. Somasundaram 1 & R. Siva Shankar
Hybrid Approach for MRI Human Head Scans Classification using HTT based SFTA Texture Feature Extraction Technique
 Volume 118 No. 17 2018, 691-701 ISSN: 1311-8080 (printed version); ISSN: 1314-3395 (on-line version) url: http://www.ijpam.eu ijpam.eu Hybrid Approach for MRI Human Head Scans Classification using HTT
Volume 118 No. 17 2018, 691-701 ISSN: 1311-8080 (printed version); ISSN: 1314-3395 (on-line version) url: http://www.ijpam.eu ijpam.eu Hybrid Approach for MRI Human Head Scans Classification using HTT
DEVELOPMENT OF REALISTIC HEAD MODELS FOR ELECTRO- MAGNETIC SOURCE IMAGING OF THE HUMAN BRAIN
 DEVELOPMENT OF REALISTIC HEAD MODELS FOR ELECTRO- MAGNETIC SOURCE IMAGING OF THE HUMAN BRAIN Z. "! #$! Acar, N.G. Gençer Department of Electrical and Electronics Engineering, Middle East Technical University,
DEVELOPMENT OF REALISTIC HEAD MODELS FOR ELECTRO- MAGNETIC SOURCE IMAGING OF THE HUMAN BRAIN Z. "! #$! Acar, N.G. Gençer Department of Electrical and Electronics Engineering, Middle East Technical University,
Tumor Detection and classification of Medical MRI UsingAdvance ROIPropANN Algorithm
 International Journal of Engineering Research and Advanced Technology (IJERAT) DOI:http://dx.doi.org/10.31695/IJERAT.2018.3273 E-ISSN : 2454-6135 Volume.4, Issue 6 June -2018 Tumor Detection and classification
International Journal of Engineering Research and Advanced Technology (IJERAT) DOI:http://dx.doi.org/10.31695/IJERAT.2018.3273 E-ISSN : 2454-6135 Volume.4, Issue 6 June -2018 Tumor Detection and classification
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge
 Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
A hybrid approach to the skull stripping problem in MRI
 A hybrid approach to the skull stripping problem in MRI F. Ségonne, a,b, * A.M. Dale, a,b E. Busa, b M. Glessner, b D. Salat, b H.K. Hahn, c and B. Fischl a,b a MIT C.S.A.I. Laboratory, Cambridge, MA 02139,
A hybrid approach to the skull stripping problem in MRI F. Ségonne, a,b, * A.M. Dale, a,b E. Busa, b M. Glessner, b D. Salat, b H.K. Hahn, c and B. Fischl a,b a MIT C.S.A.I. Laboratory, Cambridge, MA 02139,
EE795: Computer Vision and Intelligent Systems
 EE795: Computer Vision and Intelligent Systems Spring 2012 TTh 17:30-18:45 WRI C225 Lecture 04 130131 http://www.ee.unlv.edu/~b1morris/ecg795/ 2 Outline Review Histogram Equalization Image Filtering Linear
EE795: Computer Vision and Intelligent Systems Spring 2012 TTh 17:30-18:45 WRI C225 Lecture 04 130131 http://www.ee.unlv.edu/~b1morris/ecg795/ 2 Outline Review Histogram Equalization Image Filtering Linear
Automatic Caudate Segmentation by Hybrid Generative/Discriminative Models
 Automatic Caudate Segmentation by Hybrid Generative/Discriminative Models Zhuowen Tu, Mubeena Mirza, Ivo Dinov, and Arthur W. Toga Lab of Neuro Imaging, School of Medicine University of California, Los
Automatic Caudate Segmentation by Hybrid Generative/Discriminative Models Zhuowen Tu, Mubeena Mirza, Ivo Dinov, and Arthur W. Toga Lab of Neuro Imaging, School of Medicine University of California, Los
A novel Skull Stripping Method for T1 Coronal and T2 Axial Magnetic Resonance Images of Human Head Scans Based on Resonance Principle
 A novel Skull Stripping Method for T1 Coronal and T2 Axial Magnetic Resonance Images of Human Head Scans Based on Resonance Principle K.Somasundaram 1, R. Siva Shankar 2 1&2 Image Processing Lab, Department
A novel Skull Stripping Method for T1 Coronal and T2 Axial Magnetic Resonance Images of Human Head Scans Based on Resonance Principle K.Somasundaram 1, R. Siva Shankar 2 1&2 Image Processing Lab, Department
Automated segmentation methods for liver analysis in oncology applications
 University of Szeged Department of Image Processing and Computer Graphics Automated segmentation methods for liver analysis in oncology applications Ph. D. Thesis László Ruskó Thesis Advisor Dr. Antal
University of Szeged Department of Image Processing and Computer Graphics Automated segmentation methods for liver analysis in oncology applications Ph. D. Thesis László Ruskó Thesis Advisor Dr. Antal
Automated MR Image Analysis Pipelines
 Automated MR Image Analysis Pipelines Andy Simmons Centre for Neuroimaging Sciences, Kings College London Institute of Psychiatry. NIHR Biomedical Research Centre for Mental Health at IoP & SLAM. Neuroimaging
Automated MR Image Analysis Pipelines Andy Simmons Centre for Neuroimaging Sciences, Kings College London Institute of Psychiatry. NIHR Biomedical Research Centre for Mental Health at IoP & SLAM. Neuroimaging
Sampling-Based Ensemble Segmentation against Inter-operator Variability
 Sampling-Based Ensemble Segmentation against Inter-operator Variability Jing Huo 1, Kazunori Okada, Whitney Pope 1, Matthew Brown 1 1 Center for Computer vision and Imaging Biomarkers, Department of Radiological
Sampling-Based Ensemble Segmentation against Inter-operator Variability Jing Huo 1, Kazunori Okada, Whitney Pope 1, Matthew Brown 1 1 Center for Computer vision and Imaging Biomarkers, Department of Radiological
Fast Interactive Region of Interest Selection for Volume Visualization
 Fast Interactive Region of Interest Selection for Volume Visualization Dominik Sibbing and Leif Kobbelt Lehrstuhl für Informatik 8, RWTH Aachen, 20 Aachen Email: {sibbing,kobbelt}@informatik.rwth-aachen.de
Fast Interactive Region of Interest Selection for Volume Visualization Dominik Sibbing and Leif Kobbelt Lehrstuhl für Informatik 8, RWTH Aachen, 20 Aachen Email: {sibbing,kobbelt}@informatik.rwth-aachen.de
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes
 Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
GLIRT: Groupwise and Longitudinal Image Registration Toolbox
 Software Release (1.0.1) Last updated: March. 30, 2011. GLIRT: Groupwise and Longitudinal Image Registration Toolbox Guorong Wu 1, Qian Wang 1,2, Hongjun Jia 1, and Dinggang Shen 1 1 Image Display, Enhancement,
Software Release (1.0.1) Last updated: March. 30, 2011. GLIRT: Groupwise and Longitudinal Image Registration Toolbox Guorong Wu 1, Qian Wang 1,2, Hongjun Jia 1, and Dinggang Shen 1 1 Image Display, Enhancement,
Global Thresholding Techniques to Classify Dead Cells in Diffusion Weighted Magnetic Resonant Images
 Global Thresholding Techniques to Classify Dead Cells in Diffusion Weighted Magnetic Resonant Images Ravi S 1, A. M. Khan 2 1 Research Student, Department of Electronics, Mangalore University, Karnataka
Global Thresholding Techniques to Classify Dead Cells in Diffusion Weighted Magnetic Resonant Images Ravi S 1, A. M. Khan 2 1 Research Student, Department of Electronics, Mangalore University, Karnataka
UCLA Advanced Neuroimaging Summer Program David W. Shattuck, PhD
 BrainSuite UCLA Advanced Neuroimaging Summer Program Presented 18 July 2013 David W. Shattuck, PhD Associate Professor Department of Neurology David Geffen School of Medicine at UCLA http://www.loni.ucla.edu/~shattuck/
BrainSuite UCLA Advanced Neuroimaging Summer Program Presented 18 July 2013 David W. Shattuck, PhD Associate Professor Department of Neurology David Geffen School of Medicine at UCLA http://www.loni.ucla.edu/~shattuck/
N.Priya. Keywords Compass mask, Threshold, Morphological Operators, Statistical Measures, Text extraction
 Volume, Issue 8, August ISSN: 77 8X International Journal of Advanced Research in Computer Science and Software Engineering Research Paper Available online at: www.ijarcsse.com A Combined Edge-Based Text
Volume, Issue 8, August ISSN: 77 8X International Journal of Advanced Research in Computer Science and Software Engineering Research Paper Available online at: www.ijarcsse.com A Combined Edge-Based Text
Fuzzy-Based Extraction of Vascular Structures from Time-of-Flight MR Images
 816 Medical Informatics in a United and Healthy Europe K.-P. Adlassnig et al. (Eds.) IOS Press, 2009 2009 European Federation for Medical Informatics. All rights reserved. doi:10.3233/978-1-60750-044-5-816
816 Medical Informatics in a United and Healthy Europe K.-P. Adlassnig et al. (Eds.) IOS Press, 2009 2009 European Federation for Medical Informatics. All rights reserved. doi:10.3233/978-1-60750-044-5-816
NIH Public Access Author Manuscript Proc Soc Photo Opt Instrum Eng. Author manuscript; available in PMC 2014 October 07.
 NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
Abstract. 1. Introduction
 A New Automated Method for Three- Dimensional Registration of Medical Images* P. Kotsas, M. Strintzis, D.W. Piraino Department of Electrical and Computer Engineering, Aristotelian University, 54006 Thessaloniki,
A New Automated Method for Three- Dimensional Registration of Medical Images* P. Kotsas, M. Strintzis, D.W. Piraino Department of Electrical and Computer Engineering, Aristotelian University, 54006 Thessaloniki,
Impact of acquisition protocols and processing streams on tissue segmentation of T1 weighted MR images
 www.elsevier.com/locate/ynimg NeuroImage 29 (2006) 185 202 Impact of acquisition protocols and processing streams on tissue segmentation of T1 weighted MR images Kristi A. Clark, a, * Roger P. Woods, a
www.elsevier.com/locate/ynimg NeuroImage 29 (2006) 185 202 Impact of acquisition protocols and processing streams on tissue segmentation of T1 weighted MR images Kristi A. Clark, a, * Roger P. Woods, a
MARS: Multiple Atlases Robust Segmentation
 Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Global Journal of Engineering Science and Research Management
 ADVANCED K-MEANS ALGORITHM FOR BRAIN TUMOR DETECTION USING NAIVE BAYES CLASSIFIER Veena Bai K*, Dr. Niharika Kumar * MTech CSE, Department of Computer Science and Engineering, B.N.M. Institute of Technology,
ADVANCED K-MEANS ALGORITHM FOR BRAIN TUMOR DETECTION USING NAIVE BAYES CLASSIFIER Veena Bai K*, Dr. Niharika Kumar * MTech CSE, Department of Computer Science and Engineering, B.N.M. Institute of Technology,
NIH Public Access Author Manuscript Hum Brain Mapp. Author manuscript; available in PMC 2008 June 2.
 NIH Public Access Author Manuscript Published in final edited form as: Hum Brain Mapp. 2006 February ; 27(2): 99 113. Quantitative Evaluation of Automated Skull-Stripping Methods Applied to Contemporary
NIH Public Access Author Manuscript Published in final edited form as: Hum Brain Mapp. 2006 February ; 27(2): 99 113. Quantitative Evaluation of Automated Skull-Stripping Methods Applied to Contemporary
Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry
 Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry Nivedita Agarwal, MD Nivedita.agarwal@apss.tn.it Nivedita.agarwal@unitn.it Volume and surface morphometry Brain volume White matter
Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry Nivedita Agarwal, MD Nivedita.agarwal@apss.tn.it Nivedita.agarwal@unitn.it Volume and surface morphometry Brain volume White matter
Ensemble registration: Combining groupwise registration and segmentation
 PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
Computers in Biology and Medicine
 Computers in Biology and Medicine 41 (2011) 716 725 Contents lists available at ScienceDirect Computers in Biology and Medicine journal homepage: www.elsevier.com/locate/cbm Automatic brain extraction
Computers in Biology and Medicine 41 (2011) 716 725 Contents lists available at ScienceDirect Computers in Biology and Medicine journal homepage: www.elsevier.com/locate/cbm Automatic brain extraction
Automatic Optimization of Segmentation Algorithms Through Simultaneous Truth and Performance Level Estimation (STAPLE)
 Automatic Optimization of Segmentation Algorithms Through Simultaneous Truth and Performance Level Estimation (STAPLE) Mahnaz Maddah, Kelly H. Zou, William M. Wells, Ron Kikinis, and Simon K. Warfield
Automatic Optimization of Segmentation Algorithms Through Simultaneous Truth and Performance Level Estimation (STAPLE) Mahnaz Maddah, Kelly H. Zou, William M. Wells, Ron Kikinis, and Simon K. Warfield
3D Surface Reconstruction of the Brain based on Level Set Method
 3D Surface Reconstruction of the Brain based on Level Set Method Shijun Tang, Bill P. Buckles, and Kamesh Namuduri Department of Computer Science & Engineering Department of Electrical Engineering University
3D Surface Reconstruction of the Brain based on Level Set Method Shijun Tang, Bill P. Buckles, and Kamesh Namuduri Department of Computer Science & Engineering Department of Electrical Engineering University
Automatic Generation of Training Data for Brain Tissue Classification from MRI
 MICCAI-2002 1 Automatic Generation of Training Data for Brain Tissue Classification from MRI Chris A. Cocosco, Alex P. Zijdenbos, and Alan C. Evans McConnell Brain Imaging Centre, Montreal Neurological
MICCAI-2002 1 Automatic Generation of Training Data for Brain Tissue Classification from MRI Chris A. Cocosco, Alex P. Zijdenbos, and Alan C. Evans McConnell Brain Imaging Centre, Montreal Neurological
ISSN: X Impact factor: 4.295
 ISSN: 2454-132X Impact factor: 4.295 (Volume3, Issue1) Available online at: www.ijariit.com Performance Analysis of Image Clustering Algorithm Applied to Brain MRI Kalyani R.Mandlik 1, Dr. Suresh S. Salankar
ISSN: 2454-132X Impact factor: 4.295 (Volume3, Issue1) Available online at: www.ijariit.com Performance Analysis of Image Clustering Algorithm Applied to Brain MRI Kalyani R.Mandlik 1, Dr. Suresh S. Salankar
Processing math: 100% Intensity Normalization
 Intensity Normalization Overall Pipeline 2/21 Intensity normalization Conventional MRI intensites (T1-w, T2-w, PD, FLAIR) are acquired in arbitrary units Images are not comparable across scanners, subjects,
Intensity Normalization Overall Pipeline 2/21 Intensity normalization Conventional MRI intensites (T1-w, T2-w, PD, FLAIR) are acquired in arbitrary units Images are not comparable across scanners, subjects,
Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging
 1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
Detecting Changes In Non-Isotropic Images
 Detecting Changes In Non-Isotropic Images K.J. Worsley 1, M. Andermann 1, T. Koulis 1, D. MacDonald, 2 and A.C. Evans 2 August 4, 1999 1 Department of Mathematics and Statistics, 2 Montreal Neurological
Detecting Changes In Non-Isotropic Images K.J. Worsley 1, M. Andermann 1, T. Koulis 1, D. MacDonald, 2 and A.C. Evans 2 August 4, 1999 1 Department of Mathematics and Statistics, 2 Montreal Neurological
Final Review. Image Processing CSE 166 Lecture 18
 Final Review Image Processing CSE 166 Lecture 18 Topics covered Basis vectors Matrix based transforms Wavelet transform Image compression Image watermarking Morphological image processing Segmentation
Final Review Image Processing CSE 166 Lecture 18 Topics covered Basis vectors Matrix based transforms Wavelet transform Image compression Image watermarking Morphological image processing Segmentation
NeuroQLab A Software Assistant for Neurosurgical Planning and Quantitative Image Analysis
 NeuroQLab A Software Assistant for Neurosurgical Planning and Quantitative Image Analysis Florian Weiler 1, Jan Rexilius 2, Jan Klein 1, Horst K. Hahn 1 1 Fraunhofer MEVIS, Universitätsallee 29, 28359
NeuroQLab A Software Assistant for Neurosurgical Planning and Quantitative Image Analysis Florian Weiler 1, Jan Rexilius 2, Jan Klein 1, Horst K. Hahn 1 1 Fraunhofer MEVIS, Universitätsallee 29, 28359
MRI Brain Image Analysis and Classification for Computer-Assisted Diagnosis
 MRI Brain Image Analysis and Classification for Computer-Assisted Diagnosis MADINA HAMIANE Department of Telecommunication Engineering Ahlia University Manama, Kingdom of Bahrain mhamiane@ahlia.edu.bh
MRI Brain Image Analysis and Classification for Computer-Assisted Diagnosis MADINA HAMIANE Department of Telecommunication Engineering Ahlia University Manama, Kingdom of Bahrain mhamiane@ahlia.edu.bh
Morphometric Analysis of Biomedical Images. Sara Rolfe 10/9/17
 Morphometric Analysis of Biomedical Images Sara Rolfe 10/9/17 Morphometric Analysis of Biomedical Images Object surface contours Image difference features Compact representation of feature differences
Morphometric Analysis of Biomedical Images Sara Rolfe 10/9/17 Morphometric Analysis of Biomedical Images Object surface contours Image difference features Compact representation of feature differences
Skull Segmentation of MR images based on texture features for attenuation correction in PET/MR
 Skull Segmentation of MR images based on texture features for attenuation correction in PET/MR CHAIBI HASSEN, NOURINE RACHID ITIO Laboratory, Oran University Algeriachaibih@yahoo.fr, nourine@yahoo.com
Skull Segmentation of MR images based on texture features for attenuation correction in PET/MR CHAIBI HASSEN, NOURINE RACHID ITIO Laboratory, Oran University Algeriachaibih@yahoo.fr, nourine@yahoo.com
Chapter 7 UNSUPERVISED LEARNING TECHNIQUES FOR MAMMOGRAM CLASSIFICATION
 UNSUPERVISED LEARNING TECHNIQUES FOR MAMMOGRAM CLASSIFICATION Supervised and unsupervised learning are the two prominent machine learning algorithms used in pattern recognition and classification. In this
UNSUPERVISED LEARNING TECHNIQUES FOR MAMMOGRAM CLASSIFICATION Supervised and unsupervised learning are the two prominent machine learning algorithms used in pattern recognition and classification. In this
Extraction and Features of Tumour from MR brain images
 IOSR Journal of Electronics and Communication Engineering (IOSR-JECE) e-issn: 2278-2834,p- ISSN: 2278-8735.Volume 13, Issue 2, Ver. I (Mar. - Apr. 2018), PP 67-71 www.iosrjournals.org Sai Prasanna M 1,
IOSR Journal of Electronics and Communication Engineering (IOSR-JECE) e-issn: 2278-2834,p- ISSN: 2278-8735.Volume 13, Issue 2, Ver. I (Mar. - Apr. 2018), PP 67-71 www.iosrjournals.org Sai Prasanna M 1,
Automatic Detection and Segmentation of Kidneys in Magnetic Resonance Images Using Image Processing Techniques
 Biomedical Statistics and Informatics 2017; 2(1): 22-26 http://www.sciencepublishinggroup.com/j/bsi doi: 10.11648/j.bsi.20170201.15 Automatic Detection and Segmentation of Kidneys in Magnetic Resonance
Biomedical Statistics and Informatics 2017; 2(1): 22-26 http://www.sciencepublishinggroup.com/j/bsi doi: 10.11648/j.bsi.20170201.15 Automatic Detection and Segmentation of Kidneys in Magnetic Resonance
Comparison Study of Clinical 3D MRI Brain Segmentation Evaluation
 Comparison Study of Clinical 3D MRI Brain Segmentation Evaluation Ting Song 1, Elsa D. Angelini 2, Brett D. Mensh 3, Andrew Laine 1 1 Heffner Biomedical Imaging Laboratory Department of Biomedical Engineering,
Comparison Study of Clinical 3D MRI Brain Segmentation Evaluation Ting Song 1, Elsa D. Angelini 2, Brett D. Mensh 3, Andrew Laine 1 1 Heffner Biomedical Imaging Laboratory Department of Biomedical Engineering,
09/11/2017. Morphological image processing. Morphological image processing. Morphological image processing. Morphological image processing (binary)
 Towards image analysis Goal: Describe the contents of an image, distinguishing meaningful information from irrelevant one. Perform suitable transformations of images so as to make explicit particular shape
Towards image analysis Goal: Describe the contents of an image, distinguishing meaningful information from irrelevant one. Perform suitable transformations of images so as to make explicit particular shape
An MRI-based Attenuation Correction Method for Combined PET/MRI Applications
 An MRI-based Attenuation Correction Method for Combined PET/MRI Applications Baowei Fei *, Xiaofeng Yang, Hesheng Wang Departments of Radiology, Emory University, Atlanta, GA Department of Biomedical Engineering
An MRI-based Attenuation Correction Method for Combined PET/MRI Applications Baowei Fei *, Xiaofeng Yang, Hesheng Wang Departments of Radiology, Emory University, Atlanta, GA Department of Biomedical Engineering
Local or Global Minima: Flexible Dual-Front Active Contours
 Local or Global Minima: Flexible Dual-Front Active Contours Hua Li 1,2 and Anthony Yezzi 1 1 School of ECE, Georgia Institute of Technology, Atlanta, GA, USA 2 Dept. of Elect. & Info. Eng., Huazhong Univ.
Local or Global Minima: Flexible Dual-Front Active Contours Hua Li 1,2 and Anthony Yezzi 1 1 School of ECE, Georgia Institute of Technology, Atlanta, GA, USA 2 Dept. of Elect. & Info. Eng., Huazhong Univ.
BrainSuite. presented at the UCLA/NITP Advanced Neuroimaging Summer Program 29 July 2014
 BrainSuite presented at the UCLA/NITP Advanced Neuroimaging Summer Program 29 July 2014 David Shattuck Ahmanson-Lovelace Brain Mapping Center Department of Neurology David Geffen School of Medicine at
BrainSuite presented at the UCLA/NITP Advanced Neuroimaging Summer Program 29 July 2014 David Shattuck Ahmanson-Lovelace Brain Mapping Center Department of Neurology David Geffen School of Medicine at
Advanced Visual Medicine: Techniques for Visual Exploration & Analysis
 Advanced Visual Medicine: Techniques for Visual Exploration & Analysis Interactive Visualization of Multimodal Volume Data for Neurosurgical Planning Felix Ritter, MeVis Research Bremen Multimodal Neurosurgical
Advanced Visual Medicine: Techniques for Visual Exploration & Analysis Interactive Visualization of Multimodal Volume Data for Neurosurgical Planning Felix Ritter, MeVis Research Bremen Multimodal Neurosurgical
Research Article Image Segmentation Using Gray-Scale Morphology and Marker-Controlled Watershed Transformation
 Discrete Dynamics in Nature and Society Volume 2008, Article ID 384346, 8 pages doi:10.1155/2008/384346 Research Article Image Segmentation Using Gray-Scale Morphology and Marker-Controlled Watershed Transformation
Discrete Dynamics in Nature and Society Volume 2008, Article ID 384346, 8 pages doi:10.1155/2008/384346 Research Article Image Segmentation Using Gray-Scale Morphology and Marker-Controlled Watershed Transformation
An Introduction To Automatic Tissue Classification Of Brain MRI. Colm Elliott Mar 2014
 An Introduction To Automatic Tissue Classification Of Brain MRI Colm Elliott Mar 2014 Tissue Classification Tissue classification is part of many processing pipelines. We often want to classify each voxel
An Introduction To Automatic Tissue Classification Of Brain MRI Colm Elliott Mar 2014 Tissue Classification Tissue classification is part of many processing pipelines. We often want to classify each voxel
A Review on Label Image Constrained Multiatlas Selection
 A Review on Label Image Constrained Multiatlas Selection Ms. VAIBHAVI NANDKUMAR JAGTAP 1, Mr. SANTOSH D. KALE 2 1PG Scholar, Department of Electronics and Telecommunication, SVPM College of Engineering,
A Review on Label Image Constrained Multiatlas Selection Ms. VAIBHAVI NANDKUMAR JAGTAP 1, Mr. SANTOSH D. KALE 2 1PG Scholar, Department of Electronics and Telecommunication, SVPM College of Engineering,
UNC 4D Infant Cortical Surface Atlases, from Neonate to 6 Years of Age
 UNC 4D Infant Cortical Surface Atlases, from Neonate to 6 Years of Age Version 1.0 UNC 4D infant cortical surface atlases from neonate to 6 years of age contain 11 time points, including 1, 3, 6, 9, 12,
UNC 4D Infant Cortical Surface Atlases, from Neonate to 6 Years of Age Version 1.0 UNC 4D infant cortical surface atlases from neonate to 6 years of age contain 11 time points, including 1, 3, 6, 9, 12,
Babu Madhav Institute of Information Technology Years Integrated M.Sc.(IT)(Semester - 7)
 5 Years Integrated M.Sc.(IT)(Semester - 7) 060010707 Digital Image Processing UNIT 1 Introduction to Image Processing Q: 1 Answer in short. 1. What is digital image? 1. Define pixel or picture element?
5 Years Integrated M.Sc.(IT)(Semester - 7) 060010707 Digital Image Processing UNIT 1 Introduction to Image Processing Q: 1 Answer in short. 1. What is digital image? 1. Define pixel or picture element?
Open Topology: A Toolkit for Brain Isosurface Correction
 Open Topology: A Toolkit for Brain Isosurface Correction Sylvain Jaume 1, Patrice Rondao 2, and Benoît Macq 2 1 National Institute of Research in Computer Science and Control, INRIA, France, sylvain@mit.edu,
Open Topology: A Toolkit for Brain Isosurface Correction Sylvain Jaume 1, Patrice Rondao 2, and Benoît Macq 2 1 National Institute of Research in Computer Science and Control, INRIA, France, sylvain@mit.edu,
Fiber Selection from Diffusion Tensor Data based on Boolean Operators
 Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Scene-Based Segmentation of Multiple Muscles from MRI in MITK
 Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
Local Independent Projection Based Classification Using Fuzzy Clustering
 www.ijecs.in International Journal Of Engineering And Computer Science ISSN:2319-7242 Volume 4 Issue 6 June 2015, Page No. 12306-12311 Local Independent Projection Based Classification Using Fuzzy Clustering
www.ijecs.in International Journal Of Engineering And Computer Science ISSN:2319-7242 Volume 4 Issue 6 June 2015, Page No. 12306-12311 Local Independent Projection Based Classification Using Fuzzy Clustering
Model-based segmentation and recognition from range data
 Model-based segmentation and recognition from range data Jan Boehm Institute for Photogrammetry Universität Stuttgart Germany Keywords: range image, segmentation, object recognition, CAD ABSTRACT This
Model-based segmentation and recognition from range data Jan Boehm Institute for Photogrammetry Universität Stuttgart Germany Keywords: range image, segmentation, object recognition, CAD ABSTRACT This
Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit
 Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit John Melonakos 1, Ramsey Al-Hakim 1, James Fallon 2 and Allen Tannenbaum 1 1 Georgia Institute of Technology, Atlanta GA 30332,
Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit John Melonakos 1, Ramsey Al-Hakim 1, James Fallon 2 and Allen Tannenbaum 1 1 Georgia Institute of Technology, Atlanta GA 30332,
Journal of Industrial Engineering Research
 IWNEST PUBLISHER Journal of Industrial Engineering Research (ISSN: 2077-4559) Journal home page: http://www.iwnest.com/aace/ Mammogram Image Segmentation Using voronoi Diagram Properties Dr. J. Subash
IWNEST PUBLISHER Journal of Industrial Engineering Research (ISSN: 2077-4559) Journal home page: http://www.iwnest.com/aace/ Mammogram Image Segmentation Using voronoi Diagram Properties Dr. J. Subash
Math in image processing
 Math in image processing Math in image processing Nyquist theorem Math in image processing Discrete Fourier Transformation Math in image processing Image enhancement: scaling Math in image processing Image
Math in image processing Math in image processing Nyquist theorem Math in image processing Discrete Fourier Transformation Math in image processing Image enhancement: scaling Math in image processing Image
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION
 CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
A Genus Bound for Digital Image Boundaries
 A Genus Bound for Digital Image Boundaries Lowell Abrams and Donniell E. Fishkind March 9, 2005 Abstract Shattuck and Leahy [4] conjectured and Abrams, Fishkind, and Priebe [1],[2] proved that the boundary
A Genus Bound for Digital Image Boundaries Lowell Abrams and Donniell E. Fishkind March 9, 2005 Abstract Shattuck and Leahy [4] conjectured and Abrams, Fishkind, and Priebe [1],[2] proved that the boundary
Histograms. h(r k ) = n k. p(r k )= n k /NM. Histogram: number of times intensity level rk appears in the image
 Histograms h(r k ) = n k Histogram: number of times intensity level rk appears in the image p(r k )= n k /NM normalized histogram also a probability of occurence 1 Histogram of Image Intensities Create
Histograms h(r k ) = n k Histogram: number of times intensity level rk appears in the image p(r k )= n k /NM normalized histogram also a probability of occurence 1 Histogram of Image Intensities Create
Engineering Problem and Goal
 Engineering Problem and Goal Engineering Problem: Traditional active contour models can not detect edges or convex regions in noisy images. Engineering Goal: The goal of this project is to design an algorithm
Engineering Problem and Goal Engineering Problem: Traditional active contour models can not detect edges or convex regions in noisy images. Engineering Goal: The goal of this project is to design an algorithm
Brain Tumour Detection Using Neural Network Classifier and K-Means Clustering Algorithm for Classification and Segmentation
 European Journal of Applied Sciences 9 (2): 66-71, 2017 ISSN 2079-2077 IDOSI Publications, 2017 DOI: 10.5829/idosi.ejas.2017.66.71 Brain Tumour Detection Using Neural Network Classifier and K-Means Clustering
European Journal of Applied Sciences 9 (2): 66-71, 2017 ISSN 2079-2077 IDOSI Publications, 2017 DOI: 10.5829/idosi.ejas.2017.66.71 Brain Tumour Detection Using Neural Network Classifier and K-Means Clustering
SURFACE RECONSTRUCTION OF EX-VIVO HUMAN V1 THROUGH IDENTIFICATION OF THE STRIA OF GENNARI USING MRI AT 7T
 SURFACE RECONSTRUCTION OF EX-VIVO HUMAN V1 THROUGH IDENTIFICATION OF THE STRIA OF GENNARI USING MRI AT 7T Oliver P. Hinds 1, Jonathan R. Polimeni 2, Megan L. Blackwell 3, Christopher J. Wiggins 3, Graham
SURFACE RECONSTRUCTION OF EX-VIVO HUMAN V1 THROUGH IDENTIFICATION OF THE STRIA OF GENNARI USING MRI AT 7T Oliver P. Hinds 1, Jonathan R. Polimeni 2, Megan L. Blackwell 3, Christopher J. Wiggins 3, Graham
Lilla Zöllei A.A. Martinos Center, MGH; Boston, MA
 Lilla Zöllei lzollei@nmr.mgh.harvard.edu A.A. Martinos Center, MGH; Boston, MA Bruce Fischl Gheorghe Postelnicu Jean Augustinack Anastasia Yendiki Allison Stevens Kristen Huber Sita Kakonoori + the FreeSurfer
Lilla Zöllei lzollei@nmr.mgh.harvard.edu A.A. Martinos Center, MGH; Boston, MA Bruce Fischl Gheorghe Postelnicu Jean Augustinack Anastasia Yendiki Allison Stevens Kristen Huber Sita Kakonoori + the FreeSurfer
MR IMAGE SEGMENTATION
 MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION
 ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
Automatic Segmentation of Neonatal Brain MRI
 Automatic Segmentation of Neonatal Brain MRI 1 Marcel Prastawa, 2 John Gilmore, 3 Weili Lin, and 1,2 Guido Gerig Department of 1 Computer Science, 2 Psychiatry, and 3 Radiology University of North Carolina,
Automatic Segmentation of Neonatal Brain MRI 1 Marcel Prastawa, 2 John Gilmore, 3 Weili Lin, and 1,2 Guido Gerig Department of 1 Computer Science, 2 Psychiatry, and 3 Radiology University of North Carolina,
Automated Segmentation of Brain Parts from MRI Image Slices
 Volume 114 No. 11 2017, 39-46 ISSN: 1311-8080 (printed version); ISSN: 1314-3395 (on-line version) url: http://www.ijpam.eu ijpam.eu Automated Segmentation of Brain Parts from MRI Image Slices 1 N. Madhesh
Volume 114 No. 11 2017, 39-46 ISSN: 1311-8080 (printed version); ISSN: 1314-3395 (on-line version) url: http://www.ijpam.eu ijpam.eu Automated Segmentation of Brain Parts from MRI Image Slices 1 N. Madhesh
Morphological Image Processing
 Morphological Image Processing Megha Goyal Dept. of ECE, Doaba Institute of Engineering and Technology, Kharar, Mohali, Punjab, India Abstract The purpose of this paper is to provide readers with an in-depth
Morphological Image Processing Megha Goyal Dept. of ECE, Doaba Institute of Engineering and Technology, Kharar, Mohali, Punjab, India Abstract The purpose of this paper is to provide readers with an in-depth
CHAPTER 6 DETECTION OF MASS USING NOVEL SEGMENTATION, GLCM AND NEURAL NETWORKS
 130 CHAPTER 6 DETECTION OF MASS USING NOVEL SEGMENTATION, GLCM AND NEURAL NETWORKS A mass is defined as a space-occupying lesion seen in more than one projection and it is described by its shapes and margin
130 CHAPTER 6 DETECTION OF MASS USING NOVEL SEGMENTATION, GLCM AND NEURAL NETWORKS A mass is defined as a space-occupying lesion seen in more than one projection and it is described by its shapes and margin
Ischemic Stroke Lesion Segmentation Proceedings 5th October 2015 Munich, Germany
 0111010001110001101000100101010111100111011100100011011101110101101012 Ischemic Stroke Lesion Segmentation www.isles-challenge.org Proceedings 5th October 2015 Munich, Germany Preface Stroke is the second
0111010001110001101000100101010111100111011100100011011101110101101012 Ischemic Stroke Lesion Segmentation www.isles-challenge.org Proceedings 5th October 2015 Munich, Germany Preface Stroke is the second
Automatic Registration-Based Segmentation for Neonatal Brains Using ANTs and Atropos
 Automatic Registration-Based Segmentation for Neonatal Brains Using ANTs and Atropos Jue Wu and Brian Avants Penn Image Computing and Science Lab, University of Pennsylvania, Philadelphia, USA Abstract.
Automatic Registration-Based Segmentation for Neonatal Brains Using ANTs and Atropos Jue Wu and Brian Avants Penn Image Computing and Science Lab, University of Pennsylvania, Philadelphia, USA Abstract.
CHAPTER 2. Morphometry on rodent brains. A.E.H. Scheenstra J. Dijkstra L. van der Weerd
 CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
Level Set Evolution with Region Competition: Automatic 3-D Segmentation of Brain Tumors
 1 Level Set Evolution with Region Competition: Automatic 3-D Segmentation of Brain Tumors 1 Sean Ho, 2 Elizabeth Bullitt, and 1,3 Guido Gerig 1 Department of Computer Science, 2 Department of Surgery,
1 Level Set Evolution with Region Competition: Automatic 3-D Segmentation of Brain Tumors 1 Sean Ho, 2 Elizabeth Bullitt, and 1,3 Guido Gerig 1 Department of Computer Science, 2 Department of Surgery,
Measuring longitudinal brain changes in humans and small animal models. Christos Davatzikos
 Measuring longitudinal brain changes in humans and small animal models Christos Davatzikos Section of Biomedical Image Analysis University of Pennsylvania (Radiology) http://www.rad.upenn.edu/sbia Computational
Measuring longitudinal brain changes in humans and small animal models Christos Davatzikos Section of Biomedical Image Analysis University of Pennsylvania (Radiology) http://www.rad.upenn.edu/sbia Computational
Feature Extraction and Image Processing, 2 nd Edition. Contents. Preface
 , 2 nd Edition Preface ix 1 Introduction 1 1.1 Overview 1 1.2 Human and Computer Vision 1 1.3 The Human Vision System 3 1.3.1 The Eye 4 1.3.2 The Neural System 7 1.3.3 Processing 7 1.4 Computer Vision
, 2 nd Edition Preface ix 1 Introduction 1 1.1 Overview 1 1.2 Human and Computer Vision 1 1.3 The Human Vision System 3 1.3.1 The Eye 4 1.3.2 The Neural System 7 1.3.3 Processing 7 1.4 Computer Vision
Mathematical Morphology and Distance Transforms. Robin Strand
 Mathematical Morphology and Distance Transforms Robin Strand robin.strand@it.uu.se Morphology Form and structure Mathematical framework used for: Pre-processing Noise filtering, shape simplification,...
Mathematical Morphology and Distance Transforms Robin Strand robin.strand@it.uu.se Morphology Form and structure Mathematical framework used for: Pre-processing Noise filtering, shape simplification,...
Organ Surface Reconstruction using B-Splines and Hu Moments
 Organ Surface Reconstruction using B-Splines and Hu Moments Andrzej Wytyczak-Partyka Institute of Computer Engineering Control and Robotics, Wroclaw University of Technology, 27 Wybrzeze Wyspianskiego
Organ Surface Reconstruction using B-Splines and Hu Moments Andrzej Wytyczak-Partyka Institute of Computer Engineering Control and Robotics, Wroclaw University of Technology, 27 Wybrzeze Wyspianskiego
Object Shape Recognition in Image for Machine Vision Application
 Object Shape Recognition in Image for Machine Vision Application Mohd Firdaus Zakaria, Hoo Seng Choon, and Shahrel Azmin Suandi Abstract Vision is the most advanced of our senses, so it is not surprising
Object Shape Recognition in Image for Machine Vision Application Mohd Firdaus Zakaria, Hoo Seng Choon, and Shahrel Azmin Suandi Abstract Vision is the most advanced of our senses, so it is not surprising
Smallest Intersecting Circle for a Set of Polygons
 Smallest Intersecting Circle for a Set of Polygons Peter Otfried Joachim Christian Marc Esther René Michiel Antoine Alexander 31st August 2005 1 Introduction Motivated by automated label placement of groups
Smallest Intersecting Circle for a Set of Polygons Peter Otfried Joachim Christian Marc Esther René Michiel Antoine Alexander 31st August 2005 1 Introduction Motivated by automated label placement of groups
Brain extraction from cerebral MRI volume using a hybrid level set based active contour neighborhood model
 Jiang et al. BioMedical Engineering OnLine 2013, 12:31 RESEARCH Open Access Brain extraction from cerebral MRI volume using a hybrid level set based active contour neighborhood model Shaofeng Jiang *,
Jiang et al. BioMedical Engineering OnLine 2013, 12:31 RESEARCH Open Access Brain extraction from cerebral MRI volume using a hybrid level set based active contour neighborhood model Shaofeng Jiang *,
SCIENCE & TECHNOLOGY
 Pertanika J. Sci. & Technol. 26 (1): 309-316 (2018) SCIENCE & TECHNOLOGY Journal homepage: http://www.pertanika.upm.edu.my/ Application of Active Contours Driven by Local Gaussian Distribution Fitting
Pertanika J. Sci. & Technol. 26 (1): 309-316 (2018) SCIENCE & TECHNOLOGY Journal homepage: http://www.pertanika.upm.edu.my/ Application of Active Contours Driven by Local Gaussian Distribution Fitting
Comparison of Vessel Segmentations Using STAPLE
 Comparison of Vessel Segmentations Using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, The University of North Carolina at Chapel Hill, Department
Comparison of Vessel Segmentations Using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, The University of North Carolina at Chapel Hill, Department
Tissue Tracking: Applications for Brain MRI Classification
 Tissue Tracking: Applications for Brain MRI Classification John Melonakos a and Yi Gao a and Allen Tannenbaum a a Georgia Institute of Technology, 414 Ferst Dr, Atlanta, GA, USA; ABSTRACT Bayesian classification
Tissue Tracking: Applications for Brain MRI Classification John Melonakos a and Yi Gao a and Allen Tannenbaum a a Georgia Institute of Technology, 414 Ferst Dr, Atlanta, GA, USA; ABSTRACT Bayesian classification
Automatic Quantification of DTI Parameters along Fiber Bundles
 Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein 1, Simon Hermann 1, Olaf Konrad 1, Horst K. Hahn 1, and Heinz-Otto Peitgen 1 1 MeVis Research, 28359 Bremen Email: klein@mevis.de
Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein 1, Simon Hermann 1, Olaf Konrad 1, Horst K. Hahn 1, and Heinz-Otto Peitgen 1 1 MeVis Research, 28359 Bremen Email: klein@mevis.de
Image Segmentation Techniques for Object-Based Coding
 Image Techniques for Object-Based Coding Junaid Ahmed, Joseph Bosworth, and Scott T. Acton The Oklahoma Imaging Laboratory School of Electrical and Computer Engineering Oklahoma State University {ajunaid,bosworj,sacton}@okstate.edu
Image Techniques for Object-Based Coding Junaid Ahmed, Joseph Bosworth, and Scott T. Acton The Oklahoma Imaging Laboratory School of Electrical and Computer Engineering Oklahoma State University {ajunaid,bosworj,sacton}@okstate.edu
