Segmentation of multiple organs in non-contrast 3D abdominal CT images
|
|
|
- Meryl Roberts
- 5 years ago
- Views:
Transcription
1 Int J CARS (2007) 2: DOI /s z ORIGINAL ARTICLE Segmentation of multiple organs in non-contrast 3D abdominal CT images Akinobu Shimizu Rena Ohno Takaya Ikegami Hidefumi Kobatake Shigeru Nawano Daniel Smutek Received: 11 April 2007 / Accepted: 31 October 2007 / Published online: 27 November 2007 CARS 2007 Abstract Objective We propose a simultaneous extraction method for 12 organs from non-contrast three-dimensional abdominal CT images. Materials and methods The proposed method uses an abdominal cavity standardization process and atlas guided segmentation incorporating parameter estimation with the EM algorithm to deal with the large fluctuations in the feature distribution parameters between subjects. Segmentation is then performed using multiple level sets, which minimize the energy function that considers the hierarchy and exclusiveness between organs as well as uniformity of grey values in organs. To assess the performance of the proposed method, ten non-contrast 3D CT volumes were used. Results The accuracy of the feature distribution parameter estimation was slightly improved using the proposed EM method, resulting in better performance of the segmentation process. Nine organs out of twelve were statistically improved compared with the results without the proposed parameter estimation process. The proposed multiple level sets also boosted the performance of the segmentation by 7.2 points on average compared with the atlas guided This study was originally presented at CARS 2006 and supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan. A. Shimizu (B) R. Ohno T. Ikegami H. Kobatake Tokyo University of Agriculture and Technology, Koganei, Tokyo, Japan simiz@cc.tuat.ac.jp S. Nawano International University of Health and Welfare, Minato-ku, Tokyo, Japan D. Smutek 1st Medical Faculty, Charles University, Prague, Czech Republic segmentation. Nine out of twelve organs were confirmed to be statistically improved compared with the atlas guided method. Conclusion The proposed method was statistically proved to have better performance in the segmentation of 3D CT volumes. Keywords Computer-assisted diagnosis Three-dimensional image Computed tomography Segmentation of abdominal organs Abbreviations MAP Maximum a posteriori CAD Computer-assisted diagnosis EM Expectation maximization LSM Level set method 3D Three-dimensional IVC Inferior vena cava PV Portal vein Introduction The spatial range and resolution of medical images improved markedly upon the development of multi-slice CT scanners. Now, the entire abdominal area can be scanned with less than a 1-mm-slice interval within a single breath-hold. Although such volume images do aid the diagnosis of multiple organs, a large number of slice images need to be interpreted, leading to lesions occasionally being missed by radiologists. In order to assist radiologists, we have been developing computer-assisted diagnosis (CAD) systems for lesions in the multiple abdominal organs [1]. Accurate segmentation of the multiple abdominal organs is very important for such CAD systems from the viewpoint of diagnostic accuracy and computational cost. To this end,
2 136 Int J CARS (2007) 2: Fig. 1 Organs to be extracted. Boundaries of different organs are shown in different colours. To evaluate the performance of the proposed method, all the organ boundaries were manually defined by the authors and approved by a medical doctor extraction algorithms that can process the multiple abdominal organs have been proposed. For example, Park et al. [2] proposed an atlas-based segmentation of abdominal organs using a combination of the probabilistic atlas and the maximum a posteriori (MAP) approach, where they demonstrated successful integration of the atlas information into the segmentation. However, segmentation results have irregular shapes due to the large variation in shape and grey value of organs among different human bodies. Recently, there have also been significant efforts to use the spatial relationship between regions to boost the performance of multiple objects segmentation [3], constraints by neighbouring regions [4], and interactions between neighbouring organs [5], all of which are very effective in extracting the multiple structures in the brain and pelvis in MRI. Although these methods can be applied to abdominal organs, less attention has been paid to actually segmenting the multiple abdominal organs in non-contrast CT images, as there are several important differences between brain and abdominal images. Namely, abdominal organs are mainly surrounded by soft tissues and show large variations in their positions and shapes, which is a different case from the brain within the solid skull. Consequently, it has become necessary to develop a new algorithm that can reduce these variations and consider the spatial relationship between the abdominal organs to obtain fine segmentation results. Park et al. proposed the standardisation process for four organs liver, kidneys and spinal cord. They used the thin-plate spline based method to align the input data with the target reference geometry. Such nonrigid registration is effective in reducing variation not only in position but also in size and shape of organs. However, the non-linear registration technique may cause unfavourable distortion in the region where the non-continuous transformation occurs, such as sliding between neighbouring organs, which makes the segmentation task more difficult. In this study, we focus on 12 organs with such a non-continuous deformation. To prevent excessive distortion, we employed linear transformation to standardize the abdominal cavity. The proposed standardization method is based on automatically detected landmarks. Subsequently, it performs an atlas guided segmentation algorithm incorporating an expectation maximisation (EM) algorithm for the extended-mixture Gaussian distribution in which the mixture ratio is defined at each voxel. We also propose a multiple level set method (LSM), which minimises the energy function that considers the hierarchy and exclusiveness between organs as well as the uniformity of grey values in organs. The LSM is implemented in a clustered computer environment using message passing interface to reduce the computational cost. We report here the experimental results of applying the algorithm to 12 organs using actual non-contrast 3D CT images. Materials and methods Materials Ten abdominal non-contrast 3D CT images taken from ten males were used to assess the performance of the proposed method, where the average number of slices per case was 184, the slice interval was 1 mm and the spatial resolution in the axial slices was mm. The images were converted to isotropic voxels and image size was reduced to one-third for the following experiments. Target organs were the liver, spleen, left and right kidneys, heart, gallbladder, pancreas, portal vein (PV) between the spleen and liver, stomach wall, oesophagus, abdominal aorta and inferior vena cava (IVC) (Fig. 1). Segmentation of multiple organs from abdominal CT images Figure 2 shows a flowchart of the proposed segmentation process consisting of standardisation of the abdominal cavity, rough extraction and fine segmentation. Standardization of abdominal cavity Although many papers have described standardization of the brain [6], not much attention has been paid to the
3 Int J CARS (2007) 2: D CT image Atlas guided rough extraction Fine segmentation based on multiple level set method Segmented regions Probabilistic atlas Standardization of abdominal cavity Feature database Fig. 2 Outline of the proposed algorithm, which has three steps: standardization of the abdominal cavity, rough segmentation based on a digital atlas of human anatomy, and fine segmentation using the multiple level sets standardization of the abdominal cavity. Since abdominal organs are mainly surrounded by soft tissues, there are large variations in the positions and shapes of organs. Park et al. proposed a standardisation process based on the non-linear registration technique, which is effective in reducing variation in the location of organs as well as in their shape and size. However, such a flexible method might cause excessive distortion in the region where non-continuous transformation occurs and result in decreasing the performance of the segmentation. Therefore, we used an affine transformation to normalize the abdominal cavity. The abdominal cavity includes several landmark points, which are relatively easy to detect and help to normalize the abdominal cavity, such as the tops of the kidneys. Therefore, we have focused on a landmark-based standardization process and employed five landmarks that are relatively easy to detect and are widely distributed in the abdominal cavity. The landmarks are the top voxels of the diaphragm and kidneys, as well as the lower end voxels of the liver and spleen (Fig. 3). First, the top of the diaphragm is determined by the detector ensemble, each part of which estimates the position using the changes in the lung area in axial slices, z coordinates of the right lung base, and directional difference in CT values along the body axis around the right lung base. The median of the estimated positions by these three detectors is defined as the top of the diaphragm. Next, the remaining four landmarks are detected independently using the template matching technique, which maximises the correlation coefficient between the input image and the average templates of the kidneys, liver, and spleen. It exhaustively searches within the restricted area, determined statistically from the training dataset, and then the positions with maximum correlation are defined. Finally, we apply an affine transformation, which minimizes the difference between the detected landmarks of the input image and those in the reference image. The minimization process is performed using Powell s method [7]. Atlas guided segmentation Inputs for this atlas-based method are 3D abdominal CT images and a digital atlas of human anatomy, which consists of a probabilistic atlas [2] and feature database including feature distribution parameters, such as average and covariance. Here the probabilistic atlas is computed by applying the same standardisation to each manually segmented image. Then, for each voxel in the reference volume, the fractional percentage of data that has a label at that reference voxel location is calculated. Figure 4 shows volume rendering of the probabilistic atlas of liver generated from manually segmented liver regions in 3D CT images obtained from ten patients. In the following equations, the label space is denoted L and the observed data (= feature vector) is denoted as V. Sample realizations of L and V are represented as lower case letters. The atlas guided segmentation is realized using the maximum a posteriori (MAP) algorithm. The purpose of this algorithm is to estimate the label L that best explains the observation V by maximizing the a posteriori probability given V. ˆl = arg maxp(l = l V = v) (1) l Fig. 3 Five landmarks used for standardisation of the abdominal cavity
4 138 Int J CARS (2007) 2: Fig. 4 Examples of input images. Left CT images, right probabilistic atlas of liver Using Bayes formula, the a posteriori probability p(l v) is replaced as follows: P(L = l V = v) = p(v l)p(l) (2) p(v) The elements of V were (x, y, z) coordinates at the voxel of interest and the CT values of the voxel; p(l) shows a priori probability of organ l, which is derived from the probabilistic atlas. In order to cope with the changes of the distribution parameters of the feature vector V between subjects, our method estimates the parameters by the EM algorithm for the extended mixture Gaussian distributions as follows: p(v; θ) = 1 N M l=1 n=1 N α l (n)n(v; µ l, l ) (3) where M and N are the number of organs and voxels in the image, respectively. N(v; µ l, l ) is a Gaussian distribution with average µ l and covariance l for organ l. Compared with the conventional mixture model, the mixture ratio α l (n) is extended to be defined at each voxel, which is consistent with the fact that each organ occupies a limited area. Moreover, such extension allows us to use the prior probability of organs in the digital atlas of human anatomy, resulting in higher accuracy in parameter estimation than in conventional mixture distribution, which will be demonstrated in the next section. We have derived the update equation of the mixture ratio α l (n) by introducing the Lagrange multiplier λ with the constraint M Nn=1 l=1 α l (n) = N resulting in ( α (t+1) l (n) = p l x n,θ (t)) (4) Note that there are no changes from the update equations for µ l and l of the conventional mixture distribution [8]. Finally, the proposed method applies morphological operations or opening and closing with the structure element whose size was optimized experimentally to eliminate unnecessary protrusions, holes and cavities. We performed the experiment to determine the optimal size for each organ. We applied the structure elements with different sizes to each organ extracted by the atlas-guided segmentation and selected the size when the accuracy (Eq. (9) in the next section) of the extracted regions from the training dataset was maximal. Note that the optimal size depends on organ but not patient. So it is not necessary to change the size when you apply the morphological operations to different patients. Fine segmentation based on the multiple level set method Although the above process can roughly delineate the organs borders, details of the boundaries are missed. Thus, the proposed method sophisticates the boundaries using the level set method (LSM). The introduction of constraints from neighbouring organs is known to be effective in obtaining fine segmentation [3 5]. Moreover, variation in the CT value of the exactly extracted region is usually lower than that of incorrect regions, which include several organs of different CT values. Consequently, we have developed a new method for extracting abdominal organs, which minimises the energy function that considers the hierarchy and exclusiveness between organs and uniformity of grey values in the organs. Energy relating to the hierarchy of the organs is defined as overlapping volume with outside regions of the abdominal cavity (Fig. 5) extracted with an active cylinder model [9]. Energy associated with exclusiveness is designed based on overlapping volume with neighbouring organs. Deviation in grey values I (x, y, z) is also introduced into the energy function by referring [10]. The proposed energy function
5 Int J CARS (2007) 2: c = (G σ I) where G σ is a Gaussian operator with standard deviation σ. Results Fig. 5 Estimated outside region of abdominal cavity using active cylinder model [9] expressed using regularized delta and Heaviside functions follows: M M E = α ij (1 H ε (φ i )) i=1 ω ij j=1, j =i (1 H ε (φ j ))dxdydz +β i (I(x, y, z) µ i ) 2 (1 H ε (φ i ))dxdydz (5) σ 2 i where ω ij,α ij and β i are weights and φ i is a level set function of organ i. shows an image domain, and µ i and i are the estimated average and variance of CT values. We deduced the associated Euler Lagrange equation, which minimizes the energy function with respect to φ i, while keeping µ i and i fixed. Parameterizing the descent direction by artificial time t(>0), the evolution equations in φ i (i = 1, 2,...,N) are φ i M [ ( ( ))] = δ ε (φ i ) ω ij αij 1 Hε φ j t j=1, j =i ] (I(x, y, z) µ i ) 2 +β i σi 2 (6) In this study, we linearly combine the function with the following conventional evolution equation [11] in order to consider the edges and the curvature in the process of evolution: φ i = δ ε (φ i )[c(κ + V 0 ) φ + c φ] (7) t where κ is a mean curvature of a surface of organ and V 0 is a constant value that makes a level set function go up or down with a constant speed. The number of Gaussian distributions assumed in the atlasguided segmentation is 13, including 12 organs and another tissue class corresponding to the non-target organ, such as bowel. Note that the non-organ regions, such as fat and air, as well as the lungs and bones are extracted in advance using conventional image processing, or binarisation followed by morphological processing. The LSM was implemented in a clustered computer environment that included 16 CPUs, each of which had a 2.8 GHz clock speed with 4 GB memory and was assigned to one of the organs to be extracted. The validation test was done by the re-substitution method where the dataset for learning is the same as that for the test. Estimation accuracy of the feature distribution parameters (average µ l and covariance l ) in the atlas guided segmentation process was evaluated by error = estimated value true value (8) where the true values were defined using the true regions of organs manually segmented by authors and approved by a medical doctor. Note that the µ l and l in Eq. (3) depend on organs but not on the location n. So the error of Eq. (8) was evaluated at each organ. Figure 6 demonstrates that estimation accuracy was improved by the proposed estimation process denoted as EM-ex: EM algorithm for extended mixture Gaussian distribution. In the case of liver, both the conventional and proposed estimation processes gave good results. The errors were less than two. For other organs, however, the errors of the proposed process were smaller than those of the conventional one. The errors were 32 points lower on average than the errors with the EM algorithm for conventional mixture distribution, which significantly failed to estimate the parameters of the small or elongated organs, such as the pancreas PV and IVC. The number of points in the above sentence was computed by dividing the difference between the errors of the two processes by the error of the conventional one. Compared to the errors of the parameters stored in the feature database (denoted as DB in the figure), the number of errors in the proposed method was 23 points fewer on average. Figure 7 presents examples of the extracted boundaries by MAP segmentation, which uses the estimated parameters. This figure shows that, although, most of the organs were roughly extracted, underestimation and overestimation still occurred compared with true regions when focusing on regions near boundaries.
6 140 Int J CARS (2007) 2: Error Liver Spleen L. Kidney R. Kidney Heart Gallbladder Pancreas PV Stomach Oesophagus Aorta DB EM EM-ex IVC Fig. 6 Mean and standard deviation of errors between the estimated average and true average. DB indicates errors between parameters in the database and true parameters. EM and EM-ex show results of the EM algorithm for the conventional mixture model and the extended mixture model, respectively Fig. 7 Examples of extracted boundaries by atlas guided segmentation Fig. 8 Left and middle figures show results without/with the new energy term, respectively. Please note that only the boundaries of liver and right kidney are indicated for sake of simplicity. The right figure depicts a 3D view of the extracted regions of all organs by the proposed method We evaluated the performance of MAP segmentation based on the degree of correspondence between the results and the corresponding manually segmented true regions, as defined by degree of correspondence (%) = #(extracted region true region) 100 (9) #(extracted region true region) where #(region) is a function that computes the number of voxels in the region. The degree of correspondence ranged from 0 (completely different) to 100 (completely overlapped). Using this validation test, we found that the degree of correspondence of the proposed method was four points better on average than the segmentation results using the parameters in the database. It was still higher by 2 points on average than the segmentation results with the EM algorithm for conventional mixture distribution.
7 Int J CARS (2007) 2: Degree of correspondence MAP conventional energy proposed energy 0 Liver Spleen L. Kidney R. Kidney Heart Gallbladder Pancreas PV Stomach Oesophagus Aorta IVC Fig. 9 Performance of the segmentation process for each organ. MAP gives initial boundary of the level set method. This figure shows that the proposed energy function outperforms the conventional energy function Fig. 10 Example of the segmentation by atlas guided segmentation. Left figure shows the true boundaries manually defined by authors. Middle one corresponds to the result based on the conventional parameter estimation algorithm. The right one shows the result using the proposed algorithm The subsequent LSM refined the boundaries of the organs. Figure 8 shows resultant examples without/with the proposed evolution equation. The red lines show the resultant boundaries of the liver, while the orange lines correspond to those of the right kidney. The boundaries in the middle figure successfully demonstrate the improvement by the proposed LSM. The degree of correspondence computed from Eq. (9) was improved by six points for liver and two points for the right kidney. Figure 9 presents the degree of correspondence for each organ. Based on this figure, the degree of correspondence was increased by 3.7 points on average compared with MAP based segmentation and 3.5 points better by introducing the new energy term. However, the boundaries of elongated organs with blurred boundaries, such as the pancreas, oesophagus, IVC and PV were poorly defined. The average degree of correspondence of these four organs with the stomach wall was 32.5%. In contrast, the average performance of the remaining seven organs (liver, spleen, left and right kidneys, heart, gallbladder and abdominal aorta) was 78.8%, which is acceptable for a subsequent diagnosis process. Discussion In this paper, we have presented a novel algorithm for the simultaneous extraction of 12 organs from non-contrast 3D abdominal CT images. The main contributions of this paper are the extension of the MAP based segmentation algorithm to be applicable to 12 organs in non-contrast CT images and the proposal of EM algorithm for extended mixture Gaussian distribution. Moreover the concatenation of MAP based segmentation to LSM with mutual interactions between neighbouring level sets is another important proposal. With the proposed EM algorithm, unfavourable fluctuation of the features between subjects was reduced. A typical example of the atlas-guided segmentation is presented in Fig. 10 to show the advantage of using the proposed parameter estimation algorithm. The regions denoted by yellow arrows suggested to us that the segmentation algorithm based on the proposed EM algorithm outperformed the conventional EM algorithm. The accuracy improvement in the parameter estimation process resulted in a high accuracy estimation of the regions. To statistically assess the difference in performance (= degree of correspondence defined
8 142 Int J CARS (2007) 2: Fig. 11 Example of the segmentation by LSM. Left figure shows the true boundaries. Middle one corresponds to the result based on the conventional energy, and the right one shows the result using the proposed energy by Eq. (9)) statistically, we carried out a single-tailed paired t-test. The null hypothesis was no improvement in performance. The significance level was When comparing the segmentation results using the parameters in the database with that of the proposed algorithm, we found that the improvement was statistically significant for nine organs and no difference was observed for three organs. On the contrary, when using conventional algorithm, there were two organs whose performance was significantly decreased, while eight organs were improved. The same statistical test was carried out to evaluate the improvement by the proposed LSM. Comparing the degree of correspondence in Fig. 9 of the LSM using the conventional energy with those of the atlas-guided segmentation, accuracy in the segmentation of five organs was improved, but performance of oesophagus segmentation was decreased significantly. In contrast, the LSM based on the new energy increased the performance in segmentation of nine organs, and no deterioration was observed. The statistical difference between the results using the conventional energy and those of the proposed energy was found in seven organs. An example is presented in Fig. 11 where mutual interaction improved the accuracy in segmentation of organs as indicated by the yellow arrows. However, such interaction occasionally caused error in segmentation. The failure to extract the abdominal cavity s boundary led to the error in segmentation of the heart as indicated by the red arrow. As shown in Fig. 9, the proposed method was not sufficiently accurate in the segmentation of the small and elongated organs, such as the oesophagus, PV and pancreas, even though it is difficult for radiologists to define these boundaries on non-contrast CT images. When segmenting such structures, a priori knowledge of the shapes and spatial relationships between organs is very helpful [3 5,12]. Inthe future, a novel a posteriori probability that takes into account the probabilities of neighbouring organs will be introduced into the atlas-guided segmentation process. We also plan to develop a new energy term to incorporate the shape a priori information to recover the 3D shapes. Testing the performance, using a large-scale database and extension of the method to be applicable to the whole body are other exciting and challenging future projects. References 1. Kobatake H (2007) Future CAD in multi-dimensional medical images - project on multi-organ, multi-disease CAD system - Comput Med Imaging Graph 31(4 5): Park H, Bland PH, Meyer CR (2003) Construction of an abdominal probabilistic atlas its application in segmentation. IEEE Trans Med Imaging 22(4): Yang J, Staib L, Duncan J (2004) Neighbor-constrained segmentation with level set based 3-D deformable models. IEEE Trans Med Imaging 23(8): Tsai A, Wells W, Tempany C et al (2003) Coupled multi-shape model and mutual information for medical image segmentation. Inf Process Med Imaging 18: Yan P, Shen W, Kassim AA et al (2005) Segmentation of neighboring organs in medical image with model competition. In: Proceedings of MICCAI 2005, LNCS3749, pp Maintz JBA, Viergever MA (1998) A survey of medical image registration. Med Image Anal 2: Press WH, Flannery BP, Teukolsky SA et al (1995) Numerical recipes in C, 2nd edn. Cambridge University Press, Kluwer, London, Dordrecht, pp Ghahramami Z, Jordan M (1995) Learning from incomplete data. Technical Report AI Lab Memo no. 1509, CBCL Paper no. 108, MIT AI Lab 9. Okumura T, Yamamoto S, Matsumoto M et al (1998) The lung region extraction in the chest CT images by the active cylinder model. Med Imaging Technol 16(1): Chan TF, Vese LF (2001) Active contours without edges. IEEE Trans Image Process 10(2): Caselles V, Kimmel R, Sapiro G (1997) Geodesic active contours. Int J Comput Vis 22(1): Yao C, Wada T, Shimizu A et al (2006) Probabilistic atlas-guided Eigen-organ method for simultaneous bounding box estimation of multiple organs in volumetric CT images. Med Imaging Technol 24(3):
3D Surface Reconstruction of the Brain based on Level Set Method
 3D Surface Reconstruction of the Brain based on Level Set Method Shijun Tang, Bill P. Buckles, and Kamesh Namuduri Department of Computer Science & Engineering Department of Electrical Engineering University
3D Surface Reconstruction of the Brain based on Level Set Method Shijun Tang, Bill P. Buckles, and Kamesh Namuduri Department of Computer Science & Engineering Department of Electrical Engineering University
Machine Learning for Medical Image Analysis. A. Criminisi
 Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
A Study of Medical Image Analysis System
 Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
Deformable Segmentation using Sparse Shape Representation. Shaoting Zhang
 Deformable Segmentation using Sparse Shape Representation Shaoting Zhang Introduction Outline Our methods Segmentation framework Sparse shape representation Applications 2D lung localization in X-ray 3D
Deformable Segmentation using Sparse Shape Representation Shaoting Zhang Introduction Outline Our methods Segmentation framework Sparse shape representation Applications 2D lung localization in X-ray 3D
Hierarchical Multi structure Segmentation Guided by Anatomical Correlations
 Hierarchical Multi structure Segmentation Guided by Anatomical Correlations Oscar Alfonso Jiménez del Toro oscar.jimenez@hevs.ch Henning Müller henningmueller@hevs.ch University of Applied Sciences Western
Hierarchical Multi structure Segmentation Guided by Anatomical Correlations Oscar Alfonso Jiménez del Toro oscar.jimenez@hevs.ch Henning Müller henningmueller@hevs.ch University of Applied Sciences Western
Annales UMCS Informatica AI 1 (2003) UMCS. Registration of CT and MRI brain images. Karol Kuczyński, Paweł Mikołajczak
 Annales Informatica AI 1 (2003) 149-156 Registration of CT and MRI brain images Karol Kuczyński, Paweł Mikołajczak Annales Informatica Lublin-Polonia Sectio AI http://www.annales.umcs.lublin.pl/ Laboratory
Annales Informatica AI 1 (2003) 149-156 Registration of CT and MRI brain images Karol Kuczyński, Paweł Mikołajczak Annales Informatica Lublin-Polonia Sectio AI http://www.annales.umcs.lublin.pl/ Laboratory
Modeling and preoperative planning for kidney surgery
 Modeling and preoperative planning for kidney surgery Refael Vivanti Computer Aided Surgery and Medical Image Processing Lab Hebrew University of Jerusalem, Israel Advisor: Prof. Leo Joskowicz Clinical
Modeling and preoperative planning for kidney surgery Refael Vivanti Computer Aided Surgery and Medical Image Processing Lab Hebrew University of Jerusalem, Israel Advisor: Prof. Leo Joskowicz Clinical
Prostate Detection Using Principal Component Analysis
 Prostate Detection Using Principal Component Analysis Aamir Virani (avirani@stanford.edu) CS 229 Machine Learning Stanford University 16 December 2005 Introduction During the past two decades, computed
Prostate Detection Using Principal Component Analysis Aamir Virani (avirani@stanford.edu) CS 229 Machine Learning Stanford University 16 December 2005 Introduction During the past two decades, computed
Comparison of Vessel Segmentations Using STAPLE
 Comparison of Vessel Segmentations Using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, The University of North Carolina at Chapel Hill, Department
Comparison of Vessel Segmentations Using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, The University of North Carolina at Chapel Hill, Department
Comparison of Vessel Segmentations using STAPLE
 Comparison of Vessel Segmentations using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel Hill, Department
Comparison of Vessel Segmentations using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel Hill, Department
A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images
 A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images Jianhua Yao 1, Russell Taylor 2 1. Diagnostic Radiology Department, Clinical Center,
A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images Jianhua Yao 1, Russell Taylor 2 1. Diagnostic Radiology Department, Clinical Center,
Extraction and recognition of the thoracic organs based on 3D CT images and its application
 1 Extraction and recognition of the thoracic organs based on 3D CT images and its application Xiangrong Zhou, PhD a, Takeshi Hara, PhD b, Hiroshi Fujita, PhD b, Yoshihiro Ida, RT c, Kazuhiro Katada, MD
1 Extraction and recognition of the thoracic organs based on 3D CT images and its application Xiangrong Zhou, PhD a, Takeshi Hara, PhD b, Hiroshi Fujita, PhD b, Yoshihiro Ida, RT c, Kazuhiro Katada, MD
Rule-Based Ventral Cavity Multi-organ Automatic Segmentation in CT Scans
 Rule-Based Ventral Cavity Multi-organ Automatic Segmentation in CT Scans Assaf B. Spanier (B) and Leo Joskowicz The Rachel and Selim Benin School of Computer Science and Engineering, The Hebrew University
Rule-Based Ventral Cavity Multi-organ Automatic Segmentation in CT Scans Assaf B. Spanier (B) and Leo Joskowicz The Rachel and Selim Benin School of Computer Science and Engineering, The Hebrew University
Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs
 Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs Yonghong Shi 1 and Dinggang Shen 2,*1 1 Digital Medical Research Center, Fudan University, Shanghai, 232, China
Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs Yonghong Shi 1 and Dinggang Shen 2,*1 1 Digital Medical Research Center, Fudan University, Shanghai, 232, China
College of Engineering, Trivandrum.
 Analysis of CT Liver Images Using Level Sets with Bayesian Analysis-A Hybrid Approach Sajith A.G 1, Dr. Hariharan.S 2 1 Research Scholar, 2 Professor, Department of Electrical&Electronics Engineering College
Analysis of CT Liver Images Using Level Sets with Bayesian Analysis-A Hybrid Approach Sajith A.G 1, Dr. Hariharan.S 2 1 Research Scholar, 2 Professor, Department of Electrical&Electronics Engineering College
arxiv: v1 [cs.cv] 6 Jun 2017
![arxiv: v1 [cs.cv] 6 Jun 2017 arxiv: v1 [cs.cv] 6 Jun 2017](/thumbs/96/128307097.jpg) Volume Calculation of CT lung Lesions based on Halton Low-discrepancy Sequences Liansheng Wang a, Shusheng Li a, and Shuo Li b a Department of Computer Science, Xiamen University, Xiamen, China b Dept.
Volume Calculation of CT lung Lesions based on Halton Low-discrepancy Sequences Liansheng Wang a, Shusheng Li a, and Shuo Li b a Department of Computer Science, Xiamen University, Xiamen, China b Dept.
Automated segmentation methods for liver analysis in oncology applications
 University of Szeged Department of Image Processing and Computer Graphics Automated segmentation methods for liver analysis in oncology applications Ph. D. Thesis László Ruskó Thesis Advisor Dr. Antal
University of Szeged Department of Image Processing and Computer Graphics Automated segmentation methods for liver analysis in oncology applications Ph. D. Thesis László Ruskó Thesis Advisor Dr. Antal
3D Volume Mesh Generation of Human Organs Using Surface Geometries Created from the Visible Human Data Set
 3D Volume Mesh Generation of Human Organs Using Surface Geometries Created from the Visible Human Data Set John M. Sullivan, Jr., Ziji Wu, and Anand Kulkarni Worcester Polytechnic Institute Worcester,
3D Volume Mesh Generation of Human Organs Using Surface Geometries Created from the Visible Human Data Set John M. Sullivan, Jr., Ziji Wu, and Anand Kulkarni Worcester Polytechnic Institute Worcester,
Global-to-Local Shape Matching for Liver Segmentation in CT Imaging
 Global-to-Local Shape Matching for Liver Segmentation in CT Imaging Kinda Anna Saddi 1,2, Mikaël Rousson 1, Christophe Chefd hotel 1, and Farida Cheriet 2 1 Department of Imaging and Visualization, Siemens
Global-to-Local Shape Matching for Liver Segmentation in CT Imaging Kinda Anna Saddi 1,2, Mikaël Rousson 1, Christophe Chefd hotel 1, and Farida Cheriet 2 1 Department of Imaging and Visualization, Siemens
Universities of Leeds, Sheffield and York
 promoting access to White Rose research papers Universities of Leeds, Sheffield and York http://eprints.whiterose.ac.uk/ This is an author produced version of a paper published in Lecture Notes in Computer
promoting access to White Rose research papers Universities of Leeds, Sheffield and York http://eprints.whiterose.ac.uk/ This is an author produced version of a paper published in Lecture Notes in Computer
CHAPTER 2. Morphometry on rodent brains. A.E.H. Scheenstra J. Dijkstra L. van der Weerd
 CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
Computational Neuroanatomy
 Computational Neuroanatomy John Ashburner john@fil.ion.ucl.ac.uk Smoothing Motion Correction Between Modality Co-registration Spatial Normalisation Segmentation Morphometry Overview fmri time-series kernel
Computational Neuroanatomy John Ashburner john@fil.ion.ucl.ac.uk Smoothing Motion Correction Between Modality Co-registration Spatial Normalisation Segmentation Morphometry Overview fmri time-series kernel
A multi-atlas approach for prostate segmentation in MR images
 A multi-atlas approach for prostate segmentation in MR images Geert Litjens, Nico Karssemeijer, and Henkjan Huisman Diagnostic Image Analysis Group, Radboud University Nijmegen Medical Centre, Nijmegen,
A multi-atlas approach for prostate segmentation in MR images Geert Litjens, Nico Karssemeijer, and Henkjan Huisman Diagnostic Image Analysis Group, Radboud University Nijmegen Medical Centre, Nijmegen,
Medical Image Segmentation
 Medical Image Segmentation Xin Yang, HUST *Collaborated with UCLA Medical School and UCSB Segmentation to Contouring ROI Aorta & Kidney 3D Brain MR Image 3D Abdominal CT Image Liver & Spleen Caudate Nucleus
Medical Image Segmentation Xin Yang, HUST *Collaborated with UCLA Medical School and UCSB Segmentation to Contouring ROI Aorta & Kidney 3D Brain MR Image 3D Abdominal CT Image Liver & Spleen Caudate Nucleus
Manifold Learning-based Data Sampling for Model Training
 Manifold Learning-based Data Sampling for Model Training Shuqing Chen 1, Sabrina Dorn 2, Michael Lell 3, Marc Kachelrieß 2,Andreas Maier 1 1 Pattern Recognition Lab, FAU Erlangen-Nürnberg 2 German Cancer
Manifold Learning-based Data Sampling for Model Training Shuqing Chen 1, Sabrina Dorn 2, Michael Lell 3, Marc Kachelrieß 2,Andreas Maier 1 1 Pattern Recognition Lab, FAU Erlangen-Nürnberg 2 German Cancer
Computational Radiology Lab, Children s Hospital, Harvard Medical School, Boston, MA.
 Shape prior integration in discrete optimization segmentation algorithms M. Freiman Computational Radiology Lab, Children s Hospital, Harvard Medical School, Boston, MA. Email: moti.freiman@childrens.harvard.edu
Shape prior integration in discrete optimization segmentation algorithms M. Freiman Computational Radiology Lab, Children s Hospital, Harvard Medical School, Boston, MA. Email: moti.freiman@childrens.harvard.edu
A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation
 A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation Xiahai Zhuang (PhD) Centre for Medical Image Computing University College London Fields-MITACS Conference on Mathematics
A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation Xiahai Zhuang (PhD) Centre for Medical Image Computing University College London Fields-MITACS Conference on Mathematics
Edge-Preserving Denoising for Segmentation in CT-Images
 Edge-Preserving Denoising for Segmentation in CT-Images Eva Eibenberger, Anja Borsdorf, Andreas Wimmer, Joachim Hornegger Lehrstuhl für Mustererkennung, Friedrich-Alexander-Universität Erlangen-Nürnberg
Edge-Preserving Denoising for Segmentation in CT-Images Eva Eibenberger, Anja Borsdorf, Andreas Wimmer, Joachim Hornegger Lehrstuhl für Mustererkennung, Friedrich-Alexander-Universität Erlangen-Nürnberg
Conference Biomedical Engineering
 Automatic Medical Image Analysis for Measuring Bone Thickness and Density M. Kovalovs *, A. Glazs Image Processing and Computer Graphics Department, Riga Technical University, Latvia * E-mail: mihails.kovalovs@rtu.lv
Automatic Medical Image Analysis for Measuring Bone Thickness and Density M. Kovalovs *, A. Glazs Image Processing and Computer Graphics Department, Riga Technical University, Latvia * E-mail: mihails.kovalovs@rtu.lv
3D Statistical Shape Model Building using Consistent Parameterization
 3D Statistical Shape Model Building using Consistent Parameterization Matthias Kirschner, Stefan Wesarg Graphisch Interaktive Systeme, TU Darmstadt matthias.kirschner@gris.tu-darmstadt.de Abstract. We
3D Statistical Shape Model Building using Consistent Parameterization Matthias Kirschner, Stefan Wesarg Graphisch Interaktive Systeme, TU Darmstadt matthias.kirschner@gris.tu-darmstadt.de Abstract. We
Scene-Based Segmentation of Multiple Muscles from MRI in MITK
 Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
3-D Compounding of B-Scan Ultrasound Images
 3-D Compounding of B-Scan Ultrasound Images Jochen F. Krücker, Charles R. Meyer, Theresa A. Tuthill, Gerald L. LeCarpentier, J. Brian Fowlkes, Paul L. Carson University of Michigan, Dept. of Radiology,
3-D Compounding of B-Scan Ultrasound Images Jochen F. Krücker, Charles R. Meyer, Theresa A. Tuthill, Gerald L. LeCarpentier, J. Brian Fowlkes, Paul L. Carson University of Michigan, Dept. of Radiology,
Segmenting the Left Ventricle in 3D Using a Coupled ASM and a Learned Non-Rigid Spatial Model
 Segmenting the Left Ventricle in 3D Using a Coupled ASM and a Learned Non-Rigid Spatial Model Stephen O Brien, Ovidiu Ghita, and Paul F. Whelan Centre for Image Processing and Analysis, Dublin City University,
Segmenting the Left Ventricle in 3D Using a Coupled ASM and a Learned Non-Rigid Spatial Model Stephen O Brien, Ovidiu Ghita, and Paul F. Whelan Centre for Image Processing and Analysis, Dublin City University,
Adaptive Local Multi-Atlas Segmentation: Application to Heart Segmentation in Chest CT Scans
 Adaptive Local Multi-Atlas Segmentation: Application to Heart Segmentation in Chest CT Scans Eva M. van Rikxoort, Ivana Isgum, Marius Staring, Stefan Klein and Bram van Ginneken Image Sciences Institute,
Adaptive Local Multi-Atlas Segmentation: Application to Heart Segmentation in Chest CT Scans Eva M. van Rikxoort, Ivana Isgum, Marius Staring, Stefan Klein and Bram van Ginneken Image Sciences Institute,
Automated Segmentation Using a Fast Implementation of the Chan-Vese Models
 Automated Segmentation Using a Fast Implementation of the Chan-Vese Models Huan Xu, and Xiao-Feng Wang,,3 Intelligent Computation Lab, Hefei Institute of Intelligent Machines, Chinese Academy of Science,
Automated Segmentation Using a Fast Implementation of the Chan-Vese Models Huan Xu, and Xiao-Feng Wang,,3 Intelligent Computation Lab, Hefei Institute of Intelligent Machines, Chinese Academy of Science,
Computer-Aided Diagnosis in Abdominal and Cardiac Radiology Using Neural Networks
 Computer-Aided Diagnosis in Abdominal and Cardiac Radiology Using Neural Networks Du-Yih Tsai, Masaru Sekiya and Yongbum Lee Department of Radiological Technology, School of Health Sciences, Faculty of
Computer-Aided Diagnosis in Abdominal and Cardiac Radiology Using Neural Networks Du-Yih Tsai, Masaru Sekiya and Yongbum Lee Department of Radiological Technology, School of Health Sciences, Faculty of
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy
 The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
Application of level set based method for segmentation of blood vessels in angiography images
 Lodz University of Technology Faculty of Electrical, Electronic, Computer and Control Engineering Institute of Electronics PhD Thesis Application of level set based method for segmentation of blood vessels
Lodz University of Technology Faculty of Electrical, Electronic, Computer and Control Engineering Institute of Electronics PhD Thesis Application of level set based method for segmentation of blood vessels
Automatic Vascular Tree Formation Using the Mahalanobis Distance
 Automatic Vascular Tree Formation Using the Mahalanobis Distance Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, Department of Radiology The University
Automatic Vascular Tree Formation Using the Mahalanobis Distance Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, Department of Radiology The University
Robust Linear Registration of CT images using Random Regression Forests
 Robust Linear Registration of CT images using Random Regression Forests Ender Konukoglu a Antonio Criminisi a Sayan Pathak b Duncan Robertson a Steve White b David Haynor c Khan Siddiqui b a Microsoft
Robust Linear Registration of CT images using Random Regression Forests Ender Konukoglu a Antonio Criminisi a Sayan Pathak b Duncan Robertson a Steve White b David Haynor c Khan Siddiqui b a Microsoft
Fully Automatic Model Creation for Object Localization utilizing the Generalized Hough Transform
 Fully Automatic Model Creation for Object Localization utilizing the Generalized Hough Transform Heike Ruppertshofen 1,2,3, Cristian Lorenz 2, Peter Beyerlein 4, Zein Salah 3, Georg Rose 3, Hauke Schramm
Fully Automatic Model Creation for Object Localization utilizing the Generalized Hough Transform Heike Ruppertshofen 1,2,3, Cristian Lorenz 2, Peter Beyerlein 4, Zein Salah 3, Georg Rose 3, Hauke Schramm
Marginal Space Learning for Efficient Detection of 2D/3D Anatomical Structures in Medical Images
 Marginal Space Learning for Efficient Detection of 2D/3D Anatomical Structures in Medical Images Yefeng Zheng, Bogdan Georgescu, and Dorin Comaniciu Integrated Data Systems Department, Siemens Corporate
Marginal Space Learning for Efficient Detection of 2D/3D Anatomical Structures in Medical Images Yefeng Zheng, Bogdan Georgescu, and Dorin Comaniciu Integrated Data Systems Department, Siemens Corporate
Comparison of Different Metrics for Appearance-model-based 2D/3D-registration with X-ray Images
 Comparison of Different Metrics for Appearance-model-based 2D/3D-registration with X-ray Images Philipp Steininger 1, Karl D. Fritscher 1, Gregor Kofler 1, Benedikt Schuler 1, Markus Hänni 2, Karsten Schwieger
Comparison of Different Metrics for Appearance-model-based 2D/3D-registration with X-ray Images Philipp Steininger 1, Karl D. Fritscher 1, Gregor Kofler 1, Benedikt Schuler 1, Markus Hänni 2, Karsten Schwieger
Automated segmentations of skin, soft-tissue, and skeleton from torso CT images
 Automated segmentations of skin, soft-tissue, and skeleton from torso CT images Xiangrong Zhou* a, Takeshi Hara a, Hiroshi Fujita a, Ryujiro Yokoyama b, Takuji Kiryu b, and Hiroaki Hoshi b a Department
Automated segmentations of skin, soft-tissue, and skeleton from torso CT images Xiangrong Zhou* a, Takeshi Hara a, Hiroshi Fujita a, Ryujiro Yokoyama b, Takuji Kiryu b, and Hiroaki Hoshi b a Department
Image Segmentation and Registration
 Image Segmentation and Registration Dr. Christine Tanner (tanner@vision.ee.ethz.ch) Computer Vision Laboratory, ETH Zürich Dr. Verena Kaynig, Machine Learning Laboratory, ETH Zürich Outline Segmentation
Image Segmentation and Registration Dr. Christine Tanner (tanner@vision.ee.ethz.ch) Computer Vision Laboratory, ETH Zürich Dr. Verena Kaynig, Machine Learning Laboratory, ETH Zürich Outline Segmentation
TUBULAR SURFACES EXTRACTION WITH MINIMAL ACTION SURFACES
 TUBULAR SURFACES EXTRACTION WITH MINIMAL ACTION SURFACES XIANGJUN GAO Department of Computer and Information Technology, Shangqiu Normal University, Shangqiu 476000, Henan, China ABSTRACT This paper presents
TUBULAR SURFACES EXTRACTION WITH MINIMAL ACTION SURFACES XIANGJUN GAO Department of Computer and Information Technology, Shangqiu Normal University, Shangqiu 476000, Henan, China ABSTRACT This paper presents
Joint CI-JAI advanced accelerator lecture series Imaging and detectors for medical physics Lecture 1: Medical imaging
 Joint CI-JAI advanced accelerator lecture series Imaging and detectors for medical physics Lecture 1: Medical imaging Dr Barbara Camanzi barbara.camanzi@stfc.ac.uk Course layout Day AM 09.30 11.00 PM 15.30
Joint CI-JAI advanced accelerator lecture series Imaging and detectors for medical physics Lecture 1: Medical imaging Dr Barbara Camanzi barbara.camanzi@stfc.ac.uk Course layout Day AM 09.30 11.00 PM 15.30
Advanced Image Reconstruction Methods for Photoacoustic Tomography
 Advanced Image Reconstruction Methods for Photoacoustic Tomography Mark A. Anastasio, Kun Wang, and Robert Schoonover Department of Biomedical Engineering Washington University in St. Louis 1 Outline Photoacoustic/thermoacoustic
Advanced Image Reconstruction Methods for Photoacoustic Tomography Mark A. Anastasio, Kun Wang, and Robert Schoonover Department of Biomedical Engineering Washington University in St. Louis 1 Outline Photoacoustic/thermoacoustic
Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion
 Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion Mattias P. Heinrich Julia A. Schnabel, Mark Jenkinson, Sir Michael Brady 2 Clinical
Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion Mattias P. Heinrich Julia A. Schnabel, Mark Jenkinson, Sir Michael Brady 2 Clinical
2D-3D Registration using Gradient-based MI for Image Guided Surgery Systems
 2D-3D Registration using Gradient-based MI for Image Guided Surgery Systems Yeny Yim 1*, Xuanyi Chen 1, Mike Wakid 1, Steve Bielamowicz 2, James Hahn 1 1 Department of Computer Science, The George Washington
2D-3D Registration using Gradient-based MI for Image Guided Surgery Systems Yeny Yim 1*, Xuanyi Chen 1, Mike Wakid 1, Steve Bielamowicz 2, James Hahn 1 1 Department of Computer Science, The George Washington
Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information
 Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information Andreas Biesdorf 1, Stefan Wörz 1, Hans-Jürgen Kaiser 2, Karl Rohr 1 1 University of Heidelberg, BIOQUANT, IPMB,
Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information Andreas Biesdorf 1, Stefan Wörz 1, Hans-Jürgen Kaiser 2, Karl Rohr 1 1 University of Heidelberg, BIOQUANT, IPMB,
Basic principles of MR image analysis. Basic principles of MR image analysis. Basic principles of MR image analysis
 Basic principles of MR image analysis Basic principles of MR image analysis Julien Milles Leiden University Medical Center Terminology of fmri Brain extraction Registration Linear registration Non-linear
Basic principles of MR image analysis Basic principles of MR image analysis Julien Milles Leiden University Medical Center Terminology of fmri Brain extraction Registration Linear registration Non-linear
2 Michael E. Leventon and Sarah F. F. Gibson a b c d Fig. 1. (a, b) Two MR scans of a person's knee. Both images have high resolution in-plane, but ha
 Model Generation from Multiple Volumes using Constrained Elastic SurfaceNets Michael E. Leventon and Sarah F. F. Gibson 1 MIT Artificial Intelligence Laboratory, Cambridge, MA 02139, USA leventon@ai.mit.edu
Model Generation from Multiple Volumes using Constrained Elastic SurfaceNets Michael E. Leventon and Sarah F. F. Gibson 1 MIT Artificial Intelligence Laboratory, Cambridge, MA 02139, USA leventon@ai.mit.edu
MRA Image Segmentation with Capillary Active Contour
 MRA Image Segmentation with Capillary Active Contour Pingkun Yan and Ashraf A. Kassim Department of Electrical & Computer Engineering, National University of Singapore {pingkun,ashraf}@nus.edu.sg Abstract.
MRA Image Segmentation with Capillary Active Contour Pingkun Yan and Ashraf A. Kassim Department of Electrical & Computer Engineering, National University of Singapore {pingkun,ashraf}@nus.edu.sg Abstract.
Object Identification in Ultrasound Scans
 Object Identification in Ultrasound Scans Wits University Dec 05, 2012 Roadmap Introduction to the problem Motivation Related Work Our approach Expected Results Introduction Nowadays, imaging devices like
Object Identification in Ultrasound Scans Wits University Dec 05, 2012 Roadmap Introduction to the problem Motivation Related Work Our approach Expected Results Introduction Nowadays, imaging devices like
Automated segmentation of 3D anatomical structures on CT images by using a deep convolutional network based on end-to-end learning approach
 Automated segmentation of 3D anatomical structures on CT images by using a deep convolutional network based on end-to-end learning approach Xiangrong Zhou* a, Ryosuke Takayama a, Song Wang b, Xinxin Zhou
Automated segmentation of 3D anatomical structures on CT images by using a deep convolutional network based on end-to-end learning approach Xiangrong Zhou* a, Ryosuke Takayama a, Song Wang b, Xinxin Zhou
ISSN: X International Journal of Advanced Research in Electronics and Communication Engineering (IJARECE) Volume 6, Issue 8, August 2017
 ENTROPY BASED CONSTRAINT METHOD FOR IMAGE SEGMENTATION USING ACTIVE CONTOUR MODEL M.Nirmala Department of ECE JNTUA college of engineering, Anantapuramu Andhra Pradesh,India Abstract: Over the past existing
ENTROPY BASED CONSTRAINT METHOD FOR IMAGE SEGMENTATION USING ACTIVE CONTOUR MODEL M.Nirmala Department of ECE JNTUA college of engineering, Anantapuramu Andhra Pradesh,India Abstract: Over the past existing
City, University of London Institutional Repository
 City Research Online City, University of London Institutional Repository Citation: Doan, H., Slabaugh, G.G., Unal, G.B. & Fang, T. (2006). Semi-Automatic 3-D Segmentation of Anatomical Structures of Brain
City Research Online City, University of London Institutional Repository Citation: Doan, H., Slabaugh, G.G., Unal, G.B. & Fang, T. (2006). Semi-Automatic 3-D Segmentation of Anatomical Structures of Brain
Three-dimensional Image Processing Techniques to Perform Landmarking and Segmentation of Computed Tomographic Images
 Three-dimensional Image Processing Techniques to Perform Landmarking and Segmentation of Computed Tomographic Images Rangaraj M. Rangayyan, Shantanu Banik, Graham S. Boag Department of Electrical and Computer
Three-dimensional Image Processing Techniques to Perform Landmarking and Segmentation of Computed Tomographic Images Rangaraj M. Rangayyan, Shantanu Banik, Graham S. Boag Department of Electrical and Computer
Corso di laurea in Fisica A.A Fisica Medica 4 TC
 Corso di laurea in Fisica A.A. 2007-2008 Fisica Medica 4 TC Computed Tomography Principles 1. Projection measurement 2. Scanner systems 3. Scanning modes Basic Tomographic Principle The internal structure
Corso di laurea in Fisica A.A. 2007-2008 Fisica Medica 4 TC Computed Tomography Principles 1. Projection measurement 2. Scanner systems 3. Scanning modes Basic Tomographic Principle The internal structure
Methods for data preprocessing
 Methods for data preprocessing John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. Overview Voxel-Based Morphometry Morphometry in general Volumetrics VBM preprocessing
Methods for data preprocessing John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. Overview Voxel-Based Morphometry Morphometry in general Volumetrics VBM preprocessing
Hierarchical Patch Generation for Multi-level Statistical Shape Analysis by Principal Factor Analysis Decomposition
 Hierarchical Patch Generation for Multi-level Statistical Shape Analysis by Principal Factor Analysis Decomposition Mauricio Reyes 1, Miguel A. González Ballester 2, Nina Kozic 1, Jesse K. Sandberg 3,
Hierarchical Patch Generation for Multi-level Statistical Shape Analysis by Principal Factor Analysis Decomposition Mauricio Reyes 1, Miguel A. González Ballester 2, Nina Kozic 1, Jesse K. Sandberg 3,
CT NOISE POWER SPECTRUM FOR FILTERED BACKPROJECTION AND ITERATIVE RECONSTRUCTION
 CT NOISE POWER SPECTRUM FOR FILTERED BACKPROJECTION AND ITERATIVE RECONSTRUCTION Frank Dong, PhD, DABR Diagnostic Physicist, Imaging Institute Cleveland Clinic Foundation and Associate Professor of Radiology
CT NOISE POWER SPECTRUM FOR FILTERED BACKPROJECTION AND ITERATIVE RECONSTRUCTION Frank Dong, PhD, DABR Diagnostic Physicist, Imaging Institute Cleveland Clinic Foundation and Associate Professor of Radiology
Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging
 1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
AN essential part of any computer-aided surgery is planning
 1 A Model Based Validation Scheme for Organ Segmentation in CT Scan Volumes Hossein Badakhshannoory, Student Member, IEEE, and Parvaneh Saeedi, Member, IEEE Abstract In this work, we propose a novel approach
1 A Model Based Validation Scheme for Organ Segmentation in CT Scan Volumes Hossein Badakhshannoory, Student Member, IEEE, and Parvaneh Saeedi, Member, IEEE Abstract In this work, we propose a novel approach
Image Registration + Other Stuff
 Image Registration + Other Stuff John Ashburner Pre-processing Overview fmri time-series Motion Correct Anatomical MRI Coregister m11 m 21 m 31 m12 m13 m14 m 22 m 23 m 24 m 32 m 33 m 34 1 Template Estimate
Image Registration + Other Stuff John Ashburner Pre-processing Overview fmri time-series Motion Correct Anatomical MRI Coregister m11 m 21 m 31 m12 m13 m14 m 22 m 23 m 24 m 32 m 33 m 34 1 Template Estimate
Learning-based Neuroimage Registration
 Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge
 Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
MR IMAGE SEGMENTATION
 MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
Automatic Lung Surface Registration Using Selective Distance Measure in Temporal CT Scans
 Automatic Lung Surface Registration Using Selective Distance Measure in Temporal CT Scans Helen Hong 1, Jeongjin Lee 2, Kyung Won Lee 3, and Yeong Gil Shin 2 1 School of Electrical Engineering and Computer
Automatic Lung Surface Registration Using Selective Distance Measure in Temporal CT Scans Helen Hong 1, Jeongjin Lee 2, Kyung Won Lee 3, and Yeong Gil Shin 2 1 School of Electrical Engineering and Computer
NIH Public Access Author Manuscript Proc Soc Photo Opt Instrum Eng. Author manuscript; available in PMC 2014 October 07.
 NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
Automatized & Interactive. Muscle tissues characterization using. Na MRI
 Automatized & Interactive Human Skeletal Muscle Segmentation Muscle tissues characterization using 23 Na MRI Noura Azzabou 30 April 2013 What is muscle segmentation? Axial slice of the thigh of a healthy
Automatized & Interactive Human Skeletal Muscle Segmentation Muscle tissues characterization using 23 Na MRI Noura Azzabou 30 April 2013 What is muscle segmentation? Axial slice of the thigh of a healthy
SCIENCE & TECHNOLOGY
 Pertanika J. Sci. & Technol. 26 (1): 309-316 (2018) SCIENCE & TECHNOLOGY Journal homepage: http://www.pertanika.upm.edu.my/ Application of Active Contours Driven by Local Gaussian Distribution Fitting
Pertanika J. Sci. & Technol. 26 (1): 309-316 (2018) SCIENCE & TECHNOLOGY Journal homepage: http://www.pertanika.upm.edu.my/ Application of Active Contours Driven by Local Gaussian Distribution Fitting
PART-LEVEL OBJECT RECOGNITION
 PART-LEVEL OBJECT RECOGNITION Jaka Krivic and Franc Solina University of Ljubljana Faculty of Computer and Information Science Computer Vision Laboratory Tržaška 25, 1000 Ljubljana, Slovenia {jakak, franc}@lrv.fri.uni-lj.si
PART-LEVEL OBJECT RECOGNITION Jaka Krivic and Franc Solina University of Ljubljana Faculty of Computer and Information Science Computer Vision Laboratory Tržaška 25, 1000 Ljubljana, Slovenia {jakak, franc}@lrv.fri.uni-lj.si
Spiral CT. Protocol Optimization & Quality Assurance. Ge Wang, Ph.D. Department of Radiology University of Iowa Iowa City, Iowa 52242, USA
 Spiral CT Protocol Optimization & Quality Assurance Ge Wang, Ph.D. Department of Radiology University of Iowa Iowa City, Iowa 52242, USA Spiral CT Protocol Optimization & Quality Assurance Protocol optimization
Spiral CT Protocol Optimization & Quality Assurance Ge Wang, Ph.D. Department of Radiology University of Iowa Iowa City, Iowa 52242, USA Spiral CT Protocol Optimization & Quality Assurance Protocol optimization
Fast 3D Mean Shift Filter for CT Images
 Fast 3D Mean Shift Filter for CT Images Gustavo Fernández Domínguez, Horst Bischof, and Reinhard Beichel Institute for Computer Graphics and Vision, Graz University of Technology Inffeldgasse 16/2, A-8010,
Fast 3D Mean Shift Filter for CT Images Gustavo Fernández Domínguez, Horst Bischof, and Reinhard Beichel Institute for Computer Graphics and Vision, Graz University of Technology Inffeldgasse 16/2, A-8010,
doi: /
 Shuang Liu ; Mary Salvatore ; David F. Yankelevitz ; Claudia I. Henschke ; Anthony P. Reeves; Segmentation of the whole breast from low-dose chest CT images. Proc. SPIE 9414, Medical Imaging 2015: Computer-Aided
Shuang Liu ; Mary Salvatore ; David F. Yankelevitz ; Claudia I. Henschke ; Anthony P. Reeves; Segmentation of the whole breast from low-dose chest CT images. Proc. SPIE 9414, Medical Imaging 2015: Computer-Aided
Discriminative Dictionary Learning for Abdominal Multi-Organ Segmentation
 Discriminative Dictionary Learning for Abdominal Multi-Organ Segmentation Tong Tong a,, Robin Wolz a, Zehan Wang a, Qinquan Gao a, Kazunari Misawa b, Michitaka Fujiwara c, Kensaku Mori d, Joseph V. Hajnal
Discriminative Dictionary Learning for Abdominal Multi-Organ Segmentation Tong Tong a,, Robin Wolz a, Zehan Wang a, Qinquan Gao a, Kazunari Misawa b, Michitaka Fujiwara c, Kensaku Mori d, Joseph V. Hajnal
8/3/2017. Contour Assessment for Quality Assurance and Data Mining. Objective. Outline. Tom Purdie, PhD, MCCPM
 Contour Assessment for Quality Assurance and Data Mining Tom Purdie, PhD, MCCPM Objective Understand the state-of-the-art in contour assessment for quality assurance including data mining-based techniques
Contour Assessment for Quality Assurance and Data Mining Tom Purdie, PhD, MCCPM Objective Understand the state-of-the-art in contour assessment for quality assurance including data mining-based techniques
Edge and local feature detection - 2. Importance of edge detection in computer vision
 Edge and local feature detection Gradient based edge detection Edge detection by function fitting Second derivative edge detectors Edge linking and the construction of the chain graph Edge and local feature
Edge and local feature detection Gradient based edge detection Edge detection by function fitting Second derivative edge detectors Edge linking and the construction of the chain graph Edge and local feature
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION
 ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
Lesion Segmentation and Bias Correction in Breast Ultrasound B-mode Images Including Elastography Information
 Lesion Segmentation and Bias Correction in Breast Ultrasound B-mode Images Including Elastography Information Gerard Pons a, Joan Martí a, Robert Martí a, Mariano Cabezas a, Andrew di Battista b, and J.
Lesion Segmentation and Bias Correction in Breast Ultrasound B-mode Images Including Elastography Information Gerard Pons a, Joan Martí a, Robert Martí a, Mariano Cabezas a, Andrew di Battista b, and J.
Using Probability Maps for Multi organ Automatic Segmentation
 Using Probability Maps for Multi organ Automatic Segmentation Ranveer Joyseeree 1,2, Óscar Jiménez del Toro1, and Henning Müller 1,3 1 University of Applied Sciences Western Switzerland (HES SO), Sierre,
Using Probability Maps for Multi organ Automatic Segmentation Ranveer Joyseeree 1,2, Óscar Jiménez del Toro1, and Henning Müller 1,3 1 University of Applied Sciences Western Switzerland (HES SO), Sierre,
Shape-based Diffeomorphic Registration on Hippocampal Surfaces Using Beltrami Holomorphic Flow
 Shape-based Diffeomorphic Registration on Hippocampal Surfaces Using Beltrami Holomorphic Flow Abstract. Finding meaningful 1-1 correspondences between hippocampal (HP) surfaces is an important but difficult
Shape-based Diffeomorphic Registration on Hippocampal Surfaces Using Beltrami Holomorphic Flow Abstract. Finding meaningful 1-1 correspondences between hippocampal (HP) surfaces is an important but difficult
STATISTICAL ATLAS-BASED SUB-VOXEL SEGMENTATION OF 3D BRAIN MRI
 STATISTICA ATAS-BASED SUB-VOXE SEGMENTATION OF 3D BRAIN MRI Marcel Bosc 1,2, Fabrice Heitz 1, Jean-Paul Armspach 2 (1) SIIT UMR-7005 CNRS / Strasbourg I University, 67400 Illkirch, France (2) IPB UMR-7004
STATISTICA ATAS-BASED SUB-VOXE SEGMENTATION OF 3D BRAIN MRI Marcel Bosc 1,2, Fabrice Heitz 1, Jean-Paul Armspach 2 (1) SIIT UMR-7005 CNRS / Strasbourg I University, 67400 Illkirch, France (2) IPB UMR-7004
Automatic Generation of Training Data for Brain Tissue Classification from MRI
 MICCAI-2002 1 Automatic Generation of Training Data for Brain Tissue Classification from MRI Chris A. Cocosco, Alex P. Zijdenbos, and Alan C. Evans McConnell Brain Imaging Centre, Montreal Neurological
MICCAI-2002 1 Automatic Generation of Training Data for Brain Tissue Classification from MRI Chris A. Cocosco, Alex P. Zijdenbos, and Alan C. Evans McConnell Brain Imaging Centre, Montreal Neurological
US 1.
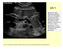 US 1 Sample image: Normal pancreas seen on sonogram. Looking up from abdomen toward the head of the patient. The liver is in front of the pancreas. A vein draining the spleen is behind the pancreas http://www.radiologyinfo.org/photocat/photos.cfm?image=abdo-us-pancr.jpg&&subcategory=abdomen&&stop=9
US 1 Sample image: Normal pancreas seen on sonogram. Looking up from abdomen toward the head of the patient. The liver is in front of the pancreas. A vein draining the spleen is behind the pancreas http://www.radiologyinfo.org/photocat/photos.cfm?image=abdo-us-pancr.jpg&&subcategory=abdomen&&stop=9
Interactive segmentation of vascular structures in CT images for liver surgery planning
 Interactive segmentation of vascular structures in CT images for liver surgery planning L. Wang¹, C. Hansen¹, S.Zidowitz¹, H. K. Hahn¹ ¹ Fraunhofer MEVIS, Institute for Medical Image Computing, Bremen,
Interactive segmentation of vascular structures in CT images for liver surgery planning L. Wang¹, C. Hansen¹, S.Zidowitz¹, H. K. Hahn¹ ¹ Fraunhofer MEVIS, Institute for Medical Image Computing, Bremen,
Medical Image Segmentation by Active Contour Improvement
 American Journal of Software Engineering and Applications 7; 6(): 3-7 http://www.sciencepublishinggroup.com//asea doi:.648/.asea.76. ISSN: 37-473 (Print); ISSN: 37-49X (Online) Medical Image Segmentation
American Journal of Software Engineering and Applications 7; 6(): 3-7 http://www.sciencepublishinggroup.com//asea doi:.648/.asea.76. ISSN: 37-473 (Print); ISSN: 37-49X (Online) Medical Image Segmentation
Whole Body MRI Intensity Standardization
 Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Segmentation of 3D CT Volume Images Using a Single 2D Atlas
 Segmentation of 3D CT Volume Images Using a Single 2D Atlas Feng Ding 1, Wee Kheng Leow 1, and Shih-Chang Wang 2 1 Dept. of Computer Science, National University of Singapore, 3 Science Drive 2, Singapore
Segmentation of 3D CT Volume Images Using a Single 2D Atlas Feng Ding 1, Wee Kheng Leow 1, and Shih-Chang Wang 2 1 Dept. of Computer Science, National University of Singapore, 3 Science Drive 2, Singapore
Atlas Based Segmentation of the prostate in MR images
 Atlas Based Segmentation of the prostate in MR images Albert Gubern-Merida and Robert Marti Universitat de Girona, Computer Vision and Robotics Group, Girona, Spain {agubern,marly}@eia.udg.edu Abstract.
Atlas Based Segmentation of the prostate in MR images Albert Gubern-Merida and Robert Marti Universitat de Girona, Computer Vision and Robotics Group, Girona, Spain {agubern,marly}@eia.udg.edu Abstract.
Integrated Approaches to Non-Rigid Registration in Medical Images
 Work. on Appl. of Comp. Vision, pg 102-108. 1 Integrated Approaches to Non-Rigid Registration in Medical Images Yongmei Wang and Lawrence H. Staib + Departments of Electrical Engineering and Diagnostic
Work. on Appl. of Comp. Vision, pg 102-108. 1 Integrated Approaches to Non-Rigid Registration in Medical Images Yongmei Wang and Lawrence H. Staib + Departments of Electrical Engineering and Diagnostic
Brilliance CT Big Bore.
 1 2 2 There are two methods of RCCT acquisition in widespread clinical use: cine axial and helical. In RCCT with cine axial acquisition, repeat CT images are taken each couch position while recording respiration.
1 2 2 There are two methods of RCCT acquisition in widespread clinical use: cine axial and helical. In RCCT with cine axial acquisition, repeat CT images are taken each couch position while recording respiration.
GPU Based Region Growth and Vessel Tracking. Supratik Moulik M.D. Jason Walsh
 GPU Based Region Growth and Vessel Tracking Supratik Moulik M.D. (supratik@moulik.com) Jason Walsh Conflict of Interest Dr. Supratik Moulik does not have a significant financial stake in any company, nor
GPU Based Region Growth and Vessel Tracking Supratik Moulik M.D. (supratik@moulik.com) Jason Walsh Conflict of Interest Dr. Supratik Moulik does not have a significant financial stake in any company, nor
Learning a Manifold as an Atlas Supplementary Material
 Learning a Manifold as an Atlas Supplementary Material Nikolaos Pitelis Chris Russell School of EECS, Queen Mary, University of London [nikolaos.pitelis,chrisr,lourdes]@eecs.qmul.ac.uk Lourdes Agapito
Learning a Manifold as an Atlas Supplementary Material Nikolaos Pitelis Chris Russell School of EECS, Queen Mary, University of London [nikolaos.pitelis,chrisr,lourdes]@eecs.qmul.ac.uk Lourdes Agapito
Three-Dimensional CT Image Segmentation by Combining 2D Fully Convolutional Network with 3D Majority Voting
 Three-Dimensional CT Image Segmentation by Combining 2D Fully Convolutional Network with 3D Majority Voting Xiangrong Zhou 1(&), Takaaki Ito 1, Ryosuke Takayama 1, Song Wang 2, Takeshi Hara 1, and Hiroshi
Three-Dimensional CT Image Segmentation by Combining 2D Fully Convolutional Network with 3D Majority Voting Xiangrong Zhou 1(&), Takaaki Ito 1, Ryosuke Takayama 1, Song Wang 2, Takeshi Hara 1, and Hiroshi
Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation
 Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Koen Van Leemput 3, Danial Lashkari 4 Marc-André Weber 5, Nicholas Ayache 2, and Polina
Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Koen Van Leemput 3, Danial Lashkari 4 Marc-André Weber 5, Nicholas Ayache 2, and Polina
Mirrored LH Histograms for the Visualization of Material Boundaries
 Mirrored LH Histograms for the Visualization of Material Boundaries Petr Šereda 1, Anna Vilanova 1 and Frans A. Gerritsen 1,2 1 Department of Biomedical Engineering, Technische Universiteit Eindhoven,
Mirrored LH Histograms for the Visualization of Material Boundaries Petr Šereda 1, Anna Vilanova 1 and Frans A. Gerritsen 1,2 1 Department of Biomedical Engineering, Technische Universiteit Eindhoven,
Extract Object Boundaries in Noisy Images using Level Set. Literature Survey
 Extract Object Boundaries in Noisy Images using Level Set by: Quming Zhou Literature Survey Submitted to Professor Brian Evans EE381K Multidimensional Digital Signal Processing March 15, 003 Abstract Finding
Extract Object Boundaries in Noisy Images using Level Set by: Quming Zhou Literature Survey Submitted to Professor Brian Evans EE381K Multidimensional Digital Signal Processing March 15, 003 Abstract Finding
