Users Guide: (Hemodynamic Evoked Response)
|
|
|
- Camron Flynn
- 6 years ago
- Views:
Transcription
1 Users Guide: (Hemodynamic Evoked Response) Developed by T. Huppert and D. Boas A.A. Martinos Center for Neurological Imaging Massachusetts General Hospital. Charlestown, MA Copyright 2003 MGH Questions or Comments: contact T. Huppert (thuppert@nmr.mgh.harvard.edu) Introduction to program: HomER is a data analysis program written by the PMI lab at MGH for the purpose of quick, reliable, and user-friendly analysis of near-infrared spectroscopy. Currently, HOMer supports the continious wave measurements, but both frequency domain and time-domain data can also be made to display and analyze with HomER. The features of this program are organized into several tab-style display windows. This structure is easily expandable for future versions released by us, or custom writen by the user. For programming tips, see the accompanied Intro-to-programming script. Additionally, comments and suggestions are always welcome and should be sent to T. Huppert (thuppert@nmr.mgh.harvard.edu). Data Format: A sample data file (FakeData.mat) is included in the download of this program. The structure of each data file has a mimimum of 4 basic (required) fields. ml - The measurement list. This variable serves to map the data arrays onto the probe geometry. This variable has the structure: [source position, detector position, frequency, wavelength index] ie. [ ] would imply that the data referenced by this row is for the 2 nd source/optode position (not to be confused with laser #2- source positions refer to the probe geometry), the 3 rd detector position, the freq index of 1 indicates that this is a CW measurement, and the Lambda index of 1 indicates that this is the wavelength listed in the SD.lambda variable in the first position. The SD.lambda variable is specifed in the first line of the probe geometry (i.e. SD.Lambda = [ ]; ). The ML serves to map the d array (see below) onto the probe geometry. For example, when the probe geometry plot is highlighted between src #2 and det #3, then HOMer searches the ML for the row indexed as [2 3 <> <>] and plots that data. Each src-det pair must be represented for each wavelength. Additionally, the structure of ML should appear with all the lamdaindex=1 values listed first, followed by lambda=2, etc (more then 2 wavelengths are supported).
2 d - The actual data variable. This variable has dimensions of [number measurements by- number time pts]. Rows in d are mapped by the measurement list. For example, the 5 th row of the ML should describe the 5 th row of d (etc) t- The time variable. This maps the aquisition time to index of the measurment (i.e. the slop of this variable is the aquisition frequency). This will almost always be a straight line with slope equal to the acquisition frequency. s- The stimulus variable. This variable is used to determine the impulse train for averaging. aux10- (optional) This variable specifies the recorded auxillary data. This variable is optional. Although, SPM and other physiology features will not be allowed if this variable is not present Loading New Data: New data sets should be loaded by clicking on the pull-down menu under HOMER and clicking on Open SDG. This will bring up a pop-up menu prompting for subject number (leaving this field empty will save the subject as anoynomus ) For loading the sample data, click on the SampleProbe.m file. This will load the image of the probe into the figure on the top right of HOMER. The next step is to load the data sets. To do this, click on the Open Data File option on the same pull-down menu. This will again bring up the matlab uigetfile menu. Click on the file you wish to add. (For example, click on FakeData.mat to load the sample data set). To open more then one file simply repeat this step. An unlimited number of data files may be opened for each subject- however, too many files (or really large files) will be slow in matlab. Once the data file is opened, clicking around on the SD-geometry plot will change the display of the main plot. Loading Saved Data: To load previously saved data, click on the pull-down menu FILE then select Load Subject and Load Subject (again). This will load the SDG and any files that were previously saved (including any data anaylsis done).
3 Adding More Subjects: HomER allows the analysis of more then one subject. Subjects may have different file sizes, different stimulus type, and even different geometry files (probes). For example, you can load the data for a ipsolateral finger tapping experiment as one subject and a contralateral finger tapping as another. Multiple subjects can be averaged together (as a region of interest) to find group resonses. To add another subject or to remove a subject from the list, click on Load Subject>>Add or Remove Subject. This will prompt for the subject number/name and create a new subject field. To toggle between subjects, use the pop-up menu on the bottom left of the screen (upper right in multi-subject mode). Selecting a subject from this list will change all displays and fields to the specifics of that subject. NOTE: Adding files, updating, averaging, etc is always done to currently displayed subject only. To add data to an empty subject, first select that subject from the list and then proceed as described above. Selecting Load Subject will allow a previously saved data set to be loaded. This data set is added in addition to any existing loaded subjects. Close All: The close all feature will clear ALL loaded data. To close only a single subject- use the remove subject feature. Minimize Data Set: This removes all Non-nessacary fields from the data. This is done to speed-up display and calculations. Once the data set has been minimized, certain fields will no longer be available until the data is updated again. Saving Data: To save data (for future loading as described above), click on Save Data under the File pull-down. Here you will find the options to save ALL subjects or just the current subject. Save Flags gives the options to save only certain compentents of the data. Only the checked fields will be saved (for example MagGIE in the image to the left- this is the multi-subject data). Lisa and Bart are not yet
4 implemented but will be future expansions on the tab menu on the right side of the GUI. The option of Save Minimum under HoMER will save a reduced data set (same as Minimize data set described above). All data is saved as a *.mat file with the std. PMI data structure. Updating/Filtering: The first step in data analysis (after loading data) will be to update the fields. This action creates several variables that are nessacary for other features of the program such as averaging and imaging. Updating can be done per data file (#5) or for all files associated with the current subject all at once (on HOMER>>filtering pull-down option). The Filtering menu (demonstrated to the right) specifies the filtering parameters to be applied to the data upon updating. In addition, under the pull-down menu (HOMER>>FILTERS>>Adv Options) a number of additional features can be specified. These will also be applied upon updating 1) Low Pass Filter (LPF)- This field specifies the low pass filter value (in Hz) to apply to the raw data. This is the first step in the updating process. If this value is set to zero then the LPF step is skipped. This uses the matlab filtfilt command. NOTE: Both low and high pass filtering are done on the complex data. The log-normal data is then calculated from the absolute value of this data prior to the nsv filter. This allowed the I &Q compenenets of the data to remain separate as low as possible in the data processing. NOTE: HPF is now done prior to nsv filtering. 3) High Pass Filter (HPF): After the application of the first PCA filter, the data is then HPF d to remove any drifts (etc) in the data. Data must be at least 3 times the length of the HPF applied. If not, this filtering step is skipped. 2) nsv_pre: The next step in the updating process is the application of principle-compenent analysis filters. The first of such is a PCA filter that removes the first N eigen-vector components obtained from the entire data set. The effect of this filter is to remove strong coorelates between data traces (i.e. motion artifacts). The single-value spectrum can be displayed by selecting the pull-down menu (FIGURES>>Plot SV spectrum). The top plot in this figure is the SVD of the entire data set and the eigen-values of interest here. 4) NSV-dOD: This is the second PCA filter in the updating pathway. Unlike the first PCA filter, this filter uses baseline data and removes the first [N, M] eigen-vectors of the baseline data (at Lambda 1 and 2 respectively). This filter thus removes physiological noise from the data. The
5 SV spectrum can be displayed as above. The bottom two plots are the SV spectrums from lambda 1 and 2. (without baseline data, these plots are not available) Note: In order to use this filter, baseline data MUST be specified. Note: At this step, a custom filtering step can be added to the Update path. The DOT variable is passed to this user-defined filter and the (modified) DOT variable is expected to be returned by the function. Custom filters must have the structure: function {DOT,varargout} = <My_Filter>(DOT) After the data has passed through all filtering steps, delta-concentration is then calculated. Note: In the data structure, Conc data is referenced by the Lambda=1 compenent of the ML. The data structure stores HbO, HbR, and HbT in the [1,2,3] indexes of the 3 rd dimension of the data. Advanced Options: Here, the user has the option to specify custom filters (to be applied as described above) and/or partial volume and path-length corrections. These are all applied immediately after the last filtering step described above (the custom filter is applied first). Path-length corrections are multiplied against the linear distance between Src-Det pairs (specified in cm in the probe file). These options must be checked to be applied during updating. If Pathlength corrections are not applied, concentration data has units of Molar*cm (i.e. still has the L factor included as per Beer s Law). Pruning: Pruning provides a way to remove unwanted src-det pairs from being included in averaging and/or imaging (for example noisy or low signal channels). This can be done on the basis of Src-Det seperation distances, min/max AVERAGE intensities in the RAW data, or manually by clicking on the SD-geometry (dotted-lines are src-det pairs that have been removed from the ML).
6 Only active Src-Det pairs (those displayed with a solid line in the probe geometry) will be included in averaging, image reconstruction, and ROI averaging (if that option is chosen). Single Subject Averaging: NOTE: Data files MUST be updated prior to averaging. If Avgerging multiple data files together, all data files must be updated first. Stimulus: The stimulus train is calculated from the s variable. The features to the right side of the Averaging menu deal with this aspect of averaging. This process works by first normalizing the s variable. Next, the derivative of this trace is taken. After this, all points above the specified Threshold are found as potential stimulus time points. Finally, this list is compared against the Tsep value (for Time Seperation). This value (in seconds) specifies the minimum temperal distance between adjacent stimulus points. For example, if Tsep is 2 - then any point that is above the threshold but LESS then 2seconds following the last point that also met the threshold criteria is NOT included in the stimulus list. The Reset Stim button recalculates the stimulus train from the s variable. The Stim toggle button turns on/off the display of the calculated Stim train. The View Original Stim toggle button turns on/off the display of the original s varible used to calculate the stimulus points. User Stim allows the user to specify a set of stimulus points rather then calculating them from a s variable. This button must be down and the reset stim button pressed to apply the user defined stim points. Removing individual stimulus points. HOMER also allows for the selective removal of individual stimulus points (i.e. bad data points). By clicking on the green lines (active stim points), the stimulus lines can be turned off meaning that they will not be included in any averaging (or deconvolving). Red lines are inactive
7 stimulus points/green are active. Reset Stim restores all lines to active. Note: If the ZOOM or DISPLAY AVG ALL features are down, then clicking on the stimulus lines will have no effect. Turn off both of these features to toggle the stimulus lines. Averaging: Once the stimulus points have been defined, the data can now be averaged. The pre-time field specifies the time (in seconds) prior to each stimulus point to include in each epoch. (same with the post-time). Only epochs that FULLY fit into the data will be included in the average (for example typing a post-time of 60s if the total length of the experiment is only 30 seconds will result in an error and thus such epochs are not included in the averaging). Avg: Averages ONLY the current file Avg All: Computes the average from all files associated with the current subject. (Baseline data is ignored). Deconvolve: If this toggle button is down, then a Least-squares deconvolution method is used rather then a block average. Displaying data:
8 1) Main plot field. This displays the data trace for the entire exeriment. The data that is displayed is specified by the pop-up menu (#3). In the figure above, the raw data is being displayed. The wavelength to be displayed in specified by pop-menu #4 2) This figure displays the averaged data trace. Although this is also specified by menus #3 and 4, the only options for display here are dod_cov and dconc (both described below). If any other display option is selected, then dod_cov data will be displayed (such is the case in the figure above). 3) Display Menu. This specifes which data trace to display. Raw- Data loaded directly from data file. This data has not passed through any filters. Normalized- the Log of the normalized data- This data has been subjected to the LP and HP filters. Norm-Cov- This data is Normalized data from above with additional covariance (PCA) filtering HPF- The drift correction to be applied to the data set. This is NOT the high pass filtered Data itself, but instead is the correction that was applied to high-pass filter the data. I.e. this is the drift correction. dod- Delta Optical Density calculated from filtered data HPF,LPF,Cov-reduced} dod-cov- Applies Covarience reduction (against baseline) filter to dod data for each wavelength. Only avaliable if baseline data is specified. Conc- Displays concentration traces (specified by check concentrations off pull down menus from top of figure). This data has been calculated from the modified Beer-Lambert law. If partial-volume/pathlength corrections were specified, they have been applied here. Conc-Cov- Not yet implimented Aux10- Display Auxillary data (if any). 4) Wavelength selection. Choose which wavelegth to display. Note: Since wavelength doesn t make sense for concentration, the wavelength display must be the first index to display dconc (this has to do with the structure of the Conc variable). HOMER should automatically correct for this. 5) This allows the scale of the Average plot (#2) to be manually specified. Type 999 to autoscale 6) Display Even/Odd- Displays Averages on only the even and odd stimulus points seperately.
9 Display ErrorBars- Toggles on/off the display of error bars on the average plots. Advanced Averaging Options (on pull-down menu) allows the choice of Std Dev or Std Error Zoom- allows zooming on any of the three plots (main data, avg, and SD-geometry) Display All- Plots time traces (along x-axis) for the entire measurement list (along the y-axis). Magnitude is given in color scale Display Avg All: This toggles between displaying the average trace for ALL the data files (for current subject) or only the active one. This is only available when more then one data file is open (and averaged). 7) Probe- All displays will reflect the currently selected src-det pairs from this plot. Only active lines will be displayed. Additional Figures: FIGURES>NEW FIGURE: This will create a new figure displaying the plots from #1,2, and #3 from above FIGURES>>PLOT PSD This displays the power spectrum for the currently diplayed data (selected from #3 and #4) FIGURES>>PLOT ALL This displays the data reflecting its position in the SD-geometry. For dod, a black line is used for the Lambda Index #1 and the blue line is Lambda #2. (Red is used for #3 etc). For dconc, Black is HbO, Blue is HbR and Green is HbT.
10 Additionally, by left (mouse) clicking on any of the figures, the option to plot new plot can be selected. This plots that figure in a new window. Physiology: Currently not used in this version of HOMER To be implemented in the SPM add-on (coming soon) Imaging: NOTE: Averaging must be done before image reconstructions can be preformed. The basic fields in the imaging menu allow one to specify the Musp (scattering) and Mua (absorption) coefficients. These can be entered as a single value (i.e 10 cm-1) which then is taken to be the same for both wavelengths, or entered for each wavelength separately (i.e. [10 9] ) which is then applied to the two (or more) wavelengths independently. The units are in inverse cm. The X/Y/Z voxel fields are the range for the reconstruction (in cm). The optimal voxel size is automatically computed upon loading the geometry file. Make Matrix: This button forms the sensitivity matrix from the specified Musp and Mua coefficients. This is done using the PMI toolbox and assumes a semi-infinite slab geometry and Born approximation to generate the matrix. The matrix size is based upon the currently active ML (all the src-det currently active in the probe image). Note: Future versions will allow the control of the approximation type Invert Matrix: This button performs the regularized inversion of the sensitivity matrix. Trikonov regularization (λ=1). This inversion uses Note: Future versions will allow the regularzation parameter to be specified.
11 Make Image: This final button multiplies the sensitivity matrix against the dod_cov (active ML) variable to form an image. For reconstructing dconc images (see Adv Image options), the dod image is pixel-wise converted to a dconc matrix. Note: Partial-volume and pathlength corrections are NOT applied to the reconstructed dconc images Note: If the size of the ML is changed during image reconstruction, then the sensitivity matrix will need to be re-updated. Advance Options Imaging: This menu (found in the pull-down menu HOMER>>IMAGING) allows for several display options on the reconstructed image. dod vrs dconc: Which type of image to reconstruct. Show CNR Image: This option is only available if baseline data is specified AND a dod image is desired. To calculate noise, the image of the baseline is taken and then the standard error is calculated in the time dimension. This then becomes the image of noise, which is divided into the dod (contrast) image. Autoscale: Sets a change of zero to be green and autoscales the max/min of the image. Show Probe: Superimposes an image of the probe geometry onto the reconstructed image. Displaying the Image: The image is inheriently a 4D matrix (x/y/z/time). If reconstruction is done for multiple z-sections each of these slices will be displayed separately. In the time dimension, the image is reconstructed from a time average over the timewindow indicated in the average plot window (it is a red bar in the plot). The length/location of this bar can be moved around by clicking in the window above or below the bar. This will automatically update the displayed image to reflect the new time averaging. The display image button will recall the image reconstruction after the window has been closed. Note: While the Display Image button is down, the context menus (right-clicking features) of the Avg data plot are disabled.
12 Exporting Data: By right-clicking on any of the plot axes, the option will appear to export the data to file. This gives the option to export the currently displayed data type (i.e. dconc,dod etc) to either a *.mat or *.dat (ascii) file structure (only *.mat for the SD geometry). The structure of this exported variable is [ML x time pts]. For example, if the 3 rd row of the ML is [ ] then the 3 rd row of this data set is the data associated with the 1 st source to 2 nd detector at wavelength #2. Concentration data is stored as a three dimensional array in *.mat format (the third dimension is indexed by [HbO,HbR, and HbT] respectively). In *.dat (ascii), this data is stored as three separate varibles. MAGGIE (Multi-subject/ROI averaging) - Allows averaging across ROI for single and multiple subjects. Average All subjects: This button calculates the ROI average. The data to be included in this average is specified by the radio buttons to the right. Use all subjects vrs. Use current subject: If the current subject choice is selected, the ROI for only the currently displayed subject (shown in the pull-down menu on the right-side of the display) is displayed in the plot. For both choices, both the individual subject ROI averages AND the ROI average from ALL subjects is calculated. Toggling this radio-button between ALL and CURRENT changes the display (reflecting one or all subjects). The subject list controls which (individual) current subject to display. Use Act ML: If this option is selected, only currently active src-det s (for each subject) are used to specify the ROI. Otherwise, the entire ML is used as the ROI. Compare All: This button plots the ROI averages for all subjects as subplots on a single figure. Subjects are displayed on the same scale.
13 Show Statistics: This button brings up a pop-up window that allows the display of the zero-lag coorelation coefficients between all subjects for the specified variable. Subjects are denoted by there index on the list (i.e. the 3 rd subject on the list is the 3 rd row/column in the coorelation matrix). Programming new Features: This program was reworked to allow for easy expansion of future functions- you may have noticed that there are a few buttons on the right tab panel that don't do anything yet. This code will explain (as best I can) how to add your own functions to these tabs. HOW to add buttons to the GUI: In HOMer.fig file- after placing your button set callback to %automatic --this will make a call to this main program passing the name of the function and all GUI handles (see that code for details) The visibilty of buttons is controlled by the UserData field. "0" means that the button will be invisible; '1' means it will be visible. This is done so that visibility can be controled/referenced by the tabs and allow button access to be controlled by visibility and have that visibility state be recalled properly even when the tab is displaying a different field All default values for buttons should be: visible = false UserDate= [0 or 1] -- this sets the default state 'Tooltiptext'- "What does this button do?" Context menu- one of the ready made ones or add your own code The rest is up to you! Objects associated with the "Debug" context functions will be able to move around in Debug mode during execution (ie if the Deebug flag in HOMer.m is set to "1". This means that after you ste the position of the object You can look at it position during execution and fine-tune the size and positions. After getting it to where you want- click the display properties feature and it will tell you the size/position information. You then must go back to the main HOMer.fig file and make the appropriate changes in the property editor and save the fig file. This makes it easier to customize the fig file (expecially given it multi-layering nature). After creating a button in the HOMer.m program add the code: function your_button_name_callback(hobject, eventdata, handles) <Subprogram name>('<name of function in subprogram>',hobject, eventdata, handles, <any flags passed>); The reason to call all programs from this main program (rather then calling them directly from the callback itself is so I can allow users to change functions without giving access to the code itself and allow the program to (eventually) be compiled into a stand-alone cpp code.
14 ---- Asociating objects with a Tab button ---- Objects can be associated with a tab- i.e. their visibilty controled by a tab by simpily adding the name of the object handle to the list in the appropriate *_FUNs.m file (i.e. HOMer_FUNs.m) Whenever this tab is pressed, the main program will go through this list of objects- any objects on this list with a value of "1" for the UserData will be turned on and "0" will be left off. The current state, value, visibility, and any data associated with the object (i.e. strings/text) will be saved and recalled when switching between subjects provided the object is on one of these lists. Passing data into functions: All the data is stored on the main figure's UserData field and can be recalled using the command: data = get(handles.figure1,'userdata'); This data is stored in the Std DOT format used by this lab (same as PMI toolbox format). Data is a cell array indexed by the subect index. For currently displayed subject index type: subnum=get(handles.popupmenu_subjectlist,'value'); Data{subnum} -> data associated with subject subnum for more details- thuppert@nmr.mgh.harvard.edu
Capstone Appendix. A guide to your lab computer software
 Capstone Appendix A guide to your lab computer software Important Notes Many of the Images will look slightly different from what you will see in lab. This is because each lab setup is different and so
Capstone Appendix A guide to your lab computer software Important Notes Many of the Images will look slightly different from what you will see in lab. This is because each lab setup is different and so
SIVIC GUI Overview. SIVIC GUI Layout Overview
 SIVIC GUI Overview SIVIC GUI Layout Overview At the top of the SIVIC GUI is a row of buttons called the Toolbar. It is a quick interface for loading datasets, controlling how the mouse manipulates the
SIVIC GUI Overview SIVIC GUI Layout Overview At the top of the SIVIC GUI is a row of buttons called the Toolbar. It is a quick interface for loading datasets, controlling how the mouse manipulates the
TMSEEG Tutorial. Version 4.0. This tutorial was written by: Sravya Atluri and Matthew Frehlich. Contact:
 TMSEEG Tutorial Version 4.0 This tutorial was written by: Sravya Atluri and Matthew Frehlich Contact: faranak.farzan@sfu.ca For more detail, please see the Method article describing the TMSEEG Toolbox:
TMSEEG Tutorial Version 4.0 This tutorial was written by: Sravya Atluri and Matthew Frehlich Contact: faranak.farzan@sfu.ca For more detail, please see the Method article describing the TMSEEG Toolbox:
Tutorial BOLD Module
 m a k i n g f u n c t i o n a l M R I e a s y n o r d i c B r a i n E x Tutorial BOLD Module Please note that this tutorial is for the latest released nordicbrainex. If you are using an older version please
m a k i n g f u n c t i o n a l M R I e a s y n o r d i c B r a i n E x Tutorial BOLD Module Please note that this tutorial is for the latest released nordicbrainex. If you are using an older version please
Agilent EZChrom Elite. PDA Analysis
 Agilent EZChrom Elite PDA Analysis Notices Copyright Scientific Software, Inc 1997-2003 Agilent Technologies, Inc. 2006. No part of this manual may be reproduced in any form or by any means (including
Agilent EZChrom Elite PDA Analysis Notices Copyright Scientific Software, Inc 1997-2003 Agilent Technologies, Inc. 2006. No part of this manual may be reproduced in any form or by any means (including
Getting Started with DADiSP
 Section 1: Welcome to DADiSP Getting Started with DADiSP This guide is designed to introduce you to the DADiSP environment. It gives you the opportunity to build and manipulate your own sample Worksheets
Section 1: Welcome to DADiSP Getting Started with DADiSP This guide is designed to introduce you to the DADiSP environment. It gives you the opportunity to build and manipulate your own sample Worksheets
ERPEEG Tutorial. Version 1.0. This tutorial was written by: Sravya Atluri, Matthew Frehlich and Dr. Faranak Farzan.
 ERPEEG Tutorial Version 1.0 This tutorial was written by: Sravya Atluri, Matthew Frehlich and Dr. Faranak Farzan. Contact: faranak.farzan@sfu.ca Temerty Centre for Therapeutic Brain Stimulation Centre
ERPEEG Tutorial Version 1.0 This tutorial was written by: Sravya Atluri, Matthew Frehlich and Dr. Faranak Farzan. Contact: faranak.farzan@sfu.ca Temerty Centre for Therapeutic Brain Stimulation Centre
TracePro Tutorial Tissue Optics
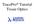 TracePro Tutorial Tissue Optics Splitting the Screen To view the System Tree, select Window Split, then drag the mouse to the right to position the vertical splitter bar. Alternatively, you can place your
TracePro Tutorial Tissue Optics Splitting the Screen To view the System Tree, select Window Split, then drag the mouse to the right to position the vertical splitter bar. Alternatively, you can place your
Physics MRI Research Centre UNIFIT VERSION User s Guide
 Physics MRI Research Centre UNIFIT VERSION 1.24 User s Guide Note: If an error occurs please quit UNIFIT and type:.reset in the IDL command line, and restart UNIFIT. Last Update November 2016 by Katie
Physics MRI Research Centre UNIFIT VERSION 1.24 User s Guide Note: If an error occurs please quit UNIFIT and type:.reset in the IDL command line, and restart UNIFIT. Last Update November 2016 by Katie
Emerge Workflow CE8 SAMPLE IMAGE. Simon Voisey July 2008
 Emerge Workflow SAMPLE IMAGE CE8 Simon Voisey July 2008 Introduction The document provides a step-by-step guide for producing Emerge predicted petrophysical volumes based on log data of the same type.
Emerge Workflow SAMPLE IMAGE CE8 Simon Voisey July 2008 Introduction The document provides a step-by-step guide for producing Emerge predicted petrophysical volumes based on log data of the same type.
EMERGE Workflow CE8R2 SAMPLE IMAGE. Simon Voisey Hampson-Russell London Office
 EMERGE Workflow SAMPLE IMAGE CE8R2 Simon Voisey Hampson-Russell London Office August 2008 Introduction The document provides a step-by-step guide for producing Emerge-predicted petrophysical p volumes
EMERGE Workflow SAMPLE IMAGE CE8R2 Simon Voisey Hampson-Russell London Office August 2008 Introduction The document provides a step-by-step guide for producing Emerge-predicted petrophysical p volumes
Assessment of Diffuse Optical Tomography Image Reconstruction Methods Using a Photon Transport Model
 Assessment of Diffuse Optical Tomography Image Reconstruction Methods Using a Photon Transport Model Murad M. Althobaiti 1 *, Hassan S. Salehi 2, and Quing Zhu 1,2 1 Department of Biomedical Engineering,
Assessment of Diffuse Optical Tomography Image Reconstruction Methods Using a Photon Transport Model Murad M. Althobaiti 1 *, Hassan S. Salehi 2, and Quing Zhu 1,2 1 Department of Biomedical Engineering,
SystemVue 2011 Fundamentals (version ) LAB EXERCISE 1. SystemVue Basics
 SystemVue 2011 Fundamentals (version 1-2012) LAB EXERCISE 1 SystemVue Basics This lab exercise introduces the basic use model of SystemVue, including using the built-in templates, running analyses and
SystemVue 2011 Fundamentals (version 1-2012) LAB EXERCISE 1 SystemVue Basics This lab exercise introduces the basic use model of SystemVue, including using the built-in templates, running analyses and
Modify Panel. Flatten Tab
 AFM Image Processing Most images will need some post acquisition processing. A typical procedure is to: i) modify the image by flattening, using a planefit, and possibly also a mask, ii) analyzing the
AFM Image Processing Most images will need some post acquisition processing. A typical procedure is to: i) modify the image by flattening, using a planefit, and possibly also a mask, ii) analyzing the
MindWare Electromyography (EMG) Analysis User Reference Guide Version Copyright 2011 by MindWare Technologies LTD. All Rights Reserved.
 MindWare Electromyography (EMG) Analysis User Reference Guide Version 3.0.12 Copyright 2011 by MindWare Technologies LTD. All Rights Reserved. MindWare EMG 3.0.12 User Guide Internet Support E-mail: sales@mindwaretech.com
MindWare Electromyography (EMG) Analysis User Reference Guide Version 3.0.12 Copyright 2011 by MindWare Technologies LTD. All Rights Reserved. MindWare EMG 3.0.12 User Guide Internet Support E-mail: sales@mindwaretech.com
Appendix A. Introduction to MATLAB. A.1 What Is MATLAB?
 Appendix A Introduction to MATLAB A.1 What Is MATLAB? MATLAB is a technical computing environment developed by The Math- Works, Inc. for computation and data visualization. It is both an interactive system
Appendix A Introduction to MATLAB A.1 What Is MATLAB? MATLAB is a technical computing environment developed by The Math- Works, Inc. for computation and data visualization. It is both an interactive system
Plotting Graphs. Error Bars
 E Plotting Graphs Construct your graphs in Excel using the method outlined in the Graphing and Error Analysis lab (in the Phys 124/144/130 laboratory manual). Always choose the x-y scatter plot. Number
E Plotting Graphs Construct your graphs in Excel using the method outlined in the Graphing and Error Analysis lab (in the Phys 124/144/130 laboratory manual). Always choose the x-y scatter plot. Number
Single Subject Demo Data Instructions 1) click "New" and answer "No" to the "spatially preprocess" question.
 (1) conn - Functional connectivity toolbox v1.0 Single Subject Demo Data Instructions 1) click "New" and answer "No" to the "spatially preprocess" question. 2) in "Basic" enter "1" subject, "6" seconds
(1) conn - Functional connectivity toolbox v1.0 Single Subject Demo Data Instructions 1) click "New" and answer "No" to the "spatially preprocess" question. 2) in "Basic" enter "1" subject, "6" seconds
Manual. User Reference Guide. Analysis Application (EMG) Electromyography Analysis
 Phone: (888) 765-9735 WWW.MINDWARETECH.COM User Reference Guide Manual Analysis Application Electromyography Analysis (EMG) Copyright 2014 by MindWare Technologies LTD. All Rights Reserved. 1 Phone: (614)
Phone: (888) 765-9735 WWW.MINDWARETECH.COM User Reference Guide Manual Analysis Application Electromyography Analysis (EMG) Copyright 2014 by MindWare Technologies LTD. All Rights Reserved. 1 Phone: (614)
To Plot a Graph in Origin. Example: Number of Counts from a Geiger- Müller Tube as a Function of Supply Voltage
 To Plot a Graph in Origin Example: Number of Counts from a Geiger- Müller Tube as a Function of Supply Voltage 1 Digression on Error Bars What entity do you use for the magnitude of the error bars? Standard
To Plot a Graph in Origin Example: Number of Counts from a Geiger- Müller Tube as a Function of Supply Voltage 1 Digression on Error Bars What entity do you use for the magnitude of the error bars? Standard
Excel Primer CH141 Fall, 2017
 Excel Primer CH141 Fall, 2017 To Start Excel : Click on the Excel icon found in the lower menu dock. Once Excel Workbook Gallery opens double click on Excel Workbook. A blank workbook page should appear
Excel Primer CH141 Fall, 2017 To Start Excel : Click on the Excel icon found in the lower menu dock. Once Excel Workbook Gallery opens double click on Excel Workbook. A blank workbook page should appear
Volocity ver (2013) Standard Operation Protocol
 Faculty Core Facility Volocity 6.3.0 (2013) SOP A-1 Volocity ver. 6.3.0 (2013) Standard Operation Protocol Faculty Core Facility Volocity 6.3.0 (2013) SOP A-2 A. Content Overview. 3 Start up. 3 Change
Faculty Core Facility Volocity 6.3.0 (2013) SOP A-1 Volocity ver. 6.3.0 (2013) Standard Operation Protocol Faculty Core Facility Volocity 6.3.0 (2013) SOP A-2 A. Content Overview. 3 Start up. 3 Change
Appendix 1: DataStudio with ScienceWorkshop Sensors Tech Tips
 Appendix 1: DataStudio with ScienceWorkshop Sensors Tech Tips Section 1: Starting an experiment 1.1 Opening a file 1. Open the File menu and select Open Activity. 2. In the Open dialog box, navigate to
Appendix 1: DataStudio with ScienceWorkshop Sensors Tech Tips Section 1: Starting an experiment 1.1 Opening a file 1. Open the File menu and select Open Activity. 2. In the Open dialog box, navigate to
LAB 2: DATA FILTERING AND NOISE REDUCTION
 NAME: LAB TIME: LAB 2: DATA FILTERING AND NOISE REDUCTION In this exercise, you will use Microsoft Excel to generate several synthetic data sets based on a simplified model of daily high temperatures in
NAME: LAB TIME: LAB 2: DATA FILTERING AND NOISE REDUCTION In this exercise, you will use Microsoft Excel to generate several synthetic data sets based on a simplified model of daily high temperatures in
Managing custom montage files Quick montages How custom montage files are applied Markers Adding markers...
 AnyWave Contents What is AnyWave?... 3 AnyWave home directories... 3 Opening a file in AnyWave... 4 Quick re-open a recent file... 4 Viewing the content of a file... 5 Choose what you want to view and
AnyWave Contents What is AnyWave?... 3 AnyWave home directories... 3 Opening a file in AnyWave... 4 Quick re-open a recent file... 4 Viewing the content of a file... 5 Choose what you want to view and
Microsoft Word for Report-Writing (2016 Version)
 Microsoft Word for Report-Writing (2016 Version) Microsoft Word is a versatile, widely-used tool for producing presentation-quality documents. Most students are well-acquainted with the program for generating
Microsoft Word for Report-Writing (2016 Version) Microsoft Word is a versatile, widely-used tool for producing presentation-quality documents. Most students are well-acquainted with the program for generating
BioFuel Graphing instructions using Microsoft Excel 2003 (Microsoft Excel 2007 instructions start on page mei-7)
 BioFuel Graphing instructions using Microsoft Excel 2003 (Microsoft Excel 2007 instructions start on page mei-7) Graph as a XY Scatter Chart, add titles for chart and axes, remove gridlines. A. Select
BioFuel Graphing instructions using Microsoft Excel 2003 (Microsoft Excel 2007 instructions start on page mei-7) Graph as a XY Scatter Chart, add titles for chart and axes, remove gridlines. A. Select
MATLAB Introduction to MATLAB Programming
 MATLAB Introduction to MATLAB Programming MATLAB Scripts So far we have typed all the commands in the Command Window which were executed when we hit Enter. Although every MATLAB command can be executed
MATLAB Introduction to MATLAB Programming MATLAB Scripts So far we have typed all the commands in the Command Window which were executed when we hit Enter. Although every MATLAB command can be executed
If you are not familiar with IMP you should download and read WhatIsImp and CoordGenManual before proceeding.
 IMP: PCAGen6- Principal Components Generation Utility PCAGen6 This software package is a Principal Components Analysis utility intended for use with landmark based morphometric data. The program loads
IMP: PCAGen6- Principal Components Generation Utility PCAGen6 This software package is a Principal Components Analysis utility intended for use with landmark based morphometric data. The program loads
CS/NEUR125 Brains, Minds, and Machines. Due: Wednesday, April 5
 CS/NEUR125 Brains, Minds, and Machines Lab 8: Using fmri to Discover Language Areas in the Brain Due: Wednesday, April 5 In this lab, you will analyze fmri data from an experiment that was designed to
CS/NEUR125 Brains, Minds, and Machines Lab 8: Using fmri to Discover Language Areas in the Brain Due: Wednesday, April 5 In this lab, you will analyze fmri data from an experiment that was designed to
Data and Function Plotting with MATLAB (Linux-10)
 Data and Function Plotting with MATLAB (Linux-10) This tutorial describes the use of MATLAB for general plotting of experimental data and equations and for special plots like histograms. (Astronomers -
Data and Function Plotting with MATLAB (Linux-10) This tutorial describes the use of MATLAB for general plotting of experimental data and equations and for special plots like histograms. (Astronomers -
Forston Labs LabNavigator
 Forston Labs LabNavigator (Order Code LabNavigator) Overview Welcome to LabNavigator. This guide will get you started with basic data collection. More information is available in the LabNavigator Reference
Forston Labs LabNavigator (Order Code LabNavigator) Overview Welcome to LabNavigator. This guide will get you started with basic data collection. More information is available in the LabNavigator Reference
Exporting data from DataStudio
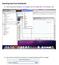 Exporting data from DataStudio 1) Select a data set by clicking on it on the graph, then click Export Data in the Display menu. 2) Save the file in the format N_# where N is a string of characters and
Exporting data from DataStudio 1) Select a data set by clicking on it on the graph, then click Export Data in the Display menu. 2) Save the file in the format N_# where N is a string of characters and
1. What specialist uses information obtained from bones to help police solve crimes?
 Mathematics: Modeling Our World Unit 4: PREDICTION HANDOUT VIDEO VIEWING GUIDE H4.1 1. What specialist uses information obtained from bones to help police solve crimes? 2.What are some things that can
Mathematics: Modeling Our World Unit 4: PREDICTION HANDOUT VIDEO VIEWING GUIDE H4.1 1. What specialist uses information obtained from bones to help police solve crimes? 2.What are some things that can
Fourier Transforms and Signal Analysis
 Fourier Transforms and Signal Analysis The Fourier transform analysis is one of the most useful ever developed in Physical and Analytical chemistry. Everyone knows that FTIR is based on it, but did one
Fourier Transforms and Signal Analysis The Fourier transform analysis is one of the most useful ever developed in Physical and Analytical chemistry. Everyone knows that FTIR is based on it, but did one
A Guide to Using Some Basic MATLAB Functions
 A Guide to Using Some Basic MATLAB Functions UNC Charlotte Robert W. Cox This document provides a brief overview of some of the essential MATLAB functionality. More thorough descriptions are available
A Guide to Using Some Basic MATLAB Functions UNC Charlotte Robert W. Cox This document provides a brief overview of some of the essential MATLAB functionality. More thorough descriptions are available
Matlab Tutorial 1: Working with variables, arrays, and plotting
 Matlab Tutorial 1: Working with variables, arrays, and plotting Setting up Matlab First of all, let's make sure we all have the same layout of the different windows in Matlab. Go to Home Layout Default.
Matlab Tutorial 1: Working with variables, arrays, and plotting Setting up Matlab First of all, let's make sure we all have the same layout of the different windows in Matlab. Go to Home Layout Default.
Zahner 12/2011. Light Spectra Display and Analysis
 Zahner 12/2011 Light Spectra Display and Analysis Light Spectra Analysis -2-1. Introduction 3 2. Light Spectra Analysis Graphic Software 4 2.1. Startup...4 2.2. Export Functions...6 2.2.1. Export ASCII
Zahner 12/2011 Light Spectra Display and Analysis Light Spectra Analysis -2-1. Introduction 3 2. Light Spectra Analysis Graphic Software 4 2.1. Startup...4 2.2. Export Functions...6 2.2.1. Export ASCII
The LabScribe Tutorial
 The LabScribe Tutorial LabScribe allows data to be accumulated, displayed and analyzed on a computer screen in a format similar to a laboratory strip chart recorder. Equipment Setup 1 Place the iworx unit
The LabScribe Tutorial LabScribe allows data to be accumulated, displayed and analyzed on a computer screen in a format similar to a laboratory strip chart recorder. Equipment Setup 1 Place the iworx unit
IgorCD Procedures. Data -> Load CD Data
 IgorCD Procedures IgorCD is a set of Igor Pro procedures for processing CD spectra and CD thermal melts. It correctly implements spectral smoothing functions and facilitates subtracting baselines (or constant
IgorCD Procedures IgorCD is a set of Igor Pro procedures for processing CD spectra and CD thermal melts. It correctly implements spectral smoothing functions and facilitates subtracting baselines (or constant
The iworx 214 and LabScribe V2.0 Tutorial. Overview
 The iworx 214 and LabScribe V2.0 Overview Figure T-1-1: The front and rear panels of IWX/214. The data acquisition unit used in the iworx teaching kits is the IWX/214 (Figure T-1-1 on page T-1-1). The
The iworx 214 and LabScribe V2.0 Overview Figure T-1-1: The front and rear panels of IWX/214. The data acquisition unit used in the iworx teaching kits is the IWX/214 (Figure T-1-1 on page T-1-1). The
Back-Scatter Cloud Probe (BCP) Module. Operator Manual
 Particle Analysis and Display System (PADS): Back-Scatter Cloud Probe (BCP) Module Operator Manual DOC-0234 A-2 PADS 2.8.2 BCP Module 2.8.0 2545 Central Avenue Boulder, CO 80301 USA C O P Y R I G H T 2
Particle Analysis and Display System (PADS): Back-Scatter Cloud Probe (BCP) Module Operator Manual DOC-0234 A-2 PADS 2.8.2 BCP Module 2.8.0 2545 Central Avenue Boulder, CO 80301 USA C O P Y R I G H T 2
Diffuse Optical Tomography, Inverse Problems, and Optimization. Mary Katherine Huffman. Undergraduate Research Fall 2011 Spring 2012
 Diffuse Optical Tomography, Inverse Problems, and Optimization Mary Katherine Huffman Undergraduate Research Fall 11 Spring 12 1. Introduction. This paper discusses research conducted in order to investigate
Diffuse Optical Tomography, Inverse Problems, and Optimization Mary Katherine Huffman Undergraduate Research Fall 11 Spring 12 1. Introduction. This paper discusses research conducted in order to investigate
Matlab Introduction. Scalar Variables and Arithmetic Operators
 Matlab Introduction Matlab is both a powerful computational environment and a programming language that easily handles matrix and complex arithmetic. It is a large software package that has many advanced
Matlab Introduction Matlab is both a powerful computational environment and a programming language that easily handles matrix and complex arithmetic. It is a large software package that has many advanced
USER's GUIDE for PLOT_FINDIF_1
 USER's GUIDE for PLOT_FINDIF_1 S. T. Bolmer & R. A. Stephen November 2004 Page 1 PLOT_FINDIF_1 Plot_findif_1.m is a MATLAB 6 script, which is used to plot output from the Woods Hole Oceanographic Institution
USER's GUIDE for PLOT_FINDIF_1 S. T. Bolmer & R. A. Stephen November 2004 Page 1 PLOT_FINDIF_1 Plot_findif_1.m is a MATLAB 6 script, which is used to plot output from the Woods Hole Oceanographic Institution
MestReC Cheat Sheet. by Monika Ivancic, July 1 st 2005
 MestReC Cheat Sheet by Monika Ivancic, July 1 st 2005 This Cheat Sheet is to be used at UW-Madison as a quick guide to processing using the MestReC NMR software. You may find more help at the MestReC homepage
MestReC Cheat Sheet by Monika Ivancic, July 1 st 2005 This Cheat Sheet is to be used at UW-Madison as a quick guide to processing using the MestReC NMR software. You may find more help at the MestReC homepage
Bayesian estimation of optical properties of the human head via 3D structural MRI p.1
 Bayesian estimation of optical properties of the human head via 3D structural MRI June 23, 2003 at ECBO 2003 Alex Barnett Courant Institute, New York University Collaborators (NMR Center, Mass. Gen. Hosp.,
Bayesian estimation of optical properties of the human head via 3D structural MRI June 23, 2003 at ECBO 2003 Alex Barnett Courant Institute, New York University Collaborators (NMR Center, Mass. Gen. Hosp.,
Using Liberty Instruments PRAXIS for Room Sound Convolution Rev 9/12/2004
 Using Liberty Instruments PRAXIS for Room Sound Convolution Rev 9/12/2004 Overview Room Sound Convolution is an operation that allows you to measure, save, and later recreate a representation of the sound
Using Liberty Instruments PRAXIS for Room Sound Convolution Rev 9/12/2004 Overview Room Sound Convolution is an operation that allows you to measure, save, and later recreate a representation of the sound
ECE 202 LAB 3 ADVANCED MATLAB
 Version 1.2 1 of 13 BEFORE YOU BEGIN PREREQUISITE LABS ECE 201 Labs EXPECTED KNOWLEDGE ECE 202 LAB 3 ADVANCED MATLAB Understanding of the Laplace transform and transfer functions EQUIPMENT Intel PC with
Version 1.2 1 of 13 BEFORE YOU BEGIN PREREQUISITE LABS ECE 201 Labs EXPECTED KNOWLEDGE ECE 202 LAB 3 ADVANCED MATLAB Understanding of the Laplace transform and transfer functions EQUIPMENT Intel PC with
Ch. 4 Physical Principles of CT
 Ch. 4 Physical Principles of CT CLRS 408: Intro to CT Department of Radiation Sciences Review: Why CT? Solution for radiography/tomography limitations Superimposition of structures Distinguishing between
Ch. 4 Physical Principles of CT CLRS 408: Intro to CT Department of Radiation Sciences Review: Why CT? Solution for radiography/tomography limitations Superimposition of structures Distinguishing between
FMRI Pre-Processing and Model- Based Statistics
 FMRI Pre-Processing and Model- Based Statistics Brief intro to FMRI experiments and analysis FMRI pre-stats image processing Simple Single-Subject Statistics Multi-Level FMRI Analysis Advanced FMRI Analysis
FMRI Pre-Processing and Model- Based Statistics Brief intro to FMRI experiments and analysis FMRI pre-stats image processing Simple Single-Subject Statistics Multi-Level FMRI Analysis Advanced FMRI Analysis
In this chapter we explain the structure and use of BestDose, with examples.
 Using BestDose In this chapter we explain the structure and use of BestDose, with examples. General description The image below shows the view you get when you start BestDose. The actual view is called
Using BestDose In this chapter we explain the structure and use of BestDose, with examples. General description The image below shows the view you get when you start BestDose. The actual view is called
The Fundamentals. Document Basics
 3 The Fundamentals Opening a Program... 3 Similarities in All Programs... 3 It's On Now What?...4 Making things easier to see.. 4 Adjusting Text Size.....4 My Computer. 4 Control Panel... 5 Accessibility
3 The Fundamentals Opening a Program... 3 Similarities in All Programs... 3 It's On Now What?...4 Making things easier to see.. 4 Adjusting Text Size.....4 My Computer. 4 Control Panel... 5 Accessibility
Data Acquisition with CP-2002/2003 Micro-GC Control
 Varian Analytical Instruments 2700 Mitchell Drive Walnut Creek, CA 94598 Star Chromatography Workstation Version 6 Data Acquisition with CP-2002/2003 Micro-GC Control Operation Manual Varian, Inc. 2002
Varian Analytical Instruments 2700 Mitchell Drive Walnut Creek, CA 94598 Star Chromatography Workstation Version 6 Data Acquisition with CP-2002/2003 Micro-GC Control Operation Manual Varian, Inc. 2002
Single-sided Magnet Diffusion Measurements
 Single-sided Magnet Diffusion Measurements Kuldeep S. Panesar April 2013 (kuldeep.panesar@gmail.com) Contents 1. Initial configuration... 2 2. Preparation for diffusion measurement... 5 3. Starting a diffusion
Single-sided Magnet Diffusion Measurements Kuldeep S. Panesar April 2013 (kuldeep.panesar@gmail.com) Contents 1. Initial configuration... 2 2. Preparation for diffusion measurement... 5 3. Starting a diffusion
Introduction to the workbook and spreadsheet
 Excel Tutorial To make the most of this tutorial I suggest you follow through it while sitting in front of a computer with Microsoft Excel running. This will allow you to try things out as you follow along.
Excel Tutorial To make the most of this tutorial I suggest you follow through it while sitting in front of a computer with Microsoft Excel running. This will allow you to try things out as you follow along.
APPENDIX B STEP-BY-STEP APPLICATIONS OF THE FEMWATER-LHS
 APPENDIX B STEP-BY-STEP APPLICATIONS OF THE FEMWATER-LHS Steady Two-Dimensional Drainage Problem 1. Double-click on the Argus ONE icon to open Argus ONE. 2. From the PIEs menu found along the top of the
APPENDIX B STEP-BY-STEP APPLICATIONS OF THE FEMWATER-LHS Steady Two-Dimensional Drainage Problem 1. Double-click on the Argus ONE icon to open Argus ONE. 2. From the PIEs menu found along the top of the
KINETICS CALCS AND GRAPHS INSTRUCTIONS
 KINETICS CALCS AND GRAPHS INSTRUCTIONS 1. Open a new Excel or Google Sheets document. I will be using Google Sheets for this tutorial, but Excel is nearly the same. 2. Enter headings across the top as
KINETICS CALCS AND GRAPHS INSTRUCTIONS 1. Open a new Excel or Google Sheets document. I will be using Google Sheets for this tutorial, but Excel is nearly the same. 2. Enter headings across the top as
Appendix I. TACTICS Toolbox v3.x. Interactive MATLAB Platform For Bioimaging informatics. User Guide TRACKING MODULE
 TACTICS Toolbox v3.x Interactive MATLAB Platform For Bioimaging informatics User Guide TRACKING MODULE -17- Cell Tracking Module 1 (user interface) Once the images were successfully segmented, the next
TACTICS Toolbox v3.x Interactive MATLAB Platform For Bioimaging informatics User Guide TRACKING MODULE -17- Cell Tracking Module 1 (user interface) Once the images were successfully segmented, the next
Matlab OTKB GUI Manual:
 Matlab OTKB GUI Manual: Preface: This is the manual for the OTKB GUI. This GUI can be used to control stage position as well as perform sensitivity and stiffness calibrations on the trap. This manual will
Matlab OTKB GUI Manual: Preface: This is the manual for the OTKB GUI. This GUI can be used to control stage position as well as perform sensitivity and stiffness calibrations on the trap. This manual will
430g Dimensions. 102mm x 84mm x 59mm Detector nm Pixels 3648 Pixel size. 8μm x 200μm Pixel well depth
 Weigth 430g Dimensions 102mm x 84mm x 59mm Detector Toshiba TCD1304DG linear array Detector range 200-1200nm Pixels 3648 Pixel size 8μm x 200μm Pixel well depth 100,000 electrons Signal-to-noise ratio
Weigth 430g Dimensions 102mm x 84mm x 59mm Detector Toshiba TCD1304DG linear array Detector range 200-1200nm Pixels 3648 Pixel size 8μm x 200μm Pixel well depth 100,000 electrons Signal-to-noise ratio
Sample Sizes: up to 1 X1 X 1/4. Scanners: 50 X 50 X 17 microns and 15 X 15 X 7 microns
 R-AFM100 For Nanotechnology Researchers Wanting to do routine scanning of nano-structures Instrument Innovators Using AFM as a platform to create a new instrument Educators Teaching students about AFM
R-AFM100 For Nanotechnology Researchers Wanting to do routine scanning of nano-structures Instrument Innovators Using AFM as a platform to create a new instrument Educators Teaching students about AFM
METRICS WIN4145. How Win4145 Stores Information. Project Files. The Win4145 Initialization File. Data and Plot Windows.
 METRICS WIN4145 REFERENCE GUIDE CONTENTS CHAPTER 1: A QUICK TOUR OF WIN4145 How Win4145 Stores Information Project Files The Win4145 Initialization File Data and Plot Windows The Menu Bars The Toolbar
METRICS WIN4145 REFERENCE GUIDE CONTENTS CHAPTER 1: A QUICK TOUR OF WIN4145 How Win4145 Stores Information Project Files The Win4145 Initialization File Data and Plot Windows The Menu Bars The Toolbar
University of Minnesota Nano Fabrication Center Standard Operating Procedure
 Equipment Name: University of Minnesota Nano Fabrication Center Coral Name: hs-scope Revision Number: 1.5 Model: HS200A Revisionist: M. Fisher Location: Bay 1 Date: 9/12/2013 1 Description The Hyphenated
Equipment Name: University of Minnesota Nano Fabrication Center Coral Name: hs-scope Revision Number: 1.5 Model: HS200A Revisionist: M. Fisher Location: Bay 1 Date: 9/12/2013 1 Description The Hyphenated
Chapter 2 Surfer Tutorial
 Chapter 2 Surfer Tutorial Overview This tutorial introduces you to some of Surfer s features and shows you the steps to take to produce maps. In addition, the tutorial will help previous Surfer users learn
Chapter 2 Surfer Tutorial Overview This tutorial introduces you to some of Surfer s features and shows you the steps to take to produce maps. In addition, the tutorial will help previous Surfer users learn
Introduction to Digital Image Processing
 Fall 2005 Image Enhancement in the Spatial Domain: Histograms, Arithmetic/Logic Operators, Basics of Spatial Filtering, Smoothing Spatial Filters Tuesday, February 7 2006, Overview (1): Before We Begin
Fall 2005 Image Enhancement in the Spatial Domain: Histograms, Arithmetic/Logic Operators, Basics of Spatial Filtering, Smoothing Spatial Filters Tuesday, February 7 2006, Overview (1): Before We Begin
Multi Time Series Rev
 Multi Time Series Rev. 4.0.12 The Multi Time Series program is designed to provide flexible programming of automated time dependent experiments. The basic programming unit is a single Time Series Block
Multi Time Series Rev. 4.0.12 The Multi Time Series program is designed to provide flexible programming of automated time dependent experiments. The basic programming unit is a single Time Series Block
Here is the data collected.
 Introduction to Scientific Analysis of Data Using Spreadsheets. Computer spreadsheets are very powerful tools that are widely used in Business, Science, and Engineering to perform calculations and record,
Introduction to Scientific Analysis of Data Using Spreadsheets. Computer spreadsheets are very powerful tools that are widely used in Business, Science, and Engineering to perform calculations and record,
Charts in Excel 2003
 Charts in Excel 2003 Contents Introduction Charts in Excel 2003...1 Part 1: Generating a Basic Chart...1 Part 2: Adding Another Data Series...3 Part 3: Other Handy Options...5 Introduction Charts in Excel
Charts in Excel 2003 Contents Introduction Charts in Excel 2003...1 Part 1: Generating a Basic Chart...1 Part 2: Adding Another Data Series...3 Part 3: Other Handy Options...5 Introduction Charts in Excel
MAT 275 Laboratory 2 Matrix Computations and Programming in MATLAB
 MATLAB sessions: Laboratory MAT 75 Laboratory Matrix Computations and Programming in MATLAB In this laboratory session we will learn how to. Create and manipulate matrices and vectors.. Write simple programs
MATLAB sessions: Laboratory MAT 75 Laboratory Matrix Computations and Programming in MATLAB In this laboratory session we will learn how to. Create and manipulate matrices and vectors.. Write simple programs
Agilent ChemStation for UV-visible Spectroscopy
 Agilent ChemStation for UV-visible Spectroscopy Understanding Your Biochemical Analysis Software Agilent Technologies Notices Agilent Technologies, Inc. 2000, 2003-2008 No part of this manual may be reproduced
Agilent ChemStation for UV-visible Spectroscopy Understanding Your Biochemical Analysis Software Agilent Technologies Notices Agilent Technologies, Inc. 2000, 2003-2008 No part of this manual may be reproduced
Still More About Matlab GUI s (v. 1.3) Popup Menus. Popup Menu Exercise. Still More GUI Info - GE /29/2012. Copyright C. S. Tritt, Ph.D.
 Still More About Matlab GUI s (v. 1.3) Dr. C. S. Tritt with slides from Dr. J. LaMack January 24, 2012 Popup Menus User selects one from a mutually exclusive list of options The String property is typically
Still More About Matlab GUI s (v. 1.3) Dr. C. S. Tritt with slides from Dr. J. LaMack January 24, 2012 Popup Menus User selects one from a mutually exclusive list of options The String property is typically
Using LoggerPro. Nothing is more terrible than to see ignorance in action. J. W. Goethe ( )
 Using LoggerPro Nothing is more terrible than to see ignorance in action. J. W. Goethe (1749-1832) LoggerPro is a general-purpose program for acquiring, graphing and analyzing data. It can accept input
Using LoggerPro Nothing is more terrible than to see ignorance in action. J. W. Goethe (1749-1832) LoggerPro is a general-purpose program for acquiring, graphing and analyzing data. It can accept input
Biometrics Technology: Image Processing & Pattern Recognition (by Dr. Dickson Tong)
 Biometrics Technology: Image Processing & Pattern Recognition (by Dr. Dickson Tong) References: [1] http://homepages.inf.ed.ac.uk/rbf/hipr2/index.htm [2] http://www.cs.wisc.edu/~dyer/cs540/notes/vision.html
Biometrics Technology: Image Processing & Pattern Recognition (by Dr. Dickson Tong) References: [1] http://homepages.inf.ed.ac.uk/rbf/hipr2/index.htm [2] http://www.cs.wisc.edu/~dyer/cs540/notes/vision.html
SYSTEM LINEARITY LAB MANUAL: 2 Modifications for P551 Fall 2013 Medical Physics Laboratory
 SYSTEM LINEARITY LAB MANUAL: 2 Modifications for P551 Fall 2013 Medical Physics Laboratory Introduction In this lab exercise, you will investigate the linearity of the DeskCAT scanner by making measurements
SYSTEM LINEARITY LAB MANUAL: 2 Modifications for P551 Fall 2013 Medical Physics Laboratory Introduction In this lab exercise, you will investigate the linearity of the DeskCAT scanner by making measurements
DAQFactory U3 Tutorial Getting Started with DAQFactory-Express and your LabJack U3 11/3/06
 DAQFactory U3 Tutorial Getting Started with DAQFactory-Express and your LabJack U3 11/3/06 Congratulations on the purchase of your new LabJack U3. Included on your installation CD is a fully licensed copy
DAQFactory U3 Tutorial Getting Started with DAQFactory-Express and your LabJack U3 11/3/06 Congratulations on the purchase of your new LabJack U3. Included on your installation CD is a fully licensed copy
SURFACE XPLORER. Version 4.2 USER MANUAL
 SURFACE XPLORER Version 4.2 USER MANUAL Table of Contents List of Figures... 4 System Requirements... 5 Introduction to SURFACE XPLORER... 6 3D Surface control... 7 Spectrum Panel... 9 Kinetic Profile
SURFACE XPLORER Version 4.2 USER MANUAL Table of Contents List of Figures... 4 System Requirements... 5 Introduction to SURFACE XPLORER... 6 3D Surface control... 7 Spectrum Panel... 9 Kinetic Profile
Exam Microscopic Measurement Techniques 4T th of April, 2008
 Exam Microscopic Measurement Techniques 4T300 29 th of April, 2008 Name / Initials: Ident. #: Education: This exam consists of 5 questions. Questions and sub questions will be rewarded with the amount
Exam Microscopic Measurement Techniques 4T300 29 th of April, 2008 Name / Initials: Ident. #: Education: This exam consists of 5 questions. Questions and sub questions will be rewarded with the amount
Product information. Hi-Tech Electronics Pte Ltd
 Product information Introduction TEMA Motion is the world leading software for advanced motion analysis. Starting with digital image sequences the operator uses TEMA Motion to track objects in images,
Product information Introduction TEMA Motion is the world leading software for advanced motion analysis. Starting with digital image sequences the operator uses TEMA Motion to track objects in images,
Insight: Measurement Tool. User Guide
 OMERO Beta v2.2: Measurement Tool User Guide - 1 - October 2007 Insight: Measurement Tool User Guide Open Microscopy Environment: http://www.openmicroscopy.org OMERO Beta v2.2: Measurement Tool User Guide
OMERO Beta v2.2: Measurement Tool User Guide - 1 - October 2007 Insight: Measurement Tool User Guide Open Microscopy Environment: http://www.openmicroscopy.org OMERO Beta v2.2: Measurement Tool User Guide
Applied Biosystems Attune Cytometric Software version 1.2 Release Notes
 Applied Biosystems Attune Cytometric Software version 1.2 Release Notes Customer ( you ) must review these notes prior to installing or operating the Attune Cytometric Software version 1.2 ( Attune Software
Applied Biosystems Attune Cytometric Software version 1.2 Release Notes Customer ( you ) must review these notes prior to installing or operating the Attune Cytometric Software version 1.2 ( Attune Software
CSE 547: Machine Learning for Big Data Spring Problem Set 2. Please read the homework submission policies.
 CSE 547: Machine Learning for Big Data Spring 2019 Problem Set 2 Please read the homework submission policies. 1 Principal Component Analysis and Reconstruction (25 points) Let s do PCA and reconstruct
CSE 547: Machine Learning for Big Data Spring 2019 Problem Set 2 Please read the homework submission policies. 1 Principal Component Analysis and Reconstruction (25 points) Let s do PCA and reconstruct
Volumetric Classification: Program pca3d
 Volumetric principle component analysis for 3D SEISMIC FACIES ANALYSIS PROGRAM pca3d Overview Principal component analysis (PCA) is widely used to reduce the redundancy and excess dimensionality of the
Volumetric principle component analysis for 3D SEISMIC FACIES ANALYSIS PROGRAM pca3d Overview Principal component analysis (PCA) is widely used to reduce the redundancy and excess dimensionality of the
Chaos Culture. Multiclip Editor / Multiclip Note preview 1.5. Edited by Jason Cowling
 Chaos Culture Introduction... 2 Important stuff... 2 Setup... 3 Editing clips... 4 Using the editor... 5 Settings... 9 Workflow settings... 10 Performance... 13 Future updates... 13 Editor 1.6.61 / Note
Chaos Culture Introduction... 2 Important stuff... 2 Setup... 3 Editing clips... 4 Using the editor... 5 Settings... 9 Workflow settings... 10 Performance... 13 Future updates... 13 Editor 1.6.61 / Note
Standardized Tests: Best Practices for the TI-Nspire CX
 The role of TI technology in the classroom is intended to enhance student learning and deepen understanding. However, efficient and effective use of graphing calculator technology on high stakes tests
The role of TI technology in the classroom is intended to enhance student learning and deepen understanding. However, efficient and effective use of graphing calculator technology on high stakes tests
Finite Element Course ANSYS Mechanical Tutorial Tutorial 3 Cantilever Beam
 Problem Specification Finite Element Course ANSYS Mechanical Tutorial Tutorial 3 Cantilever Beam Consider the beam in the figure below. It is clamped on the left side and has a point force of 8kN acting
Problem Specification Finite Element Course ANSYS Mechanical Tutorial Tutorial 3 Cantilever Beam Consider the beam in the figure below. It is clamped on the left side and has a point force of 8kN acting
LEGENDplex Data Analysis Software Version 8 User Guide
 LEGENDplex Data Analysis Software Version 8 User Guide Introduction Welcome to the user s guide for Version 8 of the LEGENDplex data analysis software for Windows based computers 1. This tutorial will
LEGENDplex Data Analysis Software Version 8 User Guide Introduction Welcome to the user s guide for Version 8 of the LEGENDplex data analysis software for Windows based computers 1. This tutorial will
Surface Xplorer. Manual
 Surface Xplorer Manual Table of contents Introduction to Surface Xplorer... 4 3D Surface... 5 Spectrum Panel... 6 Kinetic Panel... 7 File Info... 8 File Menu... 9 Open File... 9 Save File As... 9 Average
Surface Xplorer Manual Table of contents Introduction to Surface Xplorer... 4 3D Surface... 5 Spectrum Panel... 6 Kinetic Panel... 7 File Info... 8 File Menu... 9 Open File... 9 Save File As... 9 Average
SynPAnal User Manual. Eric Danielson and Sang H. Lee Medical College of Wisconsin
 SynPAnal User Manual Eric Danielson and Sang H. Lee Medical College of Wisconsin Installation Instructions Make sure the most recent version of JAVA is installed and is functioning. Unzip the SynPAnal_Installation.zip
SynPAnal User Manual Eric Danielson and Sang H. Lee Medical College of Wisconsin Installation Instructions Make sure the most recent version of JAVA is installed and is functioning. Unzip the SynPAnal_Installation.zip
2. Creating Field Maps Using the Field Map GUI (Version 2.0) in SPM5
 1. Introduction This manual describes how to use the Field Map Toolbox Version 2.0 for creating unwrapped field maps that can be used to do geometric distortion correction of EPI images in SPM5. 1. 1.
1. Introduction This manual describes how to use the Field Map Toolbox Version 2.0 for creating unwrapped field maps that can be used to do geometric distortion correction of EPI images in SPM5. 1. 1.
Dealing with Data in Excel 2013/2016
 Dealing with Data in Excel 2013/2016 Excel provides the ability to do computations and graphing of data. Here we provide the basics and some advanced capabilities available in Excel that are useful for
Dealing with Data in Excel 2013/2016 Excel provides the ability to do computations and graphing of data. Here we provide the basics and some advanced capabilities available in Excel that are useful for
Lab 1- Introduction to Motion
 Partner : Purpose Partner 2: Lab - Section: The purpose of this lab is to learn via a motion detector the relationship between position and velocity. Remember that this device measures the position of
Partner : Purpose Partner 2: Lab - Section: The purpose of this lab is to learn via a motion detector the relationship between position and velocity. Remember that this device measures the position of
REDUCED ORDER MODELING IN MULTISPECTRAL PHOTOACOUSTIC TOMOGRAPHY
 REDUCED ORDER MODELING IN MULTISPECTRAL PHOTOACOUSTIC TOMOGRAPHY Arvind Saibaba Sarah Vallélian Statistical and Applied Mathematical Sciences Institute & North Carolina State University May 26, 2016 OUTLINE
REDUCED ORDER MODELING IN MULTISPECTRAL PHOTOACOUSTIC TOMOGRAPHY Arvind Saibaba Sarah Vallélian Statistical and Applied Mathematical Sciences Institute & North Carolina State University May 26, 2016 OUTLINE
SfM_Georef v.2.2.
 SfM_Georef v.2.2 http://www.lancs.ac.uk/staff/jamesm/software/sfm_georef.htm Introduction Sfm_georef is a gui-based tool for scaling and orienting SfM point clouds to real-world coordinates, using observations
SfM_Georef v.2.2 http://www.lancs.ac.uk/staff/jamesm/software/sfm_georef.htm Introduction Sfm_georef is a gui-based tool for scaling and orienting SfM point clouds to real-world coordinates, using observations
newfasant US User Guide
 newfasant US User Guide Software Version: 6.2.10 Date: April 15, 2018 Index 1. FILE MENU 2. EDIT MENU 3. VIEW MENU 4. GEOMETRY MENU 5. MATERIALS MENU 6. SIMULATION MENU 6.1. PARAMETERS 6.2. DOPPLER 7.
newfasant US User Guide Software Version: 6.2.10 Date: April 15, 2018 Index 1. FILE MENU 2. EDIT MENU 3. VIEW MENU 4. GEOMETRY MENU 5. MATERIALS MENU 6. SIMULATION MENU 6.1. PARAMETERS 6.2. DOPPLER 7.
STIPlotDigitizer. User s Manual
 STIPlotDigitizer User s Manual Table of Contents What is STIPlotDigitizer?... 3 Installation Guide... 3 Initializing STIPlotDigitizer... 4 Project GroupBox... 4 Import Image GroupBox... 5 Exit Button...
STIPlotDigitizer User s Manual Table of Contents What is STIPlotDigitizer?... 3 Installation Guide... 3 Initializing STIPlotDigitizer... 4 Project GroupBox... 4 Import Image GroupBox... 5 Exit Button...
SEM Drift Correction. Procedure Guide
 SEM Drift Correction Procedure Guide Introduction Vic-2D includes experimental functionality to correct for both drift and geometric distortions that occur in images taken using SEMs. The correction requires
SEM Drift Correction Procedure Guide Introduction Vic-2D includes experimental functionality to correct for both drift and geometric distortions that occur in images taken using SEMs. The correction requires
Geostatistics 2D GMS 7.0 TUTORIALS. 1 Introduction. 1.1 Contents
 GMS 7.0 TUTORIALS 1 Introduction Two-dimensional geostatistics (interpolation) can be performed in GMS using the 2D Scatter Point module. The module is used to interpolate from sets of 2D scatter points
GMS 7.0 TUTORIALS 1 Introduction Two-dimensional geostatistics (interpolation) can be performed in GMS using the 2D Scatter Point module. The module is used to interpolate from sets of 2D scatter points
How to use Excel Spreadsheets for Graphing
 How to use Excel Spreadsheets for Graphing 1. Click on the Excel Program on the Desktop 2. You will notice that a screen similar to the above screen comes up. A spreadsheet is divided into Columns (A,
How to use Excel Spreadsheets for Graphing 1. Click on the Excel Program on the Desktop 2. You will notice that a screen similar to the above screen comes up. A spreadsheet is divided into Columns (A,
Creates a 1 X 1 matrix (scalar) with a value of 1 in the column 1, row 1 position and prints the matrix aaa in the command window.
 EE 350L: Signals and Transforms Lab Spring 2007 Lab #1 - Introduction to MATLAB Lab Handout Matlab Software: Matlab will be the analytical tool used in the signals lab. The laboratory has network licenses
EE 350L: Signals and Transforms Lab Spring 2007 Lab #1 - Introduction to MATLAB Lab Handout Matlab Software: Matlab will be the analytical tool used in the signals lab. The laboratory has network licenses
