Übung: Breakdown of sequence homology
|
|
|
- Solomon Parsons
- 5 years ago
- Views:
Transcription
1 Übung: Breakdown of sequence homology WS 2008/09 Übung zu Anwendungen der Strukturanalyse 4-Feb Addresses Gathering sequences Collection of your PDB sequence Your random seed Generation of random sequences Necessary functions The final program Check your program Sequence searches Assignment... 6 In the lectures, much was said about remote homology detection. The question here is where simple homology detection breaks down. Unfortunately, much of bioinformatics is empirical. Many beliefs come from observation rather than rigorous theory. The goal here is to take a protein sequence, gradually destroy it and watch as the similarity disappears. There is a small amount of programming in this Übung. There is a small amount of programming in this Übung and, in general, there should be n different programs from n different students. If you have not programmed before, write this in your report and use the program from a friend. The report should be handed in by 6 Feb Addresses Fasta server: fasta.bioch.virginia.edu 2. Gathering sequences 2.1. Collection of your PDB sequence As in a previous Übung, you have to find your personal protein. This time, you must use the initials of your father or mother. The father of the Bundeskanzlerin was Horst (H) Kasner (K)
2 Merkel (M). When the Bundeskanzlerin does this Übung, she would look for a protein 1hkm which happens to be a chitinase enzyme from humans. Visit the protein databank (PDB) and look up the protein with your name. Find the number of residues in the different chains (Click on the tab near the top of the page called "Sequence Details") Now, we want to make sure that you have an interesting sequence, so while (you have selected a protein with < 60 residues) go to next alphabetical protein (1abc -> 1abd or 1xyz->1xza) You should now have a real protein from the PDB with more than 60 residues. Go back to the initial page for the protein. Near the top, you should see a tab called "sequence details". From there, there should be a link to download the sequence in fasta format. If the protein has more than one chain, download the first chain. Store the sequence in a filename like 1kdm.fasta. You will need this later Your random seed You must have your own seed for random numbers. Take the year of your birth, your father's year of birth and your mother's year of birth and add them together. Angela Merkel (born 1954) would use = 5802 as her random seed Generation of random sequences You must write a program which can read an amino acid sequence from a file read a number from the command line - the fraction of the amino acid sequence to randomise scramble (re-order) a fraction of the amino acid sequence write the new sequence to a new file This will change the sequence, but not the amino acid composition. The random sequences will be very protein-like. You can write in any language you like. If you produce a working program in fortran77, you automatically receive a beer. If you produce a working program in assembler, you get a bottle of sekt. Your program must control its random number seed. To save time, you can hard code this. If you have more time, you can make this a command line argument. 2
3 Necessary functions Two kinds of random numbers 1. Return a floating point number between 0 and Return an integer between 1 and a number N A function to compare two sequences and measure sequence identity In pseudo code, this is very easy, but the details will depend on the language and how it represents strings. It should look something like: float identity (sequence1, sequence2, n) integer count = 0 for (i=0; i < n; i++) if (sequence1[i] == sequence2[i]) count++ return ( (float) count / n) Be careful with this simple function. You need the (float) cast in C, otherwise you will get integer division and truncation. The randomizing function We do not need to change an exact number of residues. We can do this in a probabilistic manner. If we are going to change 10 % of the residues, there is a 0.1 probability a residue changing. We are going to work by interchanging residues, so each swap will affect two residues. We begin by taking the probability of a change and dividing it by two. Say frac is the fraction of the sequence to randomise. In C-like pseudocode, the idea would be: char * randomize (sequence1, sequence2, n, frac) { frac /= 2.0; set sequence2 = sequence1; for (i = 0; i < n; i++) { int k = i; while (k == i) { /* find the position to swap with */ k = random (0..n); /* a random integer */ r = rand (0..1) /* a random float from 0 to 1 */ if (frac >= r) { tmp = sequence1[i]; sequence1[i] = sequence2[k]; sequence2[k] = tmp; return sequence2; 3
4 Note that we have a while loop so that we do not try to swap an amino acid with itself. At the top of the function, we divide frac by 2. If we want to change 30 % of the residues, we try to do swaps at 15 % of the positions. The details are very language dependent. In C, you have to pass in the size n of the array, but in perl this is not necessary. In C, it is easiest to use array notation, but in perl, you could use the substr() function. Be careful with storage. In C, it is easiest for the caller to allocate space for the string (in sequence2). In languages with garbage collection you could build sequence2 and just return it. File reading and writing A sequence file from the protein data bank contains a comment line at the start. This is not part of the sequence. Your program must recognize this. An elegant approach is to read the first line of the sequence file and ignore the first line if it begins with the ">" character. When writing the output, you can put the comment line to good use. After you have calculated the sequence identity, write it into the comment line. For example, you may have wanted to change 15 % of the residues, so one expects the sequence identity to be near 85 %. Because the method is crude, you may have % sequence identity with the input. Your output file should begin with a line like > sequence from xxxx.seq NN residues where xxxx.seq should be the input filename and NN should be the number of residues in your sequence. Do not give more than 3 significant figures in the sequence identity The final program A simple interface to the program would be like: random_seq infile outfile fraction seed where infile is the name of the file containing the sequence to be randomised outfile is the output file name fraction is a floating point number saying how much of the sequence should be randomised would ask the program to change 10 % of the residues seed is the seed for the random number generator (an integer) The final program should look like 4
5 int main (argc, argv) read_command_line_arguments set_random_seed read_input from infile randomise calculate sequence identity write the sequence identity to standard output write new sequence to outfile 3. Check your program Do the following checks. if there is an error on opening the input file, the program should stop gracefully if the input has N residues, the output should have N residues If your input sequence short like "ABCDEFGHIJ" and you change 100 % of the residues, you can see in the output if most of the residues have changed, but all the letters from A to J are present. if you ask the program to change 10 % of the residues, you should have close to 90 % sequence identity with the input. On large sequences, the error should not be too large if you ask the program to change 100 % of the residues, you would not expect 0 % sequence identity. 4. Sequence searches Our input sequence came from the protein databank so we will use fast sequences against the protein databank sequences. One could use web interface to "blast" (see Frau Willhoeft exercises), but we will use "fasta". It is fast enough and has slightly better statistics. Visit the fasta web site. Follow the link to "Protein-protein FASTA". Under "(C) Database" change the option to "NCBI PDB structures". Near the bottom of the page is a box labelled "E():". Put in a number like 1e-5 (which means ). This is just a filter to reduce the output and ask the program not to print out sequences with larger e-values. Paste in your original sequence with no changes and "Search Database". The page should show you high scoring alignments. Check very carefully the name of the best scoring protein. It should be 100 % identical, to your input, but it may not have the same name. This is not an error. 5
6 Sometimes, the same protein structure is solved several times and appears in the protein databank under different names. The database used by the "fasta" server only tells you the first name. Now generate a sequence with about 40 % of the residues changed and repeat the sequence search. Remember to specify the database. This search should also find your original sequence in first rank. Note down the e-value (the number of times you would expect to see this observation by chance). It is probably a very small positive number. Note down the sequence identity. Count the number of hits with the e-value better than the threshold. 5. Assignment Do not write in staroffice or openoffice. 1. Mail your program (source code) to torda@zbh.uni-hamburg. Send it as plain text or a tar archive. Do not hide it in html or xml. 2. Write the initials of your father or mother, the protein you worked with and the random seed you used for the biased random sequence generator. Make sure you are not using staroffice or openoffice. 3. Build a table like % protein change (input) % protein identity (output) % sequence identity from fasta e-value rank of correct protein Number of hits with e-value < 10-5 The first column is the amount of change you ask your program for. The second is the sequence identity your program calculates. Do not quote more than 3 significant digits. The rest of the columns are taken from the fasta output. When you search with the original sequence, you expect it to be at rank 1 in the list of similar proteins. As the sequence changes, it may move down the rank list. Eventually, it may not appear in the rank list. The numbers in the first column are a suggestion. For small proteins, you may have to use slightly lower numbers to see where homology detection breaks down. For larger proteins, you may still 6
7 see reasonable alignments with very low sequence identity. If you are tempted to write in staroffice or openoffice, stop now. 4. If you ask your program to change x % of the sequence, you should see that the actual percentage changed is smaller than x. (identity > 100 x %) Explain why. 5. What would be a simple change to the structure of the program to make the % change of sequence more controllable? Describe the change in careful words or pseudocode. 6. What is a word processing program that the author of this Übung does not like? 7
BLAST - Basic Local Alignment Search Tool
 Lecture for ic Bioinformatics (DD2450) April 11, 2013 Searching 1. Input: Query Sequence 2. Database of sequences 3. Subject Sequence(s) 4. Output: High Segment Pairs (HSPs) Sequence Similarity Measures:
Lecture for ic Bioinformatics (DD2450) April 11, 2013 Searching 1. Input: Query Sequence 2. Database of sequences 3. Subject Sequence(s) 4. Output: High Segment Pairs (HSPs) Sequence Similarity Measures:
Homology Modeling FABP
 Homology Modeling FABP Homology modeling is a technique used to approximate the 3D structure of a protein when no experimentally determined structure exists. It operates under the principle that protein
Homology Modeling FABP Homology modeling is a technique used to approximate the 3D structure of a protein when no experimentally determined structure exists. It operates under the principle that protein
Sequence alignment theory and applications Session 3: BLAST algorithm
 Sequence alignment theory and applications Session 3: BLAST algorithm Introduction to Bioinformatics online course : IBT Sonal Henson Learning Objectives Understand the principles of the BLAST algorithm
Sequence alignment theory and applications Session 3: BLAST algorithm Introduction to Bioinformatics online course : IBT Sonal Henson Learning Objectives Understand the principles of the BLAST algorithm
INTRODUCTION TO BIOINFORMATICS
 Molecular Biology-2019 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
Molecular Biology-2019 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
Tutorial 4 BLAST Searching the CHO Genome
 Tutorial 4 BLAST Searching the CHO Genome Accessing the CHO Genome BLAST Tool The CHO BLAST server can be accessed by clicking on the BLAST button on the home page or by selecting BLAST from the menu bar
Tutorial 4 BLAST Searching the CHO Genome Accessing the CHO Genome BLAST Tool The CHO BLAST server can be accessed by clicking on the BLAST button on the home page or by selecting BLAST from the menu bar
Basic Local Alignment Search Tool (BLAST)
 BLAST 26.04.2018 Basic Local Alignment Search Tool (BLAST) BLAST (Altshul-1990) is an heuristic Pairwise Alignment composed by six-steps that search for local similarities. The most used access point to
BLAST 26.04.2018 Basic Local Alignment Search Tool (BLAST) BLAST (Altshul-1990) is an heuristic Pairwise Alignment composed by six-steps that search for local similarities. The most used access point to
Database Searching Using BLAST
 Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
INTRODUCTION TO BIOINFORMATICS
 Molecular Biology-2017 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
Molecular Biology-2017 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
BLAST, Profile, and PSI-BLAST
 BLAST, Profile, and PSI-BLAST Jianlin Cheng, PhD School of Electrical Engineering and Computer Science University of Central Florida 26 Free for academic use Copyright @ Jianlin Cheng & original sources
BLAST, Profile, and PSI-BLAST Jianlin Cheng, PhD School of Electrical Engineering and Computer Science University of Central Florida 26 Free for academic use Copyright @ Jianlin Cheng & original sources
BLAST: Basic Local Alignment Search Tool Altschul et al. J. Mol Bio CS 466 Saurabh Sinha
 BLAST: Basic Local Alignment Search Tool Altschul et al. J. Mol Bio. 1990. CS 466 Saurabh Sinha Motivation Sequence homology to a known protein suggest function of newly sequenced protein Bioinformatics
BLAST: Basic Local Alignment Search Tool Altschul et al. J. Mol Bio. 1990. CS 466 Saurabh Sinha Motivation Sequence homology to a known protein suggest function of newly sequenced protein Bioinformatics
24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, This lecture is based on the following papers, which are all recommended reading:
 24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, 2010 3 BLAST and FASTA This lecture is based on the following papers, which are all recommended reading: D.J. Lipman and W.R. Pearson, Rapid
24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, 2010 3 BLAST and FASTA This lecture is based on the following papers, which are all recommended reading: D.J. Lipman and W.R. Pearson, Rapid
Programming assignment for the course Sequence Analysis (2006)
 Programming assignment for the course Sequence Analysis (2006) Original text by John W. Romein, adapted by Bart van Houte (bart@cs.vu.nl) Introduction Please note: This assignment is only obligatory for
Programming assignment for the course Sequence Analysis (2006) Original text by John W. Romein, adapted by Bart van Houte (bart@cs.vu.nl) Introduction Please note: This assignment is only obligatory for
As of August 15, 2008, GenBank contained bases from reported sequences. The search procedure should be
 48 Bioinformatics I, WS 09-10, S. Henz (script by D. Huson) November 26, 2009 4 BLAST and BLAT Outline of the chapter: 1. Heuristics for the pairwise local alignment of two sequences 2. BLAST: search and
48 Bioinformatics I, WS 09-10, S. Henz (script by D. Huson) November 26, 2009 4 BLAST and BLAT Outline of the chapter: 1. Heuristics for the pairwise local alignment of two sequences 2. BLAST: search and
Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence or library of DNA.
 Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence or library of DNA. Fasta is used to compare a protein or DNA sequence to all of the
Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence or library of DNA. Fasta is used to compare a protein or DNA sequence to all of the
CS313 Exercise 4 Cover Page Fall 2017
 CS313 Exercise 4 Cover Page Fall 2017 Due by the start of class on Thursday, October 12, 2017. Name(s): In the TIME column, please estimate the time you spent on the parts of this exercise. Please try
CS313 Exercise 4 Cover Page Fall 2017 Due by the start of class on Thursday, October 12, 2017. Name(s): In the TIME column, please estimate the time you spent on the parts of this exercise. Please try
BLAST. NCBI BLAST Basic Local Alignment Search Tool
 BLAST NCBI BLAST Basic Local Alignment Search Tool http://www.ncbi.nlm.nih.gov/blast/ Global versus local alignments Global alignments: Attempt to align every residue in every sequence, Most useful when
BLAST NCBI BLAST Basic Local Alignment Search Tool http://www.ncbi.nlm.nih.gov/blast/ Global versus local alignments Global alignments: Attempt to align every residue in every sequence, Most useful when
MetaPhyler Usage Manual
 MetaPhyler Usage Manual Bo Liu boliu@umiacs.umd.edu March 13, 2012 Contents 1 What is MetaPhyler 1 2 Installation 1 3 Quick Start 2 3.1 Taxonomic profiling for metagenomic sequences.............. 2 3.2
MetaPhyler Usage Manual Bo Liu boliu@umiacs.umd.edu March 13, 2012 Contents 1 What is MetaPhyler 1 2 Installation 1 3 Quick Start 2 3.1 Taxonomic profiling for metagenomic sequences.............. 2 3.2
Technical University of Munich. Exercise 8: Neural Networks
 Technical University of Munich Chair for Bioinformatics and Computational Biology Protein Prediction I for Computer Scientists SoSe 2018 Prof. B. Rost M. Bernhofer, M. Heinzinger, D. Nechaev, L. Richter
Technical University of Munich Chair for Bioinformatics and Computational Biology Protein Prediction I for Computer Scientists SoSe 2018 Prof. B. Rost M. Bernhofer, M. Heinzinger, D. Nechaev, L. Richter
Introduction to Programming in C Department of Computer Science and Engineering. Lecture No. #34. Function with pointer Argument
 Introduction to Programming in C Department of Computer Science and Engineering Lecture No. #34 Function with pointer Argument (Refer Slide Time: 00:05) So, here is the stuff that we have seen about pointers.
Introduction to Programming in C Department of Computer Science and Engineering Lecture No. #34 Function with pointer Argument (Refer Slide Time: 00:05) So, here is the stuff that we have seen about pointers.
FASTA. Besides that, FASTA package provides SSEARCH, an implementation of the optimal Smith- Waterman algorithm.
 FASTA INTRODUCTION Definition (by David J. Lipman and William R. Pearson in 1985) - Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence
FASTA INTRODUCTION Definition (by David J. Lipman and William R. Pearson in 1985) - Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence
BASIC COMPUTATION. public static void main(string [] args) Fundamentals of Computer Science I
![BASIC COMPUTATION. public static void main(string [] args) Fundamentals of Computer Science I BASIC COMPUTATION. public static void main(string [] args) Fundamentals of Computer Science I](/thumbs/81/83968367.jpg) BASIC COMPUTATION x public static void main(string [] args) Fundamentals of Computer Science I Outline Using Eclipse Data Types Variables Primitive and Class Data Types Expressions Declaration Assignment
BASIC COMPUTATION x public static void main(string [] args) Fundamentals of Computer Science I Outline Using Eclipse Data Types Variables Primitive and Class Data Types Expressions Declaration Assignment
Lecture 5 Advanced BLAST
 Introduction to Bioinformatics for Medical Research Gideon Greenspan gdg@cs.technion.ac.il Lecture 5 Advanced BLAST BLAST Recap Sequence Alignment Complexity and indexing BLASTN and BLASTP Basic parameters
Introduction to Bioinformatics for Medical Research Gideon Greenspan gdg@cs.technion.ac.il Lecture 5 Advanced BLAST BLAST Recap Sequence Alignment Complexity and indexing BLASTN and BLASTP Basic parameters
Mapping Sequence Conservation onto Structures with Chimera
 This page: www.rbvi.ucsf.edu/chimera/data/tutorials/systems/outline.html Chimera in BP205A BP205A syllabus Mapping Sequence Conservation onto Structures with Chimera Case 1: You already have a structure
This page: www.rbvi.ucsf.edu/chimera/data/tutorials/systems/outline.html Chimera in BP205A BP205A syllabus Mapping Sequence Conservation onto Structures with Chimera Case 1: You already have a structure
ENGINEERING 1020 Introduction to Computer Programming M A Y 2 6, R E Z A S H A H I D I
 ENGINEERING 1020 Introduction to Computer Programming M A Y 2 6, 2 0 1 0 R E Z A S H A H I D I Today s class Constants Assignment statement Parameters and calling functions Expressions Mixed precision
ENGINEERING 1020 Introduction to Computer Programming M A Y 2 6, 2 0 1 0 R E Z A S H A H I D I Today s class Constants Assignment statement Parameters and calling functions Expressions Mixed precision
Wilson Leung 01/03/2018 An Introduction to NCBI BLAST. Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment
 An Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at https://blast.ncbi.nlm.nih.gov/blast.cgi
An Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at https://blast.ncbi.nlm.nih.gov/blast.cgi
COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1. Database Searching
 COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1 Database Searching In database search, we typically have a large sequence database
COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1 Database Searching In database search, we typically have a large sequence database
Genome Browsers - The UCSC Genome Browser
 Genome Browsers - The UCSC Genome Browser Background The UCSC Genome Browser is a well-curated site that provides users with a view of gene or sequence information in genomic context for a specific species,
Genome Browsers - The UCSC Genome Browser Background The UCSC Genome Browser is a well-curated site that provides users with a view of gene or sequence information in genomic context for a specific species,
PFstats User Guide. Aspartate/ornithine carbamoyltransferase Case Study. Neli Fonseca
 PFstats User Guide Aspartate/ornithine carbamoyltransferase Case Study 1 Contents Overview 3 Obtaining An Alignment 3 Methods 4 Alignment Filtering............................................ 4 Reference
PFstats User Guide Aspartate/ornithine carbamoyltransferase Case Study 1 Contents Overview 3 Obtaining An Alignment 3 Methods 4 Alignment Filtering............................................ 4 Reference
COMP Primitive and Class Types. Yi Hong May 14, 2015
 COMP 110-001 Primitive and Class Types Yi Hong May 14, 2015 Review What are the two major parts of an object? What is the relationship between class and object? Design a simple class for Student How to
COMP 110-001 Primitive and Class Types Yi Hong May 14, 2015 Review What are the two major parts of an object? What is the relationship between class and object? Design a simple class for Student How to
printf( Please enter another number: ); scanf( %d, &num2);
 CIT 593 Intro to Computer Systems Lecture #13 (11/1/12) Now that we've looked at how an assembly language program runs on a computer, we're ready to move up a level and start working with more powerful
CIT 593 Intro to Computer Systems Lecture #13 (11/1/12) Now that we've looked at how an assembly language program runs on a computer, we're ready to move up a level and start working with more powerful
Work relative to other classes
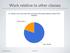 Work relative to other classes 1 Hours/week on projects 2 C BOOTCAMP DAY 1 CS3600, Northeastern University Slides adapted from Anandha Gopalan s CS132 course at Univ. of Pittsburgh Overview C: A language
Work relative to other classes 1 Hours/week on projects 2 C BOOTCAMP DAY 1 CS3600, Northeastern University Slides adapted from Anandha Gopalan s CS132 course at Univ. of Pittsburgh Overview C: A language
mpmorfsdb: A database of Molecular Recognition Features (MoRFs) in membrane proteins. Introduction
 mpmorfsdb: A database of Molecular Recognition Features (MoRFs) in membrane proteins. Introduction Molecular Recognition Features (MoRFs) are short, intrinsically disordered regions in proteins that undergo
mpmorfsdb: A database of Molecular Recognition Features (MoRFs) in membrane proteins. Introduction Molecular Recognition Features (MoRFs) are short, intrinsically disordered regions in proteins that undergo
Chapter 2 THE STRUCTURE OF C LANGUAGE
 Lecture # 5 Chapter 2 THE STRUCTURE OF C LANGUAGE 1 Compiled by SIA CHEE KIONG DEPARTMENT OF MATERIAL AND DESIGN ENGINEERING FACULTY OF MECHANICAL AND MANUFACTURING ENGINEERING Contents Introduction to
Lecture # 5 Chapter 2 THE STRUCTURE OF C LANGUAGE 1 Compiled by SIA CHEE KIONG DEPARTMENT OF MATERIAL AND DESIGN ENGINEERING FACULTY OF MECHANICAL AND MANUFACTURING ENGINEERING Contents Introduction to
Easy manual for SIRD interface
 Easy manual for SIRD interface What is SIRD interface (1) SIRDi is the web-based interface for SIRD (Structure-Interaction Relational Database) (2) You can search for protein structure models with sequence,
Easy manual for SIRD interface What is SIRD interface (1) SIRDi is the web-based interface for SIRD (Structure-Interaction Relational Database) (2) You can search for protein structure models with sequence,
CAP BLAST. BIOINFORMATICS Su-Shing Chen CISE. 8/20/2005 Su-Shing Chen, CISE 1
 CAP 5510-6 BLAST BIOINFORMATICS Su-Shing Chen CISE 8/20/2005 Su-Shing Chen, CISE 1 BLAST Basic Local Alignment Prof Search Su-Shing Chen Tool A Fast Pair-wise Alignment and Database Searching Tool 8/20/2005
CAP 5510-6 BLAST BIOINFORMATICS Su-Shing Chen CISE 8/20/2005 Su-Shing Chen, CISE 1 BLAST Basic Local Alignment Prof Search Su-Shing Chen Tool A Fast Pair-wise Alignment and Database Searching Tool 8/20/2005
MICROPROCESSOR SYSTEMS INTRODUCTION TO PYTHON
 MICROPROCESSOR SYSTEMS INTRODUCTION TO PYTHON Table of contents 2 1. Learning Outcomes 2. Introduction 3. The first program: hello world! 4. The second program: hello (your name)! 5. More data types 6.
MICROPROCESSOR SYSTEMS INTRODUCTION TO PYTHON Table of contents 2 1. Learning Outcomes 2. Introduction 3. The first program: hello world! 4. The second program: hello (your name)! 5. More data types 6.
Bioinformatics explained: BLAST. March 8, 2007
 Bioinformatics Explained Bioinformatics explained: BLAST March 8, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com Bioinformatics
Bioinformatics Explained Bioinformatics explained: BLAST March 8, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com Bioinformatics
FastA & the chaining problem
 FastA & the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem 1 Sources for this lecture: Lectures by Volker Heun, Daniel Huson and Knut Reinert,
FastA & the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem 1 Sources for this lecture: Lectures by Volker Heun, Daniel Huson and Knut Reinert,
FastA and the chaining problem, Gunnar Klau, December 1, 2005, 10:
 FastA and the chaining problem, Gunnar Klau, December 1, 2005, 10:56 4001 4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem
FastA and the chaining problem, Gunnar Klau, December 1, 2005, 10:56 4001 4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem
.. Fall 2011 CSC 570: Bioinformatics Alexander Dekhtyar..
 .. Fall 2011 CSC 570: Bioinformatics Alexander Dekhtyar.. PAM and BLOSUM Matrices Prepared by: Jason Banich and Chris Hoover Background As DNA sequences change and evolve, certain amino acids are more
.. Fall 2011 CSC 570: Bioinformatics Alexander Dekhtyar.. PAM and BLOSUM Matrices Prepared by: Jason Banich and Chris Hoover Background As DNA sequences change and evolve, certain amino acids are more
Extra-Homework Problem Set
 Extra-Homework Problem Set => Will not be graded, but might be a good idea for self-study => Solutions are posted at the end of the problem set Your adviser asks you to find out about a so far unpublished
Extra-Homework Problem Set => Will not be graded, but might be a good idea for self-study => Solutions are posted at the end of the problem set Your adviser asks you to find out about a so far unpublished
Protein structure. Enter the name of the protein: rhamnogalacturonan acetylesterase in the search field and click Go.
 In this practical you will learn how to Protein structure search the Protein Structure Databank for information critically choose the best structure, when more than one is available visualize a protein
In this practical you will learn how to Protein structure search the Protein Structure Databank for information critically choose the best structure, when more than one is available visualize a protein
Lecture 3: C Programm
 0 3 E CS 1 Lecture 3: C Programm ing Reading Quiz Note the intimidating red border! 2 A variable is: A. an area in memory that is reserved at run time to hold a value of particular type B. an area in memory
0 3 E CS 1 Lecture 3: C Programm ing Reading Quiz Note the intimidating red border! 2 A variable is: A. an area in memory that is reserved at run time to hold a value of particular type B. an area in memory
Overview.
 Overview day one 0. getting set up 1. text output and manipulation day two 2. reading and writing files 3. lists and loops today 4. writing functions 5. conditional statements day four day five day six
Overview day one 0. getting set up 1. text output and manipulation day two 2. reading and writing files 3. lists and loops today 4. writing functions 5. conditional statements day four day five day six
Lectures by Volker Heun, Daniel Huson and Knut Reinert, in particular last years lectures
 4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem 4.1 Sources for this lecture Lectures by Volker Heun, Daniel Huson and Knut
4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem 4.1 Sources for this lecture Lectures by Volker Heun, Daniel Huson and Knut
Visual C# Instructor s Manual Table of Contents
 Visual C# 2005 2-1 Chapter 2 Using Data At a Glance Instructor s Manual Table of Contents Overview Objectives s Quick Quizzes Class Discussion Topics Additional Projects Additional Resources Key Terms
Visual C# 2005 2-1 Chapter 2 Using Data At a Glance Instructor s Manual Table of Contents Overview Objectives s Quick Quizzes Class Discussion Topics Additional Projects Additional Resources Key Terms
Protein Structure and Visualization
 In this practical you will learn how to Protein Structure and Visualization By Anne Mølgaard and Thomas Holberg Blicher Search the Protein Structure Databank for information. Critically choose the best
In this practical you will learn how to Protein Structure and Visualization By Anne Mølgaard and Thomas Holberg Blicher Search the Protein Structure Databank for information. Critically choose the best
C E N T R. Introduction to bioinformatics 2007 E B I O I N F O R M A T I C S V U F O R I N T. Lecture 13 G R A T I V. Iterative homology searching,
 C E N T R E F O R I N T E G R A T I V E B I O I N F O R M A T I C S V U Introduction to bioinformatics 2007 Lecture 13 Iterative homology searching, PSI (Position Specific Iterated) BLAST basic idea use
C E N T R E F O R I N T E G R A T I V E B I O I N F O R M A T I C S V U Introduction to bioinformatics 2007 Lecture 13 Iterative homology searching, PSI (Position Specific Iterated) BLAST basic idea use
This manual describes step-by-step instructions to perform basic operations for data analysis.
 HDXanalyzer User Manual The program HDXanalyzer is available for the analysis of the deuterium exchange mass spectrometry data obtained on high-resolution mass spectrometers. Currently, the program is
HDXanalyzer User Manual The program HDXanalyzer is available for the analysis of the deuterium exchange mass spectrometry data obtained on high-resolution mass spectrometers. Currently, the program is
Other Loop Options EXAMPLE
 C++ 14 By EXAMPLE Other Loop Options Now that you have mastered the looping constructs, you should learn some loop-related statements. This chapter teaches the concepts of timing loops, which enable you
C++ 14 By EXAMPLE Other Loop Options Now that you have mastered the looping constructs, you should learn some loop-related statements. This chapter teaches the concepts of timing loops, which enable you
Programming for Engineers Introduction to C
 Programming for Engineers Introduction to C ICEN 200 Spring 2018 Prof. Dola Saha 1 Simple Program 2 Comments // Fig. 2.1: fig02_01.c // A first program in C begin with //, indicating that these two lines
Programming for Engineers Introduction to C ICEN 200 Spring 2018 Prof. Dola Saha 1 Simple Program 2 Comments // Fig. 2.1: fig02_01.c // A first program in C begin with //, indicating that these two lines
Dr. Chuck Cartledge. 15 July 2015
 Miscellanea 6.5 Fun with Fibonacci Break 1.5 Exam Conclusion References CSC-205 Computer Organization Lecture #008 Chapter 6, section 5, Chapter 1, section 5 Dr. Chuck Cartledge 15 July 2015 1/30 Table
Miscellanea 6.5 Fun with Fibonacci Break 1.5 Exam Conclusion References CSC-205 Computer Organization Lecture #008 Chapter 6, section 5, Chapter 1, section 5 Dr. Chuck Cartledge 15 July 2015 1/30 Table
MacVector for Mac OS X
 MacVector 10.6 for Mac OS X System Requirements MacVector 10.6 runs on any PowerPC or Intel Macintosh running Mac OS X 10.4 or higher. It is a Universal Binary, meaning that it runs natively on both PowerPC
MacVector 10.6 for Mac OS X System Requirements MacVector 10.6 runs on any PowerPC or Intel Macintosh running Mac OS X 10.4 or higher. It is a Universal Binary, meaning that it runs natively on both PowerPC
Biochemistry 324 Bioinformatics. Multiple Sequence Alignment (MSA)
 Biochemistry 324 Bioinformatics Multiple Sequence Alignment (MSA) Big- Οh notation Greek omicron symbol Ο The Big-Oh notation indicates the complexity of an algorithm in terms of execution speed and storage
Biochemistry 324 Bioinformatics Multiple Sequence Alignment (MSA) Big- Οh notation Greek omicron symbol Ο The Big-Oh notation indicates the complexity of an algorithm in terms of execution speed and storage
Elements of C in C++ data types if else statement for loops. random numbers rolling a die
 Elements of C in C++ 1 Types and Control Statements data types if else statement for loops 2 Simulations random numbers rolling a die 3 Functions and Pointers defining a function call by value and call
Elements of C in C++ 1 Types and Control Statements data types if else statement for loops 2 Simulations random numbers rolling a die 3 Functions and Pointers defining a function call by value and call
Bioinformatics. Sequence alignment BLAST Significance. Next time Protein Structure
 Bioinformatics Sequence alignment BLAST Significance Next time Protein Structure 1 Experimental origins of sequence data The Sanger dideoxynucleotide method F Each color is one lane of an electrophoresis
Bioinformatics Sequence alignment BLAST Significance Next time Protein Structure 1 Experimental origins of sequence data The Sanger dideoxynucleotide method F Each color is one lane of an electrophoresis
Arithmetic Expressions in C
 Arithmetic Expressions in C Arithmetic Expressions consist of numeric literals, arithmetic operators, and numeric variables. They simplify to a single value, when evaluated. Here is an example of an arithmetic
Arithmetic Expressions in C Arithmetic Expressions consist of numeric literals, arithmetic operators, and numeric variables. They simplify to a single value, when evaluated. Here is an example of an arithmetic
ACORN.COM CS 1110 SPRING 2012: ASSIGNMENT A1
 ACORN.COM CS 1110 SPRING 2012: ASSIGNMENT A1 Due to CMS by Tuesday, February 14. Social networking has caused a return of the dot-com madness. You want in on the easy money, so you have decided to make
ACORN.COM CS 1110 SPRING 2012: ASSIGNMENT A1 Due to CMS by Tuesday, February 14. Social networking has caused a return of the dot-com madness. You want in on the easy money, so you have decided to make
My First Command-Line Program
 1. Tutorial Overview My First Command-Line Program In this tutorial, you re going to create a very simple command-line application that runs in a window. Unlike a graphical user interface application where
1. Tutorial Overview My First Command-Line Program In this tutorial, you re going to create a very simple command-line application that runs in a window. Unlike a graphical user interface application where
Floating Point Puzzles. Lecture 3B Floating Point. IEEE Floating Point. Fractional Binary Numbers. Topics. IEEE Standard 754
 Floating Point Puzzles Topics Lecture 3B Floating Point IEEE Floating Point Standard Rounding Floating Point Operations Mathematical properties For each of the following C expressions, either: Argue that
Floating Point Puzzles Topics Lecture 3B Floating Point IEEE Floating Point Standard Rounding Floating Point Operations Mathematical properties For each of the following C expressions, either: Argue that
The detail of sea.c [1] #include <stdio.h> The preprocessor inserts the header file stdio.h at the point.
![The detail of sea.c [1] #include <stdio.h> The preprocessor inserts the header file stdio.h at the point. The detail of sea.c [1] #include <stdio.h> The preprocessor inserts the header file stdio.h at the point.](/thumbs/77/75082833.jpg) Chapter 1. An Overview 1.1 Programming and Preparation Operating systems : A collection of special programs Management of machine resources Text editor and C compiler C codes in text file. Source code
Chapter 1. An Overview 1.1 Programming and Preparation Operating systems : A collection of special programs Management of machine resources Text editor and C compiler C codes in text file. Source code
Part II Composition of Functions
 Part II Composition of Functions The big idea in this part of the book is deceptively simple. It s that we can take the value returned by one function and use it as an argument to another function. By
Part II Composition of Functions The big idea in this part of the book is deceptively simple. It s that we can take the value returned by one function and use it as an argument to another function. By
COMPARATIVE MICROBIAL GENOMICS ANALYSIS WORKSHOP. Exercise 2: Predicting Protein-encoding Genes, BlastMatrix, BlastAtlas
 COMPARATIVE MICROBIAL GENOMICS ANALYSIS WORKSHOP Exercise 2: Predicting Protein-encoding Genes, BlastMatrix, BlastAtlas First of all connect once again to the CBS system: Open ssh shell client. Press Quick
COMPARATIVE MICROBIAL GENOMICS ANALYSIS WORKSHOP Exercise 2: Predicting Protein-encoding Genes, BlastMatrix, BlastAtlas First of all connect once again to the CBS system: Open ssh shell client. Press Quick
Floating Point Arithmetic
 Floating Point Arithmetic Computer Systems, Section 2.4 Abstraction Anything that is not an integer can be thought of as . e.g. 391.1356 Or can be thought of as + /
Floating Point Arithmetic Computer Systems, Section 2.4 Abstraction Anything that is not an integer can be thought of as . e.g. 391.1356 Or can be thought of as + /
Lecture 4: Outline. Arrays. I. Pointers II. III. Pointer arithmetic IV. Strings
 Lecture 4: Outline I. Pointers A. Accessing data objects using pointers B. Type casting with pointers C. Difference with Java references D. Pointer pitfalls E. Use case II. Arrays A. Representation in
Lecture 4: Outline I. Pointers A. Accessing data objects using pointers B. Type casting with pointers C. Difference with Java references D. Pointer pitfalls E. Use case II. Arrays A. Representation in
What did we talk about last time? Examples switch statements
 Week 4 - Friday What did we talk about last time? Examples switch statements History of computers Hardware Software development Basic Java syntax Output with System.out.print() Mechanical Calculation
Week 4 - Friday What did we talk about last time? Examples switch statements History of computers Hardware Software development Basic Java syntax Output with System.out.print() Mechanical Calculation
Expressions and Casting. Data Manipulation. Simple Program 11/5/2013
 Expressions and Casting C# Programming Rob Miles Data Manipulation We know that programs use data storage (variables) to hold values and statements to process the data The statements are obeyed in sequence
Expressions and Casting C# Programming Rob Miles Data Manipulation We know that programs use data storage (variables) to hold values and statements to process the data The statements are obeyed in sequence
Main Program. C Programming Notes. #include <stdio.h> main() { printf( Hello ); } Comments: /* comment */ //comment. Dr. Karne Towson University
 C Programming Notes Dr. Karne Towson University Reference for C http://www.cplusplus.com/reference/ Main Program #include main() printf( Hello ); Comments: /* comment */ //comment 1 Data Types
C Programming Notes Dr. Karne Towson University Reference for C http://www.cplusplus.com/reference/ Main Program #include main() printf( Hello ); Comments: /* comment */ //comment 1 Data Types
Lecture Overview. Sequence search & alignment. Searching sequence databases. Sequence Alignment & Search. Goals: Motivations:
 Lecture Overview Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating
Lecture Overview Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating
Objectivities. Experiment 1. Lab6 Array I. Description of the Problem. Problem-Solving Tips
 Lab6 Array I Objectivities 1. Using rand to generate random numbers and using srand to seed the random-number generator. 2. Declaring, initializing and referencing arrays. 3. The follow-up questions and
Lab6 Array I Objectivities 1. Using rand to generate random numbers and using srand to seed the random-number generator. 2. Declaring, initializing and referencing arrays. 3. The follow-up questions and
Algorithms & Data Structures
 GATE- 2016-17 Postal Correspondence 1 Algorithms & Data Structures Computer Science & Information Technology (CS) 20 Rank under AIR 100 Postal Correspondence Examination Oriented Theory, Practice Set Key
GATE- 2016-17 Postal Correspondence 1 Algorithms & Data Structures Computer Science & Information Technology (CS) 20 Rank under AIR 100 Postal Correspondence Examination Oriented Theory, Practice Set Key
Introduction to Programming: Variables and Objects. HORT Lecture 7 Instructor: Kranthi Varala
 Introduction to Programming: Variables and Objects HORT 59000 Lecture 7 Instructor: Kranthi Varala What is a program? A set of instructions to the computer that perform a specified task in a specified
Introduction to Programming: Variables and Objects HORT 59000 Lecture 7 Instructor: Kranthi Varala What is a program? A set of instructions to the computer that perform a specified task in a specified
Exercise: Using Numbers
 Exercise: Using Numbers Problem: You are a spy going into an evil party to find the super-secret code phrase (made up of letters and spaces), which you will immediately send via text message to your team
Exercise: Using Numbers Problem: You are a spy going into an evil party to find the super-secret code phrase (made up of letters and spaces), which you will immediately send via text message to your team
Expressions and Casting
 Expressions and Casting C# Programming Rob Miles Data Manipulation We know that programs use data storage (variables) to hold values and statements to process the data The statements are obeyed in sequence
Expressions and Casting C# Programming Rob Miles Data Manipulation We know that programs use data storage (variables) to hold values and statements to process the data The statements are obeyed in sequence
Sequence Alignment & Search
 Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating the first version
Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating the first version
Biology 644: Bioinformatics
 Find the best alignment between 2 sequences with lengths n and m, respectively Best alignment is very dependent upon the substitution matrix and gap penalties The Global Alignment Problem tries to find
Find the best alignment between 2 sequences with lengths n and m, respectively Best alignment is very dependent upon the substitution matrix and gap penalties The Global Alignment Problem tries to find
HORIZONTAL GENE TRANSFER DETECTION
 HORIZONTAL GENE TRANSFER DETECTION Sequenzanalyse und Genomik (Modul 10-202-2207) Alejandro Nabor Lozada-Chávez Before start, the user must create a new folder or directory (WORKING DIRECTORY) for all
HORIZONTAL GENE TRANSFER DETECTION Sequenzanalyse und Genomik (Modul 10-202-2207) Alejandro Nabor Lozada-Chávez Before start, the user must create a new folder or directory (WORKING DIRECTORY) for all
Binghamton University. CS-211 Fall Syntax. What the Compiler needs to understand your program
 Syntax What the Compiler needs to understand your program 1 Pre-Processing Any line that starts with # is a pre-processor directive Pre-processor consumes that entire line Possibly replacing it with other
Syntax What the Compiler needs to understand your program 1 Pre-Processing Any line that starts with # is a pre-processor directive Pre-processor consumes that entire line Possibly replacing it with other
CSE 333 Midterm Exam 7/29/13
 Name There are 5 questions worth a total of 100 points. Please budget your time so you get to all of the questions. Keep your answers brief and to the point. The exam is closed book, closed notes, closed
Name There are 5 questions worth a total of 100 points. Please budget your time so you get to all of the questions. Keep your answers brief and to the point. The exam is closed book, closed notes, closed
ECE264 Spring 2014 Exam 2, March 11, 2014
 ECE264 Spring 2014 Exam 2, March 11, 2014 In signing this statement, I hereby certify that the work on this exam is my own and that I have not copied the work of any other student while completing it.
ECE264 Spring 2014 Exam 2, March 11, 2014 In signing this statement, I hereby certify that the work on this exam is my own and that I have not copied the work of any other student while completing it.
Introduction to C++ 2. A Simple C++ Program. A C++ program consists of: a set of data & function definitions, and the main function (or driver)
 Introduction to C++ 1. General C++ is an Object oriented extension of C which was derived from B (BCPL) Developed by Bjarne Stroustrup (AT&T Bell Labs) in early 1980 s 2. A Simple C++ Program A C++ program
Introduction to C++ 1. General C++ is an Object oriented extension of C which was derived from B (BCPL) Developed by Bjarne Stroustrup (AT&T Bell Labs) in early 1980 s 2. A Simple C++ Program A C++ program
C: How to Program. Week /Mar/05
 1 C: How to Program Week 2 2007/Mar/05 Chapter 2 - Introduction to C Programming 2 Outline 2.1 Introduction 2.2 A Simple C Program: Printing a Line of Text 2.3 Another Simple C Program: Adding Two Integers
1 C: How to Program Week 2 2007/Mar/05 Chapter 2 - Introduction to C Programming 2 Outline 2.1 Introduction 2.2 A Simple C Program: Printing a Line of Text 2.3 Another Simple C Program: Adding Two Integers
Chapter 2 Using Data. Instructor s Manual Table of Contents. At a Glance. Overview. Objectives. Teaching Tips. Quick Quizzes. Class Discussion Topics
 Java Programming, Sixth Edition 2-1 Chapter 2 Using Data At a Glance Instructor s Manual Table of Contents Overview Objectives Teaching Tips Quick Quizzes Class Discussion Topics Additional Projects Additional
Java Programming, Sixth Edition 2-1 Chapter 2 Using Data At a Glance Instructor s Manual Table of Contents Overview Objectives Teaching Tips Quick Quizzes Class Discussion Topics Additional Projects Additional
Data Mining Technologies for Bioinformatics Sequences
 Data Mining Technologies for Bioinformatics Sequences Deepak Garg Computer Science and Engineering Department Thapar Institute of Engineering & Tecnology, Patiala Abstract Main tool used for sequence alignment
Data Mining Technologies for Bioinformatics Sequences Deepak Garg Computer Science and Engineering Department Thapar Institute of Engineering & Tecnology, Patiala Abstract Main tool used for sequence alignment
Writing Functions in C
 Writing Functions in C 1 Test 2, Problem 5 b. Write a function to allocate space for a new instance of your structure, as defined in part a. Write the C code for a function to get space from the heap using
Writing Functions in C 1 Test 2, Problem 5 b. Write a function to allocate space for a new instance of your structure, as defined in part a. Write the C code for a function to get space from the heap using
Bioinformatics Programming. EE, NCKU Tien-Hao Chang (Darby Chang)
 Bioinformatics Programming EE, NCKU Tien-Hao Chang (Darby Chang) 1 Regular Expression 2 http://rp1.monday.vip.tw1.yahoo.net/res/gdsale/st_pic/0469/st-469571-1.jpg 3 Text patterns and matches A regular
Bioinformatics Programming EE, NCKU Tien-Hao Chang (Darby Chang) 1 Regular Expression 2 http://rp1.monday.vip.tw1.yahoo.net/res/gdsale/st_pic/0469/st-469571-1.jpg 3 Text patterns and matches A regular
At the end of this lecture you should be able to have a basic overview of fundamental structures in C and be ready to go into details.
 Objective of this lecture: At the end of this lecture you should be able to have a basic overview of fundamental structures in C and be ready to go into details. Fundamental Programming Structures in C
Objective of this lecture: At the end of this lecture you should be able to have a basic overview of fundamental structures in C and be ready to go into details. Fundamental Programming Structures in C
The Effect of Inverse Document Frequency Weights on Indexed Sequence Retrieval. Kevin C. O'Kane. Department of Computer Science
 The Effect of Inverse Document Frequency Weights on Indexed Sequence Retrieval Kevin C. O'Kane Department of Computer Science The University of Northern Iowa Cedar Falls, Iowa okane@cs.uni.edu http://www.cs.uni.edu/~okane
The Effect of Inverse Document Frequency Weights on Indexed Sequence Retrieval Kevin C. O'Kane Department of Computer Science The University of Northern Iowa Cedar Falls, Iowa okane@cs.uni.edu http://www.cs.uni.edu/~okane
Lesson 02 Data Types and Statements. MIT 11053, Fundamentals of Programming By: S. Sabraz Nawaz Senior Lecturer in MIT Department of MIT FMC, SEUSL
 Lesson 02 Data Types and Statements MIT 11053, Fundamentals of Programming By: S. Sabraz Nawaz Senior Lecturer in MIT Department of MIT FMC, SEUSL Topics Covered Statements Variables Data Types Arithmetic
Lesson 02 Data Types and Statements MIT 11053, Fundamentals of Programming By: S. Sabraz Nawaz Senior Lecturer in MIT Department of MIT FMC, SEUSL Topics Covered Statements Variables Data Types Arithmetic
Introduction to Computational Molecular Biology
 18.417 Introduction to Computational Molecular Biology Lecture 13: October 21, 2004 Scribe: Eitan Reich Lecturer: Ross Lippert Editor: Peter Lee 13.1 Introduction We have been looking at algorithms to
18.417 Introduction to Computational Molecular Biology Lecture 13: October 21, 2004 Scribe: Eitan Reich Lecturer: Ross Lippert Editor: Peter Lee 13.1 Introduction We have been looking at algorithms to
BIOC351: Proteins. PyMOL Laboratory #2. Objects, Distances & Images
 BIOC351: Proteins PyMOL Laboratory #2 Objects, Distances & Images Version 2.1 Valid commands in PyMOL are shown in green. Exercise A: Let s fetch 1fzc, a crystallographic dimer of a fragment of human fibrinogen
BIOC351: Proteins PyMOL Laboratory #2 Objects, Distances & Images Version 2.1 Valid commands in PyMOL are shown in green. Exercise A: Let s fetch 1fzc, a crystallographic dimer of a fragment of human fibrinogen
CE221 Programming in C++ Part 2 References and Pointers, Arrays and Strings
 CE221 Programming in C++ Part 2 References and Pointers, Arrays and Strings 19/10/2017 CE221 Part 2 1 Variables and References 1 In Java a variable of primitive type is associated with a memory location
CE221 Programming in C++ Part 2 References and Pointers, Arrays and Strings 19/10/2017 CE221 Part 2 1 Variables and References 1 In Java a variable of primitive type is associated with a memory location
Object-Oriented Principles and Practice / C++
 Object-Oriented Principles and Practice / C++ Alice E. Fischer May 4, 2013 OOPP / C++ Lecture 6... 1/34 Initialization: TestInit Disasters Failure: Symptoms, Causes, and Cures End of File and Error Handling
Object-Oriented Principles and Practice / C++ Alice E. Fischer May 4, 2013 OOPP / C++ Lecture 6... 1/34 Initialization: TestInit Disasters Failure: Symptoms, Causes, and Cures End of File and Error Handling
Utility of Sliding Window FASTA in Predicting Cross- Reactivity with Allergenic Proteins. Bob Cressman Pioneer Crop Genetics
 Utility of Sliding Window FASTA in Predicting Cross- Reactivity with Allergenic Proteins Bob Cressman Pioneer Crop Genetics The issue FAO/WHO 2001 Step 2: prepare a complete set of 80-amino acid length
Utility of Sliding Window FASTA in Predicting Cross- Reactivity with Allergenic Proteins Bob Cressman Pioneer Crop Genetics The issue FAO/WHO 2001 Step 2: prepare a complete set of 80-amino acid length
Numerical Computing in C and C++ Jamie Griffin. Semester A 2017 Lecture 2
 Numerical Computing in C and C++ Jamie Griffin Semester A 2017 Lecture 2 Visual Studio in QM PC rooms Microsoft Visual Studio Community 2015. Bancroft Building 1.15a; Queen s W207, EB7; Engineering W128.D.
Numerical Computing in C and C++ Jamie Griffin Semester A 2017 Lecture 2 Visual Studio in QM PC rooms Microsoft Visual Studio Community 2015. Bancroft Building 1.15a; Queen s W207, EB7; Engineering W128.D.
Genome Sciences 373: Genome Informatics. Quiz section #1 March 29, 2018
 Genome Sciences 373: Genome Informatics Quiz section #1 March 29, 2018 About me Email: hpliner@uw.edu Office hours: Thursday right after quiz section (2:20pm) Foege S110 Or by appointment Other help: I
Genome Sciences 373: Genome Informatics Quiz section #1 March 29, 2018 About me Email: hpliner@uw.edu Office hours: Thursday right after quiz section (2:20pm) Foege S110 Or by appointment Other help: I
Features of C. Portable Procedural / Modular Structured Language Statically typed Middle level language
 1 History C is a general-purpose, high-level language that was originally developed by Dennis M. Ritchie to develop the UNIX operating system at Bell Labs. C was originally first implemented on the DEC
1 History C is a general-purpose, high-level language that was originally developed by Dennis M. Ritchie to develop the UNIX operating system at Bell Labs. C was originally first implemented on the DEC
1 Strings (Review) CS151: Problem Solving and Programming
 1 Strings (Review) Strings are a collection of characters. quotes. this is a string "this is also a string" In python, strings can be delineated by either single or double If you use one type of quote
1 Strings (Review) Strings are a collection of characters. quotes. this is a string "this is also a string" In python, strings can be delineated by either single or double If you use one type of quote
CSE 142 Su 04 Computer Programming 1 - Java. Objects
 Objects Objects have state and behavior. State is maintained in instance variables which live as long as the object does. Behavior is implemented in methods, which can be called by other objects to request
Objects Objects have state and behavior. State is maintained in instance variables which live as long as the object does. Behavior is implemented in methods, which can be called by other objects to request
An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST
 An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST Alexander Chan 5075504 Biochemistry 218 Final Project An Analysis of Pairwise
An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST Alexander Chan 5075504 Biochemistry 218 Final Project An Analysis of Pairwise
