flowqb Automated Quadratic Characterization of Flow Cytometer Instrument Sensitivity *
|
|
|
- Sherilyn McGee
- 5 years ago
- Views:
Transcription
1 flowqb Automated Quadratic Characterization of Flow Cytometer Instrument Sensitivity * Josef Spidlen 1, Wayne Moore 2, Faysal El Khettabi, David Parks 2, Ryan Brinkman 1 1 Terry Fox Laboratory, British Columbia Cancer Agency Vancouver, BC, Canada 2 Genetics Department, Stanford University School of Medicine Stanford, California, USA January 4, Abstract This package provides methods to calculate flow cytometer s detection efficiency (Q) and background illumination (B) by analyzing LED pulses and multi-level bead sets. The method improves on previous formulations Wood (1998); Hoffman and Wood (2007); Chase and Hoffman (1998) by fitting a full quadratic model with appropriate weighting, and by providing standard errors and peak residuals as well as the fitted parameters themselves. K-means clustering is incorporated for automated peak detection of multi-peak data. Overall, this method is successful in providing good estimates of the Spe scales and backgrounds for all of the fluorescence channels on instruments with good linearity. Known photoelectron scales and measurement channel backgrounds make it possible to estimate the precision of measurements at different signal levels and the effects of compensated spectral overlap on measurement quality. 2 Introduction The basic measurement capabilities of a fluorescence cytometer measurement channel can be estimated by knowing just two values: Q, the statistical number of photoelectrons (Spe) generated per unit of dye in the sample, and B, the background light in dye equivalents that sets the minimum variance that underlies all measurements. From these the minimum detectable amount of dye and the precision of measurements at different signal levels can be calculated. Therefore, measurements of Q and B are the key to making valid comparisons between different instruments and among different channels on an instrument. * This project was supported by the Terry Fox Foundation, the Terry Fox Research Institute and the Canadian Cancer Society. Josef Spidlen is an ISAC Marylou Ingram Scholar. 1
2 We can achieve full specification of Q and B in two steps by evaluating photoelectron scales for the measurement channels of interest and relating those to measurements on dye reference samples. There are problems and challenges in implementing each step in a way that is convenient and accurate in routine use. Here we address the photoelectron scale aspect by providing reliable automated software for obtaining the needed information from LED or multi-level, multi-dye bead data. When a cell or particle moves through a sensing area on a flow cytometer, a focused laser beam excites fluorescent dyes that emit a pulse of light in all directions. The optical system collects some of this light and passes it through optical filters to the cathodes of one or more PMT detectors where a fraction of the photons generate photoelectrons. The photoelectrons are amplified through a series of dynodes to produce an output current pulse. The photoelectron production is a Poisson process, so the variance of signals at a particular level is proportional to the signal level itself. Since the amplification process through the dynodes introduces some additional variance, we introduce the term statistical photoelectrons or Spe to denote the effective Poisson number relating signal levels and their variances. Since it is difficult to measure photoelectron counts directly, the usual method for estimating photoelectron scales has been to measure uniform light signals at various levels and fit the measured means and variances to a statistical model involving the Poisson distribution expectations for the relation between them. Historically, two kinds of signal source have been used: an LED light source producing very uniform pulses at adjustable signal levels or microspheres (also called beads ) labeled with several different concentrations of a mixture of fluorescent dyes. The model can be expressed as a second-degree (quadratic) polynomial relating the observed signal means and variances to Q, B and, for microsphere samples, CV 0, a common non-statistical variability in the measurements of the different microsphere peaks. For measurements on a population of similar particles like one of the populations in a multilevel bead set, the observed variance should be the sum of the electronic noise, background light not associated with the particles, Poisson variance related to the signal levels of the particles, the variation in dye amount among the particles and illumination variations due to particles taking different flow paths through the laser beam. The electronic noise and background light combine into a variance contribution that is not dependent on the signal level. The Poisson variance is proportional to the signal level, and the dye and illumination variations combine to form CV 0 whose contribution to the variances goes with the square of the signal level. Therefore, the general equation for the observed variance can be expressed as V (M I ) = c 0 + c 1 M I + c 2 M 2 I where M I is the mean signal of the population in measurement intensity units, and V (M I ) is the observed variance of a population with mean M I. Methods implemented in this package fit sets of mean and variance data points with this equation to obtain the parameters c 0, c 1 and c 2. If we scale the signal levels in Spe, we can define B Spe as the effective background level in Spe and M Spe as the mean signal in Spe. Then, defining Q I as the Spe per intensity unit, as explained in our paper Parks et al. (2016), we can calculate B Spe = c 0 /c 2 1 Q I = 1/c 1 CV 2 0 = c 2 2
3 3 Requirements The flowqb package requires the flowcore library in order to read FCS files Spidlen et al. (2010), the stats package for its model fitting and also K-means clustering functionality, the extremevalues package to determine outliers, and it also makes use of some basic functions and classes from the methods and BiocGenerics packages. In addition, we suggest installing the flowqbdata package, which contains example data sets to demonstrate the functionality of this package. Alternatively, one can use the FlowRepositoryR package in order to work directly with data stored in FlowRepository. In addition, we recommend the xlsx package in order to parse house-keeping information from MS Excel spreadsheets. Finally, we use the RUnit package in order to implement unit tests assuring consistency and reproducibility of the flowqb functionality over time. Let s start by loading the flowqb package as well as the flowqbdata package with data to demonstrate the functionality of this package. > library("flowqb") > library("flowqbdata") 4 Fitting LED pulser data An LED light pulser is producing very uniform pulses at adjustable signal levels. White LEDs provide some signal at all visible wavelengths, but the far-red emission is weak. A given LED pulse level will generate quite different photoelectron signals on different detectors, so it is important to collect data over a wide range of LED levels to assure that the measurement series on each detector will include the low, middle and high level signals needed for optimal results in the fitting procedure. The fit_led function assumes that data generated by different LED levels are provided as separate FCS files. These files are passed to the fit_led function in the form of a vector of FCS file paths. In addition, house keeping details about the data and the way the fitting procedure should be performed need to be provided, resulting in the following list of arguments: ˆ fcs_file_path_list A vector of FCS file paths pointing to data generated by an LED pulser set to a range of LED levels; different levels generated different FCS files, all data coming from a single instrument. ˆ ignore_channels A vector of short channel names (values of the $PnN keywords) specifying channels that should not be considered for the fitting procedure. Normally, those should be all non-fluorescence channels, such as the time and the (forward and side) scatter channels. ˆ dyes A vector of dye names that you would normally use with the detectors specified below. This value does not affect the fitting, but those dyes will be highlighted in the provided results. ˆ detectors A vector of short channel names (values of the $PnN keywords) specifying channels matching to the dyes specified above. The length of this vector shall correspond to the length of the dyes vector. These channels should be all of the same type as specified by the signal_type below, i.e., area, or height of the measured signal. 3
4 ˆ signal_type The type of the signal specified as the area or height. This should match to the signal type that is being captured by the channels specified in the detectors argument. The signal type is being used in order to trigger type-specific peak validity checks. Currently, if signal type equals to height then peaks with a mean value lower than the lowest peak mean value are omitted from the fitting. In addition, peaks that are not sufficiently narrow (i.e., exceeding a specific maximum CV) are also omitted from the fitting. Currently, the maximum allowed CV is set to 0.65, but the code is designed to make this user-configurable and signal type dependent eventually. ˆ instrument_name The make/model of the instrument. The purpose if this argument is to allow for instrument-specific peak validity checks. At this point, if BD Accuri is passed as the instrument type, then peaks with a mean value lower than the lowest peak mean value are omitted from the fitting. Additional instrument-specific peak validity checks may be implemented in the future. ˆ bounds On some instruments, the lowest LED peaks may be cut off at a data baseline so that the peak statistics will not be valid. Therefore, peaks too close to the baseline need to be excluded from the fitting. Also, many instruments do not maintain good linearity to the full top of scale, so it is also important to specify a maximum level for good linearity and, on each fluorescence channel, exclude any peak that is above that maximum. The bounds argument shall provide a list specifying the minimum and maximum value for the means of valid peaks; peaks with means outsize of this range will be ignored for that particular channel. ˆ minimum_useful_peaks Different peaks may be omitted for different channels due to various validity checks described above. This argument specifies the minimal number of valid peaks required in order for the fitting procedure to be performed on a particular fluorescence channel. Generally, fitting the three quadratic parameters requires three valid points to obtain a fit at all, and 4 or more points are needed to obtain error estimates. Requiring higher values would exclude some of your data but likely produce better results. ˆ max_iterations The peaks have a wide range of variances, so unweighted least squares fitting is not appropriate, and we need to apply appropriate weights in the fitting procedure. In particular, the populations with lower variances get more weight since having the fit miss them by any particular amount is worse than missing a high variance population by the same amount. This argument specifies the maximum number of iterations for the iterative fitting approach with appropriate weight recalculations. In most cases, the fitting converges relatively fast. The iterating stops when either the maximum of iterations is used or if none of the coefficients of the model changed more than The default maximum of 10 iterations seems to be enough in most cases. You can also explore your results in order to see how many iterations were actually done for each of the all of the fitting. > ## Example is based on LED data from the flowqbdata package > fcs_directory <- system.file("extdata", "SSFF_LSRII", "LED_Series", + package="flowqbdata") 4
5 > fcs_file_path_list <- list.files(fcs_directory, "*.fcs", full.names= TRUE) > ## We are working with these FCS files: > basename(fcs_file_path_list) [1] " fcs" " fcs" " fcs" " fcs" " fcs" [6] " fcs" " fcs" " fcs" " fcs" " fcs" [11] " fcs" " fcs" " fcs" " fcs" " fcs" [16] " fcs" " fcs" " fcs" " fcs" > ## Various house keeping information > ## - Which channels should be ignored, typically the non-fluorescence > ## channels, such as the time and the scatter channels > ignore_channels <- c("time", + "FSC-A", "FSC-W", "FSC-H", + "SSC-A", "SSC-W", "SSC-H") > ## - Which dyes would you typically use with the detectors > dyes <- c("apc", "APC-Cy7", "APC-H7", "FITC", "PE", "PE-Cy7", "PerCP", + "PerCP-Cy55", "V450", "V500-C") > ## - What are the corresponding detectors, provide a vector of short channel > ## names, i.e., values of the $PnN FCS keywords. > detectors <- c("apc-a", "APC-Cy7-A", "APC-Cy7-A", "FITC-A", "PE-A", "PE-Cy7-A", + "PerCP-Cy5-5-A", "PerCP-Cy5-5-A", "Pacific Blue-A", "Aqua Amine-A") > ## - The signal type that you are looking at (Area or Height) > signal_type <- "Area" > ## - The instrument make/model > instrument_name <- 'LSRII' > ## - Set the minimum and maximum values, peaks with mean outsize of this range > ## will be ignored > bounds <- list(minimum = -100, maximum = ) > ## - The minimum number of usable peaks (represented by different FCS files > ## in case of an LED pulser) required in order for a fluorescence channel > ## to be included in the fitting. Peaks with mean expression outside of the > ## bounds specified above are omitted and therefore not considered useful. > ## Fitting the three quadratic parameters requires three valid points to obtain > ## a fit at all, and 4 or more points are needed to obtain error estimates. > minimum_fcs_files <- 3 > ## - What is the maximum number of iterations for iterative fitting with > ## weight adjustments > max_iterations <- 10 # The default 10 seems to be enough in typical cases > ## Now, let's calculate the fitting > led_results <- fit_led(fcs_file_path_list, ignore_channels, dyes, + detectors, signal_type, instrument_name, bounds = bounds, + minimum_useful_peaks = minimum_fcs_files, max_iterations = max_iterations) The results of the fit_led function is a list with the following components: 5
6 ˆ peak_stats A list with summary stats for each of the channels for all the different peaks (represented by different FCS files). For each of the channels, peak stats are captured by a data frame with rows corresponding to the different peaks (FCS files) and the following columns: N: the number of events in the peak M: the mean expression value for that peak SD: the standard deviation for that peak V: the variance for that peak (square of standard deviation) W: the weight of that peak Omit: was the peak omitted from the fitting? (TRUE or FALSE) QR: the residuals of the quadratic fitting LR: the residuals of the linear fitting QR-I: the residuals of the iterative quadratic fitting LR-I: the residuals of the iterative linear fitting The values of W, QR, LR, QR-I and LR-I will be NA if Omit is TRUE. > ## We have stats for these channels > names(led_results$peak_stats) [1] "FITC-A" "FITC-H" "PerCP-Cy5-5-A" "PerCP-Cy5-5-H" [5] "Pacific Blue-A" "Pacific Blue-H" "Aqua Amine-A" "Aqua Amine-H" [9] "Pacific Orange-A" "Pacific Orange-H" "QDot 585-A" "QDot 585-H" [13] "QDot 605-A" "QDot 605-H" "QDot 655-A" "QDot 655-H" [17] "QDot 705-A" "QDot 705-H" "QDot 800-A" "QDot 800-H" [21] "APC-A" "APC-H" "APC-Cy5-5-A" "APC-Cy5-5-H" [25] "APC-Cy7-A" "APC-Cy7-H" "PE-A" "PE-H" [29] "PE-Texas-Red-A" "PE-Texas-Red-H" "PE-Cy5-A" "PE-Cy5-H" [33] "PE-Cy5-5-A" "PE-Cy5-5-H" "PE-Cy7-A" "PE-Cy7-H" > ## Explore the peak stats for a randomly chosen channel (PE-Cy7-A) > ## Showing only the head in order to limit the output for the vignette > head(led_results$peak_stats$`pe-cy7-a`) N M SD V W Omit QR fcs FALSE fcs FALSE fcs FALSE fcs FALSE fcs FALSE fcs FALSE LR QR-I LR-I fcs
7 fcs fcs fcs fcs fcs ˆ bg_stats A data frame with background stats for each channel. These stats are a convenience way to look at peak stats of the lowest peak. The columns of the data frame correspond to the different channels, and there are the following rows: N: the number of events in the lowest peak M: the mean expression value for the lowest peak SD: the standard deviation for the lowest peak ˆ dye_bg_stats A data frame with background stats for each dye. These are the same stats as bg_stats described above, but the columns will only be restricted to the detectors/dyes that were listed as arguments of the fit_led call. The column headings are converted from channel names (detectors) to dyes as per the mapping supplied by the detectors and dyes arguments. The rows of the data frame are the same as with the bg_stats described above. > ## Explore bg_stats > led_results$bg_stats FITC-A FITC-H PerCP-Cy5-5-A PerCP-Cy5-5-H Pacific Blue-A N M SD Pacific Blue-H Aqua Amine-A Aqua Amine-H Pacific Orange-A Pacific Orange-H N M SD QDot 585-A QDot 585-H QDot 605-A QDot 605-H QDot 655-A N M SD QDot 655-H QDot 705-A QDot 705-H QDot 800-A QDot 800-H APC-A N M SD APC-H APC-Cy5-5-A APC-Cy5-5-H APC-Cy7-A APC-Cy7-H N M SD PE-A PE-H PE-Texas-Red-A PE-Texas-Red-H PE-Cy5-A N
8 M SD PE-Cy5-H PE-Cy5-5-A PE-Cy5-5-H PE-Cy7-A PE-Cy7-H N M SD > ## Explore dye_bg_stats > led_results$dye_bg_stats APC APC-Cy7 APC-H7 FITC PE N M SD PE-Cy7 PerCP PerCP-Cy55 V450 V500-C N M SD ˆ fits For the LED analysis, results are reported for both quadratic fitting and linear fitting (effectively fixing CV 0 = 0). Since the uniformity of LED signal outputs is likely to be better than the ability of the cytometer electronics to evaluate them, the c 2 term in a quadratic fit should be very close to zero with a small standard error. If the results of the quadratic fit are consistent with CV 0 = 0, we can rely on the linear fit results whose standard errors on c 1 will generally be smaller than the c 1 standard errors of the quadratic fit. The fits item contains a data frame with fits for each of the channels. The columns of the data frame correspond to the different channels. The rows of the data frame capture the coefficients of both, quadratic fitting and linear fitting as follows: c0: value of the c 0 coefficient of the quadratic fitting c0 SE: standard error of the c 0 coefficient of the quadratic fitting c0 P: the P-value for the c 0 coefficient of the quadratic fitting c1: value of the c 1 coefficient of the quadratic fitting c1 SE: standard error of the c 1 coefficient of the quadratic fitting c1 P: the P-value for the c 1 coefficient of the quadratic fitting c2: value of the c 2 coefficient of the quadratic fitting c2 SE: standard error of the c 2 coefficient of the quadratic fitting c2 P: the P-value for the c 2 coefficient of the quadratic fitting c0 : value of the c 0 coefficient of the linear fitting c0 SE: standard error of the c 0 coefficient of the linear fitting c0 P: the P-value for the c 0 coefficient of the linear fitting 8
9 c1 : value of the c 1 coefficient of the linear fitting c1 SE: standard error of the c 1 coefficient of the linear fitting c1 P: the P-value for the c 1 coefficient of the linear fitting ˆ dye_fits A data frame with fits for each dye. These are the same stats as fits described above, but the columns will only be restricted to the detectors/dyes that were listed as arguments of the fit_led call. The column headings are converted from channel names (detectors) to dyes as per the mapping supplied by the detectors and dyes arguments. The rows of the data frame are the same as with the fits described above. ˆ iterated_fits A data frame with fits for each of the channels in the same way as for the fits described above, but based on iterative fitting with weights adjustments. ˆ iterated_dye_fits A data frame with fits for each of the dyes in the same way as for the dye_fits described above, ut based on iterative fitting with weights adjustments. > ## Explore dye_fits > ## fits are the same rows but columns corresponding to all channels > led_results$dye_fits APC APC-Cy7 APC-H7 FITC PE c e e e e e+01 c0 SE e e e e e-01 c0 P e e e e e-24 c e e e e e+00 c1 SE e e e e e-03 c1 P e e e e e-24 c e e e e e-06 c2 SE e e e e e-07 c2 P e e e e e-05 c0' e e e e e+01 c0' SE e e e e e+00 c0' P e e e e e-22 c1' e e e e e+00 c1' SE e e e e e-02 c1' P e e e e e-24 PE-Cy7 PerCP PerCP-Cy55 V450 V500-C c e e e e e+02 c0 SE e e e e e+00 c0 P e e e e e-22 c e e e e e+00 c1 SE e e e e e-02 c1 P e e e e e-21 c e e e e e-06 c2 SE e e e e e-06 c2 P e e e e e-01 9
10 c0' e e e e e+02 c0' SE e e e e e+00 c0' P e e e e e-24 c1' e e e e e+00 c1' SE e e e e e-02 c1' P e e e e e-24 > ## Explore iterated_dye_fits > ## iterated_fits are the same rows but columns corresponding to all channels > led_results$iterated_dye_fits APC APC-Cy7 APC-H7 FITC PE c e e e e e+01 c0 SE e e e e e-01 c0 P e e e e e-24 c e e e e e+00 c1 SE e e e e e-03 c1 P e e e e e-24 c e e e e e-06 c2 SE e e e e e-07 c2 P e e e e e-05 c0' e e e e e+01 c0' SE e e e e e+00 c0' P e e e e e-22 c1' e e e e e+00 c1' SE e e e e e-02 c1' P e e e e e-24 PE-Cy7 PerCP PerCP-Cy55 V450 V500-C c e e e e e+02 c0 SE e e e e e+00 c0 P e e e e e-22 c e e e e e+00 c1 SE e e e e e-02 c1 P e e e e e-21 c e e e e e-06 c2 SE e e e e e-06 c2 P e e e e e-01 c0' e e e e e+02 c0' SE e e e e e+00 c0' P e e e e e-24 c1' e e e e e+00 c1' SE e e e e e-02 c1' P e e e e e-24 The Q, B and intrinsic CV 0 can be extracted from the fits using the equations shown in the introduction. For your convenience, this is implemented in the qb_from_fits 10
11 function as shown below in this vignette. ˆ iteration_numbers A data frame with rows corresponding to all the different channels and 2 columns, Q and L, showing the number of iterations used for the quadratic and linear fitting, resp. This data frame can be reviewed in order to make sure the fitting is converging fast enough and the max_iterations parameter is set large enough. > ## Explore iteration numbers > ## Showing only the head in order to limit the output for the vignette > head(led_results$iteration_numbers) Q L FITC-A 3 2 FITC-H 2 2 PerCP-Cy5-5-A 3 2 PerCP-Cy5-5-H 3 3 Pacific Blue-A 3 2 Pacific Blue-H Fitting bead data The fit_beads function performs quadratic fitting for multi-level, multi-dye bead sets. In addition, the fit_spherotech function performs fitting for the Sph8 particle sets from Spherotech, and the fit_thermo_fisher function performs fitting for the 6-level (TF6) Thermo Fisher set. Internally, this is the same fit_beads function except that the number of expected peaks is predefined to 8 and 6, resp. The parameters for the bead data fitting functions are similar to those required for the LED fitting. The main difference is that a single FCS file (supplied by the fcs_file_path argument) is expected because the bead sets are provided as a mixture of the different populations and therefore, acquiring data from a single sample will naturally result in all the peaks contained within a single FCS file. All the beads are expected to have the same (or very similar) light scatter properties. Therefore, we perform automated gating on the forward and side scatter channels in order to isolate the main population. In order to do that, the method requires a scatter_channels argument that specifies which 2 channels shall be used for the scatter gating. After the main population is isolated, we use K-means clustering to separate the expression peaks generated by different beads. The number of clusters is pre-defined as 8 for the fit_spherotech function, 6 for the fit_thermo_fisher function, and provided by the user in the form of the N_peaks argument in case of the fit_beads function. This clustering is performed on data transformed with the Logicle transformation Parks et al. (2006); Moore and Parks (2012). Generally, the Logicle width (w parameter) of 1.0 has been working well for all our data, but users can change the default by providing a different logicle_width value. The rest of the arguments is the same as with LED fitting; the complete list of arguments is a follows: ˆ fcs_file_path A character string specifying the file path to the FCS file with the acquired bead data. 11
12 ˆ scatter_channels A vector of 2 short channel names (values of the $PnN keywords) specifying the 2 channels that should not be used to gate the main bead population. The first channel should be a forward scatter channel, the second one should be a side scatter channel. ˆ ignore_channels A vector of short channel names (values of the $PnN keywords) specifying channels that should not be considered for the fitting procedure. Normally, those should be all non-fluorescence channels, such as the time and the (forward and side) scatter channels. ˆ N_peaks The number of peaks (different beads) to look for. This argument is applicable to the fit_beads function only; the fit_spherotech and fit_thermo_fisher functions have the number of peaks predefined to 8 and 6, resp. ˆ dyes A vector of dye names. This value does not affect the fitting, but those dyes will be highlighted in the provided results. ˆ detectors A vector of short channel names (values of the $PnN keywords) specifying channels matching to the dyes specified above. The length of this vector shall correspond to the length of the dyes vector. These channels should be all of the same type as specified by the signal_type below, i.e., area or height of the measured signal. ˆ signal_type The type of the signal specified as the area or height. This should match to the signal type that is being captured by the channels specified in the detectors argument. The signal type is being used in order to trigger type-specific peak validity checks. Currently, if signal type equals to height then peaks with a mean value lower than the lowest peak mean value are omitted from the fitting. In addition, peaks that are not sufficiently narrow (i.e., exceeding a specific maximum CV) are also omitted from the fitting. Currently, the maximum allowed CV is set to 0.65, but the code is designed to make this user-configurable and signal type dependent eventually. ˆ instrument_name The make/model of the instrument. The purpose if this argument is to allow for instrument-specific peak validity checks. At this point, if BD Accuri is passed as the instrument type, then peaks with a mean value lower than the lowest peak mean value are omitted from the fitting. Additional instrument-specific peak validity checks may be implemented in the future. ˆ bounds On some instruments, the lowest LED peaks may be cut off at a data baseline so that the peak statistics will not be valid. Therefore, peaks too close to the baseline need to be excluded from the fitting. Also, many instruments do not maintain good linearity to the full top of scale, so it is also important to specify a maximum level for good linearity and, on each fluorescence channel, exclude any peak that is above that maximum. The bounds argument shall provide a list specifying the minimum and maximum value for the means of valid peaks; peaks with means outsize of this range will be ignored for that particular channel. ˆ minimum_useful_peaks Different peaks may be omitted for different channels due to various validity checks described above. This argument specifies the minimal number of 12
13 valid peaks required in order for the fitting procedure to be performed on a particular fluorescence channel. ˆ max_iterations The maximum number of iterations for the iterative fitting approach with appropriate weight recalculations. ˆ logicle_width The data clustering part is performed on data transformed with the Logicle transformation Parks et al. (2006); Moore and Parks (2012). Generally, the Logicle width (w parameter) of 1.0 has been working well for all our data, but users can change the default by providing a different value. > ## Example of fitting bead data based on Sph8 particle sets from Spherotech > fcs_file_path <- system.file("extdata", "SSFF_LSRII", "Other_Tests", + " fcs", package="flowqbdata") > scatter_channels <- c("fsc-a", "SSC-A") > ## Depending on your hardware and input, this may take a few minutes, mainly > ## due to the required clustering stage of the algorithm. > spherotech_results <- fit_spherotech(fcs_file_path, scatter_channels, + ignore_channels, dyes, detectors, bounds, + signal_type, instrument_name, minimum_useful_peaks = 3, + max_iterations = max_iterations, logicle_width = 1.0) > ## This is the same as if we were running > ## fit_beads(fcs_file_path, scatter_channels, > ## ignore_channels, 8, dyes, detectors, bounds, > ## signal_type, instrument_name, minimum_useful_peaks = 3, > ## max_iterations = max_iterations, logicle_width = 1.0) Next, let s explore the results of this function. Unlike with the LED data, only the results of quadratic fitting are provided for fitting bead data by the fit_beads (and fit_spherotech, fit_thermo_fisher) functions. This is because linear fitting does not account for the intrinsic CV 0 of the beads and is not appropriate for bead data, which always has a significant non-linear component. The results of the fit_beads (and fit_spherotech, Rfunctionfit thermo fisher) functions is a list with the following components: ˆ transformed_data A flowcore s flowframe object capturing the data from the input FCS file after the Logicle transformation. This may be reviewed if one wanted to check that the Logicle transformation was appropriate for the input FCS file. ˆ peaks A list of flowcore s flowframe objects capturing the different peaks identified in the input FCS file by the K-means clustering algorithm. This may be reviewed if one wanted to check that the clustering algorithm identified the peaks correctly. ˆ peak_clusters The result of the kmeans call. This shows the assignment of cluster numbers to input data points and also provides additional details about the clustering; see the documentation for the kmeans function for additional details. 13
14 Note that you may see a warning message saying that Warning message: Quick-TRANSfer stage steps exceeded maximum (= ) According to kmeans documentation, in some cases, when some of the points (rows of x ) are extremely close, the algorithm may not converge in the Quick-Transfer stage, signaling a warning. Based on our experience, this has not negatively affected the result of the clustering. By default, we use the Hartigan and Wong algorithm. We tried other algorithms as well, but those were significantly slower. We also tried to round up the data as advised in the kmeans documentation, but that has not resolved the warning. Below, we demonstrate one way of a simple visual inspection of the clustering results. > library("flowcore") > plot( + exprs(spherotech_results$transformed_data[,"fitc-a"]), + exprs(spherotech_results$transformed_data[,"pacific Blue-A"]), + col=spherotech_results$peak_clusters$cluster, pch='.') Figure 1: Result of K-means clustering on bead data. ˆ peak_stats A list with summary stats for each of the channels for all the different peaks identified by the K-means clustering algorithm. For each of the channels, peak stats are captured by a data frame with rows corresponding to the different peaks and the following columns: 14
15 N: the number of events in the peak M: the mean expression value for that peak SD: the standard deviation for that peak V: the variance for that peak (square of standard deviation) W: the weight of that peak Omit: was the peak omitted from the fitting? (TRUE or FALSE) QR: the residuals of the quadratic fitting QR-I: the residuals of the iterative quadratic fitting The values of W, QR, and QR-I will be NA if Omit is TRUE. > ## We have stats for these channels > names(spherotech_results$peak_stats) [1] "FITC-A" "FITC-H" "PerCP-Cy5-5-A" "PerCP-Cy5-5-H" [5] "Pacific Blue-A" "Pacific Blue-H" "Aqua Amine-A" "Aqua Amine-H" [9] "Pacific Orange-A" "Pacific Orange-H" "QDot 585-A" "QDot 585-H" [13] "QDot 605-A" "QDot 605-H" "QDot 655-A" "QDot 655-H" [17] "QDot 705-A" "QDot 705-H" "QDot 800-A" "QDot 800-H" [21] "APC-A" "APC-H" "APC-Cy5-5-A" "APC-Cy5-5-H" [25] "APC-Cy7-A" "APC-Cy7-H" "PE-A" "PE-H" [29] "PE-Texas-Red-A" "PE-Texas-Red-H" "PE-Cy5-A" "PE-Cy5-H" [33] "PE-Cy5-5-A" "PE-Cy5-5-H" "PE-Cy7-A" "PE-Cy7-H" > ## Explore the peak stats for a randomly chosen channel (PE-Cy7-A) > spherotech_results$peak_stats$`pe-cy7-a` N M SD V W Omit QR e-01 FALSE e-02 FALSE e-02 FALSE e-03 FALSE e-04 FALSE e-05 FALSE e-06 FALSE e-08 FALSE QR-I
16 ˆ fits A a data frame with fits for each of the channels. The columns of the data frame correspond to the different channels. The rows of the data frame capture the coefficients of the quadratic fitting as follows: c0: value of the c 0 coefficient c0 SE: standard error of the c 0 coefficient c0 P: the P-value for the c 0 coefficient c1: value of the c 1 coefficient c1 SE: standard error of the c 1 coefficient c1 P: the P-value for the c 1 coefficient c2: value of the c 2 coefficient c2 SE: standard error of the c 2 coefficient c2 P: the P-value for the c 2 coefficient ˆ dye_fits A data frame with fits for each dye. These are the same stats as fits described above, but the columns will only be restricted to the detectors/dyes that were listed as arguments of the fit_beads (or fit_spherotech or Rfunctionfit thermo fisher) call. The column headings are converted from channel names (detectors) to dyes as per the mapping supplied by the detectors and dyes arguments. The rows of the data frame are the same as with the fits described above. ˆ iterated_fits A data frame with fits for each of the channels in the same way as for the fits described above, but based on iterative fitting with weights adjustments. ˆ iterated_dye_fits A data frame with fits for each of the dyes in the same way as for the dye_fits described above, ut based on iterative fitting with weights adjustments. > ## Explore fits > ## Selecting just a few columns to limit the output for the vignette > spherotech_results$fits[,c(1,3,5,7)] FITC-A PerCP-Cy5-5-A Pacific Blue-A Aqua Amine-A c e e e e+02 c0 SE e e e e+01 c0 P e e e e-04 c e e e e+00 c1 SE e e e e-01 c1 P e e e e-07 c e e e e-05 c2 SE e e e e-06 c2 P e e e e-04 > ## Explore iterated_fits > spherotech_results$iterated_fits[,c(1,3,5,7)] 16
17 FITC-A PerCP-Cy5-5-A Pacific Blue-A Aqua Amine-A c e e e e+02 c0 SE e e e e+01 c0 P e e e e-04 c e e e e+00 c1 SE e e e e-01 c1 P e e e e-06 c e e e e-05 c2 SE e e e e-06 c2 P e e e e-04 The Q, B and intrinsic CV 0 can be extracted from the fits using the equations shown in the introduction. For your convenience, this is implemented in the qb_from_fits function as shown below in this vignette. ˆ iteration_numbers A data frame with rows corresponding to all the different channels and a column, Q, showing the number of iterations used for the quadratic fitting. This data frame can be reviewed in order to make sure the fitting is converging fast enough and the max_iterations parameter is set large enough. > ## Explore iteration numbers > ## Showing only the head in order to limit the output for the vignette > head(spherotech_results$iteration_numbers) Q FITC-A 3 FITC-H 2 PerCP-Cy5-5-A 2 PerCP-Cy5-5-H 2 Pacific Blue-A 6 Pacific Blue-H 6 6 Calculating Q and B from fits The qb_from_fits function can be used to calculate Q and B using the equations shown in the introduction. As arguments, the qb_from_fits function can take results from both, LED data fitting (fit_led function) and bead data fitting (fit_beads, fit_spherotech function and fit_thermo_fisher functions). You can calculate based on any of the fits (fits, dye_fits, iterated_fits and iterated_dye_fits). > ## 1 QB from both quadratic and linear fits of LED data > qb_from_fits(led_results$iterated_dye_fits) APC APC-Cy7 APC-H7 FITC PE q_qi q_bspe
18 q_cv0sq e e e e e-06 l_qi l_bspe PE-Cy7 PerCP PerCP-Cy55 V450 V500-C q_qi q_bspe q_cv0sq e e e e e-06 l_qi l_bspe > ## 2 QB from quadratic fitting of bead data > qb_from_fits(spherotech_results$iterated_dye_fits) APC APC-Cy7 APC-H7 FITC PE q_qi q_bspe q_cv0sq PE-Cy7 PerCP PerCP-Cy55 V450 V500-C q_qi q_bspe q_cv0sq e e-05 The result of this function is a matrix with columns corresponding to the names of the fits used (e.g. dyes or detectors). In case of LED fits, the rows will contain the following items: ˆ q_qi The calculated Q I value of the quadratic fitting. ˆ q_bspe The calculated B Spe value of the quadratic fitting. ˆ q_cv0sq The calculated CV 2 0 value of the quadratic fitting. ˆ l_qi The calculated Q I value of the linear fitting. ˆ l_bspe The calculated B Spe value of the linear fitting. As expected, you can see that the estimated CV0 2 values for the LED data are very close to 0 and therefore, the linear model may be preferable. Note that the estimated value can also be small negative since; this is OK since those are just estimates from our model. The true CV0 2 should be very close to 0 unless there is something wrong with the instrument or LED data collection. In case of bead data fits, the rows will contain the following items: ˆ q_qi The calculated Q I value of the quadratic fitting. ˆ q_bspe The calculated B Spe value of the quadratic fitting. ˆ q_cv0sq The calculated CV 2 0 value of the quadratic fitting. 18
19 7 Parsing house-keeping information from spreadsheets We designed a simple MS Excel template in order to keep track of FCS files along with the necessary metadata to facilitate fully automated processing of bead and LED data generated by many different instruments. An example of this template is shown in the inst/extdata/140126_insteval_stanford_lsriia2.xlsx file of the flowqbdata package. This spreadsheet self-explanatory and captures details, such as the instrument name, location, data folder and file names, dyes or sample names, which channels are fluorescence based and which aren t. Below, we show an example of how the xlsx package can be used in order to process metadata from the spreadsheet and perform the fitting based on that. > ## Example of fitting based on house-keeping information in a spreadsheet > library(xlsx) > ## LED Fitting first > inst_xlsx_path <- system.file("extdata", + "140126_InstEval_Stanford_LSRIIA2.xlsx", package="flowqbdata") > xlsx <- read.xlsx(inst_xlsx_path, 1, headers=false, stringsasfactors=false) > ignore_channels_row <- 9 > ignore_channels <- vector() > i <- 1 > while(!is.na(xlsx[[i+4]][[ignore_channels_row]])) { + ignore_channels[[i]] <- xlsx[[i+4]][[ignore_channels_row]] + i <- i } > instrument_folder_row <- 9 > instrument_folder_col <- 2 > instrument_folder <- xlsx[[instrument_folder_col]][[instrument_folder_row]] > folder_column <- 18 > folder_row <- 14 > folder <- xlsx[[folder_column]][[folder_row]] > fcs_directory <- system.file("extdata", instrument_folder, + folder, package="flowqbdata") > fcs_file_path_list <- list.files(fcs_directory, "*.fcs", full.names= TRUE) > bounds_min_col <- 6 > bounds_min_row <- 7 > bounds_max_col <- 7 > bounds_max_row <- 7 > bounds <- list() > if (is.na(xlsx[[bounds_min_col]][[bounds_min_row]])) { + bounds$minimum < } else { + bounds$minimum <- as.numeric(xlsx[[bounds_min_col]][[bounds_min_row]]) + } > if (is.na(xlsx[[bounds_max_col]][[bounds_max_row]])) { + bounds$maximum < } else { 19
20 + bounds$maximum <- as.numeric(xlsx[[bounds_max_col]][[bounds_max_row]]) + } > signal_type_col <- 3 > signal_type_row <- 19 > signal_type <- xlsx[[signal_type_col]][[signal_type_row]] > instrument_name_col <- 2 > instrument_name_row <- 5 > instrument_name <- xlsx[[instrument_name_col]][[instrument_name_row]] > channel_cols <- 3:12 > dye_row <- 11 > detector_row <- 13 > dyes <- as.character(xlsx[dye_row,channel_cols]) > detectors <- as.character(xlsx[detector_row,channel_cols]) > ## Now we do the LED fitting > led_results <- fit_led(fcs_file_path_list, ignore_channels, dyes, + detectors, signal_type, instrument_name, bounds = bounds, + minimum_useful_peaks = 3, max_iterations = 10) > led_results$iterated_dye_fits APC APC-Cy7 APC-H7 FITC PE c e e e e e+01 c0 SE e e e e e-01 c0 P e e e e e-24 c e e e e e+00 c1 SE e e e e e-03 c1 P e e e e e-24 c e e e e e-06 c2 SE e e e e e-07 c2 P e e e e e-05 c0' e e e e e+01 c0' SE e e e e e+00 c0' P e e e e e-22 c1' e e e e e+00 c1' SE e e e e e-02 c1' P e e e e e-24 PE-Cy7 PerCP PerCP-Cy55 V450 V500-C c e e e e e+02 c0 SE e e e e e+00 c0 P e e e e e-22 c e e e e e+00 c1 SE e e e e e-02 c1 P e e e e e-21 c e e e e e-06 c2 SE e e e e e-06 c2 P e e e e e-01 20
21 c0' e e e e e+02 c0' SE e e e e e+00 c0' P e e e e e-24 c1' e e e e e+00 c1' SE e e e e e-02 c1' P e e e e e-24 > qb_from_fits(led_results$iterated_dye_fits) APC APC-Cy7 APC-H7 FITC PE q_qi q_bspe q_cv0sq e e e e e-06 l_qi l_bspe PE-Cy7 PerCP PerCP-Cy55 V450 V500-C q_qi q_bspe q_cv0sq e e e e e-06 l_qi l_bspe > ## Next we do the bead fitting; this example is with Thermo-fisher beads > folder_column <- 17 > folder <- xlsx[[folder_column]][[folder_row]] > filename <- xlsx[[folder_column]][[folder_row+1]] > fcs_file_path <- system.file("extdata", instrument_folder, folder, + filename, package="flowqbdata") > thermo_fisher_results <- fit_thermo_fisher(fcs_file_path, scatter_channels, + ignore_channels, dyes, detectors, bounds, + signal_type, instrument_name, minimum_useful_peaks = 3, + max_iterations = 10, logicle_width = 1.0) > ## The above is the same as this: > ## N_peaks <- 6 > ## thermo_fisher_results <- fit_beads(fcs_file_path, scatter_channels, > ## ignore_channels, N_peaks, dyes, detectors, bounds, > ## signal_type, instrument_name, minimum_useful_peaks = 3, > ## max_iterations = 10, logicle_width = 1.0) > > thermo_fisher_results$iterated_dye_fits APC APC-Cy7 APC-H7 FITC PE c e e e e e+02 c0 SE e e e e e+01 c0 P e e e e e-03 c e e e e e+00 21
Package flowqb. October 7, 2014
 Package flowqb October 7, 2014 Type Package Title Automated Quadratic Characterization of Flow Cytometer Instrument Sensitivity: Q, B and CVinstrinsic calculations. Version 1.6.0 Date 2011-11-15 Author
Package flowqb October 7, 2014 Type Package Title Automated Quadratic Characterization of Flow Cytometer Instrument Sensitivity: Q, B and CVinstrinsic calculations. Version 1.6.0 Date 2011-11-15 Author
FACScan 4 and 5 Color Upgrade Rainbow Users Guide
 FACScan 4 and 5 Color Upgrade Rainbow Users Guide For Research Use Only 1 Table of Contents Overview... 3 Hardware Specifications... 3 Rainbow Software... 4 Rainbow Preferences Window... 5 Setting up the
FACScan 4 and 5 Color Upgrade Rainbow Users Guide For Research Use Only 1 Table of Contents Overview... 3 Hardware Specifications... 3 Rainbow Software... 4 Rainbow Preferences Window... 5 Setting up the
Log into the computer using your CU Denver username and password. Log into the DIVA software using your CU and Username and the password flow
 Log into the computer using your CU Denver username and password Log into the DIVA software using your CU and Username and the password flow When DIVA opens, IF a CST Mismatch pop up box appears, then
Log into the computer using your CU Denver username and password Log into the DIVA software using your CU and Username and the password flow When DIVA opens, IF a CST Mismatch pop up box appears, then
Setup protocol for Firefly particles. On BD FACSCanto cytometers with single tube loader function
 Setup protocol for Firefly particles On BD FACSCanto cytometers with single tube loader function Version 1 Last Updated May 2016 Setup protocol for Firefly particles Contents Introduction Cytometer requirements
Setup protocol for Firefly particles On BD FACSCanto cytometers with single tube loader function Version 1 Last Updated May 2016 Setup protocol for Firefly particles Contents Introduction Cytometer requirements
Near UV Laser System Performance on the CytoFLEX S
 Near UV Laser System Performance on the CytoFLEX S APPLICATION NOTE Authors: Jeffrey Cobb James Tung Affiliation: Life Science, Beckman Coulter, Inc., Miami, FL, United States Introduction Material and
Near UV Laser System Performance on the CytoFLEX S APPLICATION NOTE Authors: Jeffrey Cobb James Tung Affiliation: Life Science, Beckman Coulter, Inc., Miami, FL, United States Introduction Material and
Setup protocol for Firefly particles. On BD LSR II and LSRFortessa cytometers with single tube loader function
 Setup protocol for Firefly particles On BD LSR II and LSRFortessa cytometers with single tube loader function Version 1 Last Updated May 2016 Setup protocol for Firefly particles Contents Introduction
Setup protocol for Firefly particles On BD LSR II and LSRFortessa cytometers with single tube loader function Version 1 Last Updated May 2016 Setup protocol for Firefly particles Contents Introduction
A Bright Cytometer Easy to use
 BriCyte E6 Flow Cytometer A Bright Cytometer Easy to use BriCyte E6 Flow Cytometer Flow cytometer is an essential tool to analyze cells, so it s used for diagnosis as well as research in laboratories worldwide.
BriCyte E6 Flow Cytometer A Bright Cytometer Easy to use BriCyte E6 Flow Cytometer Flow cytometer is an essential tool to analyze cells, so it s used for diagnosis as well as research in laboratories worldwide.
Spherotech, Inc Irma Lee Circle, Lake Forest, IL
 New flow cytometers, with fluorescent channels from the UV to Far Red, and corresponding fluorescent conjugates are now available. As a result, we have developed the Ultra Rainbow Fluorescent Particles
New flow cytometers, with fluorescent channels from the UV to Far Red, and corresponding fluorescent conjugates are now available. As a result, we have developed the Ultra Rainbow Fluorescent Particles
FACSLSRFortessa SORP QUICK REFERENCE GUIDE
 FACSLSRFortessa SORP QUICK REFERENCE GUIDE INSTRUMENT: 1. The computer is left on at all times. Note: If not Username: Administrator Password: BDIS 2. Unlock the screen with your PPMS account (UTSW username
FACSLSRFortessa SORP QUICK REFERENCE GUIDE INSTRUMENT: 1. The computer is left on at all times. Note: If not Username: Administrator Password: BDIS 2. Unlock the screen with your PPMS account (UTSW username
FloMax Software for Cytometry. Operating Manual Instrument Control and Acquisition. FloMax Operating Manual Instrument Control and Acquisition
 FloMax Software for Cytometry Operating Manual Instrument Control and Acquisition 1 Contents Introduction to FloMax Software Acquisition and Instrument Control...3 Operating Basics - Preparing FloMax for
FloMax Software for Cytometry Operating Manual Instrument Control and Acquisition 1 Contents Introduction to FloMax Software Acquisition and Instrument Control...3 Operating Basics - Preparing FloMax for
Domain, task, and dataset
 CS533C Project Proposal Visualization Tool for Flow Cytometry Project Evgeny Maksakov Department of Computer Science University of British Columbia maksakov@cs.ubc.ca The project will be carried out in
CS533C Project Proposal Visualization Tool for Flow Cytometry Project Evgeny Maksakov Department of Computer Science University of British Columbia maksakov@cs.ubc.ca The project will be carried out in
FACCalibur Users Guide
 FACCalibur Users Guide FACSCalibur Start Up Procedure If the Instrument is OFF: 1. Check the sheath and waste tanks. Open the fluidics drawer (front of instrument) and check the levels of sheath fluid
FACCalibur Users Guide FACSCalibur Start Up Procedure If the Instrument is OFF: 1. Check the sheath and waste tanks. Open the fluidics drawer (front of instrument) and check the levels of sheath fluid
Tutorial: FCAP Array Software with BD FACSArray Bioanalyzer
 Tutorial: FCAP Array Software with BD FACSArray Bioanalyzer After completing this tutorial you will be able to: Create an Experiment with the Experiment Wizard in FCAP Array software. Export the Experiment
Tutorial: FCAP Array Software with BD FACSArray Bioanalyzer After completing this tutorial you will be able to: Create an Experiment with the Experiment Wizard in FCAP Array software. Export the Experiment
CyFlow ML. Healthcare Immunology Pathology Microbiology Cell Biology. Multilaser 16 Parameter Desktop FCM System
 CyFlow ML Healthcare Immunology Pathology Microbiology Cell Biology Multilaser 16 Parameter Desktop FCM System 01 COMPANY Flow Cytometry made in Germany New sophisticated applications and increasing requirements
CyFlow ML Healthcare Immunology Pathology Microbiology Cell Biology Multilaser 16 Parameter Desktop FCM System 01 COMPANY Flow Cytometry made in Germany New sophisticated applications and increasing requirements
BD FACSDiVa Option. BD Biosciences Clontech Discovery Labware Immunocytometry Systems Pharmingen
 BD FACSDiVa Option The Digital Advantage for the BD FACSVantage SE Flow Cytometry System BD Biosciences Clontech Discovery Labware Immunocytometry Systems Pharmingen Proven Performance in Flow Cytometry
BD FACSDiVa Option The Digital Advantage for the BD FACSVantage SE Flow Cytometry System BD Biosciences Clontech Discovery Labware Immunocytometry Systems Pharmingen Proven Performance in Flow Cytometry
UNC Flow Cytometry Core Facility LSR II Operation 2018
 Reference: Startup BD FACSDiva 6 Online Course BD Training & elearning 1. Check Sheath and Waste containers: Empty Waste container in the sink Add bleach to the black mark ( ~10% final concentration).
Reference: Startup BD FACSDiva 6 Online Course BD Training & elearning 1. Check Sheath and Waste containers: Empty Waste container in the sink Add bleach to the black mark ( ~10% final concentration).
LEGENDplex Data Analysis Software Version 8 User Guide
 LEGENDplex Data Analysis Software Version 8 User Guide Introduction Welcome to the user s guide for Version 8 of the LEGENDplex data analysis software for Windows based computers 1. This tutorial will
LEGENDplex Data Analysis Software Version 8 User Guide Introduction Welcome to the user s guide for Version 8 of the LEGENDplex data analysis software for Windows based computers 1. This tutorial will
BD CellQuest Pro Acquisition Tutorial
 BD CellQuest Pro Acquisition Tutorial Introduction This tutorial guides you through a CellQuest Pro Acquisition run like the one demonstrated in the CellQuest Pro Acquisition Movie on the BD FACStation
BD CellQuest Pro Acquisition Tutorial Introduction This tutorial guides you through a CellQuest Pro Acquisition run like the one demonstrated in the CellQuest Pro Acquisition Movie on the BD FACStation
Offline Flow Cytometry Analysis Software: Importance of Scaling and Data Visualization
 WHITEPAPER Offline Flow Cytometry Analysis Software: Importance of Scaling and Data Visualization Objectives Learn about the flow cytometry open platform data structure and how it facilitates for robust
WHITEPAPER Offline Flow Cytometry Analysis Software: Importance of Scaling and Data Visualization Objectives Learn about the flow cytometry open platform data structure and how it facilitates for robust
FACSAria Fusion SORP QUICK REFERENCE GUIDE
 FACSAria Fusion SORP QUICK REFERENCE GUIDE INSTRUMENT: 1. The computer is left on at all times. Note: If not Username: Admin Password: BDIS 2. Ensure the flow cell access door is open. You will want to
FACSAria Fusion SORP QUICK REFERENCE GUIDE INSTRUMENT: 1. The computer is left on at all times. Note: If not Username: Admin Password: BDIS 2. Ensure the flow cell access door is open. You will want to
CELLQuest Acquisition Tutorial
 CELLQuest Acquisition Tutorial Introduction This tutorial guides you through a CELLQuest Acquisition run like the one demonstrated in the CELLQuest Acquisition Movie on the FACStation Overview CD-ROM.
CELLQuest Acquisition Tutorial Introduction This tutorial guides you through a CELLQuest Acquisition run like the one demonstrated in the CELLQuest Acquisition Movie on the FACStation Overview CD-ROM.
MERGE FILES TUTORIAL. Version 1.4
 MERGE FILES TUTORIAL Version 1.4 1. INTRODUCTION The merge process consists on adding two or more sample data files from the same sample or from different samples. The file resulting from the merging will
MERGE FILES TUTORIAL Version 1.4 1. INTRODUCTION The merge process consists on adding two or more sample data files from the same sample or from different samples. The file resulting from the merging will
To Plot a Graph in Origin. Example: Number of Counts from a Geiger- Müller Tube as a Function of Supply Voltage
 To Plot a Graph in Origin Example: Number of Counts from a Geiger- Müller Tube as a Function of Supply Voltage 1 Digression on Error Bars What entity do you use for the magnitude of the error bars? Standard
To Plot a Graph in Origin Example: Number of Counts from a Geiger- Müller Tube as a Function of Supply Voltage 1 Digression on Error Bars What entity do you use for the magnitude of the error bars? Standard
APS (Automatic Population Separator)
 Infinicyt provides a list of very powerful tools for research and diagnosis of haematological diseases through flow cytometry data analysis. The features that most distinguish Infinicyt software from other
Infinicyt provides a list of very powerful tools for research and diagnosis of haematological diseases through flow cytometry data analysis. The features that most distinguish Infinicyt software from other
Example how not to do it: JMP in a nutshell 1 HR, 17 Apr Subject Gender Condition Turn Reactiontime. A1 male filler
 JMP in a nutshell 1 HR, 17 Apr 2018 The software JMP Pro 14 is installed on the Macs of the Phonetics Institute. Private versions can be bought from
JMP in a nutshell 1 HR, 17 Apr 2018 The software JMP Pro 14 is installed on the Macs of the Phonetics Institute. Private versions can be bought from
DIAplex. DIAplex Pro 1.0 Software. Instructions for use. Fast Track Your Research. For research use only
 DIAplex DIAplex Pro 1.0 Software Instructions for use For research use only Fast Track Your Research Table of Contents 1. Introduction... 2 2. DIAplex Pro 1.0 installation... 2 2.1. System Requirements...
DIAplex DIAplex Pro 1.0 Software Instructions for use For research use only Fast Track Your Research Table of Contents 1. Introduction... 2 2. DIAplex Pro 1.0 installation... 2 2.1. System Requirements...
Sysmex CyFlow Cube Series
 Sysmex CyFlow Cube Series Reliable, Compact and Cost-effective Flow Cytometers For Research Use Only. Not for use in diagnostic or therapeutic procedures. Sysmex CyFlow Cube Series Compact, Economic Flow
Sysmex CyFlow Cube Series Reliable, Compact and Cost-effective Flow Cytometers For Research Use Only. Not for use in diagnostic or therapeutic procedures. Sysmex CyFlow Cube Series Compact, Economic Flow
Standard Operating Procedure for the Horiba FluroMax-4
 Standard Operating Procedure for the Horiba FluroMax-4 Adapted from Horiba Operations Manual Created by Michael Delcau, Modified by Brian Lamp The Fluoromax is capable of making a variety of measurements.
Standard Operating Procedure for the Horiba FluroMax-4 Adapted from Horiba Operations Manual Created by Michael Delcau, Modified by Brian Lamp The Fluoromax is capable of making a variety of measurements.
Customer ( you ) must review these notes prior to installing or operating the Attune NxT Software version ( Attune Software v1.1.0 ).
 Life Technologies Attune NxT Software version 1.1.0 Release Notes Customer ( you ) must review these notes prior to installing or operating the Attune NxT Software version 1.1.0 ( Attune Software v1.1.0
Life Technologies Attune NxT Software version 1.1.0 Release Notes Customer ( you ) must review these notes prior to installing or operating the Attune NxT Software version 1.1.0 ( Attune Software v1.1.0
6 th International Conference on Applied Informatics Eger, Hungary, January 27 31, MMA Technology. Robert Tornai
 6 th International Conference on Applied Informatics Eger, Hungary, January 27 31, 2004. MMA Technology Robert Tornai Department of Computer Graphics, University of Debrecen e-mail: rtornai@inf.unideb.hu
6 th International Conference on Applied Informatics Eger, Hungary, January 27 31, 2004. MMA Technology Robert Tornai Department of Computer Graphics, University of Debrecen e-mail: rtornai@inf.unideb.hu
BD CellQuest Pro Software Acquisition Tutorial
 BD CellQuest Pro Software Acquisition Tutorial This tutorial guides you through a typical acquisition using BD CellQuest Pro software. If you are already familiar with previous versions of BD CellQuest
BD CellQuest Pro Software Acquisition Tutorial This tutorial guides you through a typical acquisition using BD CellQuest Pro software. If you are already familiar with previous versions of BD CellQuest
Offline Flow Cytometry Analysis Software: Importance of Scaling and Data Visualization
 WHITEPAPER Offline Flow Cytometry Analysis Software: Importance of Scaling and Data Visualization Objectives Learn about the flow cytometry open platform data structure and how it facilitates for robust
WHITEPAPER Offline Flow Cytometry Analysis Software: Importance of Scaling and Data Visualization Objectives Learn about the flow cytometry open platform data structure and how it facilitates for robust
LSM510 Confocal Microscope Standard Operation Protocol Basic Operation
 LSM510 Confocal Microscope Standard Operation Protocol Basic Operation Please make sure that the COMPRESSED AIR has been TURNED ON prior to the use of the equipment. Kindly inform the administrator if
LSM510 Confocal Microscope Standard Operation Protocol Basic Operation Please make sure that the COMPRESSED AIR has been TURNED ON prior to the use of the equipment. Kindly inform the administrator if
6 peaks, 10 7 /ml. Figure 40 Histograms of the Ultra Rainbow
 Spherotech, Inc. 30 27845 Irma Lee Circle, Lake Forest, IL 60045 SPHERO TM Ultra Rainbow Calibration Particle Kits Consists of one bead with multiple fluorophores and intensities for calibration Ultra
Spherotech, Inc. 30 27845 Irma Lee Circle, Lake Forest, IL 60045 SPHERO TM Ultra Rainbow Calibration Particle Kits Consists of one bead with multiple fluorophores and intensities for calibration Ultra
BD Lyoplate Human Screen Analysis Instructions For analysis using FCS Express or FlowJo and heatmap representation in Excel 2007
 BD Biosciences Technical Resources Page 1 For use with the BD Lyoplate Human Cell Surface Marker Screening Panel (Cat. No. 560747). Please check that your catalog numbers for the FCS Express Excel templates
BD Biosciences Technical Resources Page 1 For use with the BD Lyoplate Human Cell Surface Marker Screening Panel (Cat. No. 560747). Please check that your catalog numbers for the FCS Express Excel templates
SOMfluor package tutorial
 SOMfluor package tutorial This tutorial serves as a guide for researchers wishing to implement Kohonen's selforganizing maps (SOM) on fluorescence data using Matlab. The following instructions and commands
SOMfluor package tutorial This tutorial serves as a guide for researchers wishing to implement Kohonen's selforganizing maps (SOM) on fluorescence data using Matlab. The following instructions and commands
FX500 Exchangeable Fluidics Cell Sorter
 FX5 Exchangeable Fluidics Cell Sorter Sony Biotechnology Inc. FX5 Cell Sorter Sorting Made Simple The FX5 is a microfluidics chip based cell sorter featuring a fully replaceable fluidics system. This lets
FX5 Exchangeable Fluidics Cell Sorter Sony Biotechnology Inc. FX5 Cell Sorter Sorting Made Simple The FX5 is a microfluidics chip based cell sorter featuring a fully replaceable fluidics system. This lets
Science is hard. Flow cytometry should be easy.
 Science is hard. Flow cytometry should be easy. CFlow User Guide TABLE OF CONTENTS 1 INTRODUCTION TO CFLOW... 1 1.1 Installing CFlow... 1 1.2 Starting CFlow... 1 1.3 CFlow Workspace... 2 1.4 Opening a
Science is hard. Flow cytometry should be easy. CFlow User Guide TABLE OF CONTENTS 1 INTRODUCTION TO CFLOW... 1 1.1 Installing CFlow... 1 1.2 Starting CFlow... 1 1.3 CFlow Workspace... 2 1.4 Opening a
North America/International Japan Europe
 North America/International Japan Europe 1730 North First Street San Jose, CA 95112 U.S.A. Voice: +1 800-275-5963 FAX: +1 408-352-4130 sales@sonybiotechnology.com http://www.sonybiotechnology.com 1-7-1,
North America/International Japan Europe 1730 North First Street San Jose, CA 95112 U.S.A. Voice: +1 800-275-5963 FAX: +1 408-352-4130 sales@sonybiotechnology.com http://www.sonybiotechnology.com 1-7-1,
flowworkspace: A Package for Importing flowjo Workspaces into R
 flowworkspace: A Package for Importing flowjo Workspaces into R Greg Finak February 24, 2014 1 Purpose The purpose of this package is to provide functionality to import relatively simple
flowworkspace: A Package for Importing flowjo Workspaces into R Greg Finak February 24, 2014 1 Purpose The purpose of this package is to provide functionality to import relatively simple
Supplementary text S6 Comparison studies on simulated data
 Supplementary text S Comparison studies on simulated data Peter Langfelder, Rui Luo, Michael C. Oldham, and Steve Horvath Corresponding author: shorvath@mednet.ucla.edu Overview In this document we illustrate
Supplementary text S Comparison studies on simulated data Peter Langfelder, Rui Luo, Michael C. Oldham, and Steve Horvath Corresponding author: shorvath@mednet.ucla.edu Overview In this document we illustrate
Phys 1020, Day 18: Questions? Cameras, Blmfld Reminders: Next Up: digital cameras finish Optics Note Final Project proposals next week!
 Lights. Action. Phys 1020, Day 18: Questions? Cameras, Blmfld 15.1 Reminders: Next Up: digital cameras finish Optics Note Final Project proposals next week! 1 What have we learned in this section: 1) Lasers
Lights. Action. Phys 1020, Day 18: Questions? Cameras, Blmfld 15.1 Reminders: Next Up: digital cameras finish Optics Note Final Project proposals next week! 1 What have we learned in this section: 1) Lasers
Data can be in the form of numbers, words, measurements, observations or even just descriptions of things.
 + What is Data? Data is a collection of facts. Data can be in the form of numbers, words, measurements, observations or even just descriptions of things. In most cases, data needs to be interpreted and
+ What is Data? Data is a collection of facts. Data can be in the form of numbers, words, measurements, observations or even just descriptions of things. In most cases, data needs to be interpreted and
cief Data Analysis Chapter Overview Chapter 12:
 page 285 Chapter 12: cief Data Analysis Chapter Overview Analysis Screen Overview Opening Run Files How Run Data is Displayed Viewing Run Data Data Notifications and Warnings Checking Your Results Group
page 285 Chapter 12: cief Data Analysis Chapter Overview Analysis Screen Overview Opening Run Files How Run Data is Displayed Viewing Run Data Data Notifications and Warnings Checking Your Results Group
Continuous Imaging Fluid Particle Analysis: A Primer
 Continuous Imaging Fluid Particle Analysis: A Primer Lew Brown Fluid Imaging Technologies Edgecomb, ME Introduction: Many scientific endeavors involve the use of particulate matter suspended within a liquid.
Continuous Imaging Fluid Particle Analysis: A Primer Lew Brown Fluid Imaging Technologies Edgecomb, ME Introduction: Many scientific endeavors involve the use of particulate matter suspended within a liquid.
Using Excel for Graphical Analysis of Data
 Using Excel for Graphical Analysis of Data Introduction In several upcoming labs, a primary goal will be to determine the mathematical relationship between two variable physical parameters. Graphs are
Using Excel for Graphical Analysis of Data Introduction In several upcoming labs, a primary goal will be to determine the mathematical relationship between two variable physical parameters. Graphs are
Unscrew and remove the cap from the waste tank
 Stanford Cytoflex HTS User Guide 11232015 Starting up 1. Check the sheath fluid level every time you use the cytometer. This ensures that you do not run out of sheath fluid during an experiment. Replenish
Stanford Cytoflex HTS User Guide 11232015 Starting up 1. Check the sheath fluid level every time you use the cytometer. This ensures that you do not run out of sheath fluid during an experiment. Replenish
flowchic - Analyze flow cytometric data of complex microbial communities based on histogram images
 flowchic - Analyze flow cytometric data of complex microbial communities based on histogram images Schumann, J., Koch, C., Fetzer, I., Müller, S. Helmholtz Centre for Environmental Research - UFZ Department
flowchic - Analyze flow cytometric data of complex microbial communities based on histogram images Schumann, J., Koch, C., Fetzer, I., Müller, S. Helmholtz Centre for Environmental Research - UFZ Department
Graphical Analysis of Data using Microsoft Excel [2016 Version]
![Graphical Analysis of Data using Microsoft Excel [2016 Version] Graphical Analysis of Data using Microsoft Excel [2016 Version]](/thumbs/72/67574169.jpg) Graphical Analysis of Data using Microsoft Excel [2016 Version] Introduction In several upcoming labs, a primary goal will be to determine the mathematical relationship between two variable physical parameters.
Graphical Analysis of Data using Microsoft Excel [2016 Version] Introduction In several upcoming labs, a primary goal will be to determine the mathematical relationship between two variable physical parameters.
A Further Study on Large Size LSO and LYSO Crystal Samples
 October 25, 2005 IEEE NSS, N12-6, Puerto-Rico 1 A Further Study on Large Size LSO and LYSO Crystal Samples Jianming Chen, Liyuan Zhang, Ren-yuan Zhu California Institute of Technology BGO, LSO & LYSO Samples
October 25, 2005 IEEE NSS, N12-6, Puerto-Rico 1 A Further Study on Large Size LSO and LYSO Crystal Samples Jianming Chen, Liyuan Zhang, Ren-yuan Zhu California Institute of Technology BGO, LSO & LYSO Samples
IQC monitoring in laboratory networks
 IQC for Networked Analysers Background and instructions for use IQC monitoring in laboratory networks Modern Laboratories continue to produce large quantities of internal quality control data (IQC) despite
IQC for Networked Analysers Background and instructions for use IQC monitoring in laboratory networks Modern Laboratories continue to produce large quantities of internal quality control data (IQC) despite
Package flowdensity. June 28, 2018
 Type Package Title Sequential Flow Cytometry Data Gating Version 1.15.0 Date 2014-10-14 Author Mehrnoush Malek,M. Jafar Taghiyar Package flowdensity June 28, 2018 Maintainer Mehrnoush Malek
Type Package Title Sequential Flow Cytometry Data Gating Version 1.15.0 Date 2014-10-14 Author Mehrnoush Malek,M. Jafar Taghiyar Package flowdensity June 28, 2018 Maintainer Mehrnoush Malek
User Operation Manual
 User Operation Manual Manual Version: 1.3 For Single Cell Bioanalyzer (SCB-402) Doc# 102-SCB-402-1.3 Table of Contents 1. General Information... 1 1.1 How to Use This Manual... 1 1.2 Operation Safety...
User Operation Manual Manual Version: 1.3 For Single Cell Bioanalyzer (SCB-402) Doc# 102-SCB-402-1.3 Table of Contents 1. General Information... 1 1.1 How to Use This Manual... 1 1.2 Operation Safety...
EPSRC Centre for Doctoral Training in Industrially Focused Mathematical Modelling
 EPSRC Centre for Doctoral Training in Industrially Focused Mathematical Modelling More Accurate Optical Measurements of the Cornea Raquel González Fariña Contents 1. Introduction... 2 Background... 2 2.
EPSRC Centre for Doctoral Training in Industrially Focused Mathematical Modelling More Accurate Optical Measurements of the Cornea Raquel González Fariña Contents 1. Introduction... 2 Background... 2 2.
FLUOSTAR Rhodamine B- encapsulating microspheres are seeding particles optimized for Particle Image Velocimetry. Technical handbook ver.
 www.ebm.vc FLUOSTAR Rhodamine B- encapsulating microspheres are seeding particles optimized for Particle Image Velocimetry Technical handbook ver.1, Feb, 2010 CONTENTS 1) Introduction of fluorescent PIV
www.ebm.vc FLUOSTAR Rhodamine B- encapsulating microspheres are seeding particles optimized for Particle Image Velocimetry Technical handbook ver.1, Feb, 2010 CONTENTS 1) Introduction of fluorescent PIV
UNDERSTANDING CALCULATION LEVEL AND ITERATIVE DECONVOLUTION
 UNDERSTANDING CALCULATION LEVEL AND ITERATIVE DECONVOLUTION Laser diffraction particle size analyzers use advanced mathematical algorithms to convert a measured scattered light intensity distribution into
UNDERSTANDING CALCULATION LEVEL AND ITERATIVE DECONVOLUTION Laser diffraction particle size analyzers use advanced mathematical algorithms to convert a measured scattered light intensity distribution into
Comparing technical specification documents
 Comparing technical specification documents Introduction Flow cytometer manufacturers provide technical specification sheets (tech specs or spec sheets) that describe the instruments key performance characteristics,
Comparing technical specification documents Introduction Flow cytometer manufacturers provide technical specification sheets (tech specs or spec sheets) that describe the instruments key performance characteristics,
Technical Note. Display of BD Accuri C6 Software- Generated FCS 3.0 Files Using FlowJo for PC and Mac v7.6. Introduction
 Display of BD Accuri C6 Software- Generated FCS 3.0 Files Using FlowJo for PC and Mac v7.6 Contents 1 Introduction 2 Before You Begin 3 Using Global FlowJo Preferences 5 Usinig the Transform Button 5 Saving
Display of BD Accuri C6 Software- Generated FCS 3.0 Files Using FlowJo for PC and Mac v7.6 Contents 1 Introduction 2 Before You Begin 3 Using Global FlowJo Preferences 5 Usinig the Transform Button 5 Saving
Data acquisition and analysis of MACSPlex Cytokine Kits using the MACSQuant Analyzer Express Modes in MACSQuantify Software version 2.
 Data acquisition and analysis of MACSPlex Cytokine Kits using the MACSQuant Analyzer Express Modes in MACSQuantify Software version 2.6 Contents 1. Description 1.1 Background information 1.2 Reagent and
Data acquisition and analysis of MACSPlex Cytokine Kits using the MACSQuant Analyzer Express Modes in MACSQuantify Software version 2.6 Contents 1. Description 1.1 Background information 1.2 Reagent and
Data acquisition and analysis of MACSPlex Exosome Kit using the MACSQuant Analyzer Express Mode in MACSQuantify Software version 2.
 Data acquisition and analysis of MACSPlex Exosome Kit using the MACSQuant Analyzer Express Mode in MACSQuantify Software version 2.8 Contents 1. Description 1.1 Background information 1.2 Reagent and instrument
Data acquisition and analysis of MACSPlex Exosome Kit using the MACSQuant Analyzer Express Mode in MACSQuantify Software version 2.8 Contents 1. Description 1.1 Background information 1.2 Reagent and instrument
STAT 311 (3 CREDITS) VARIANCE AND REGRESSION ANALYSIS ELECTIVE: ALL STUDENTS. CONTENT Introduction to Computer application of variance and regression
 STAT 311 (3 CREDITS) VARIANCE AND REGRESSION ANALYSIS ELECTIVE: ALL STUDENTS. CONTENT Introduction to Computer application of variance and regression analysis. Analysis of Variance: one way classification,
STAT 311 (3 CREDITS) VARIANCE AND REGRESSION ANALYSIS ELECTIVE: ALL STUDENTS. CONTENT Introduction to Computer application of variance and regression analysis. Analysis of Variance: one way classification,
Multivariate Calibration Quick Guide
 Last Updated: 06.06.2007 Table Of Contents 1. HOW TO CREATE CALIBRATION MODELS...1 1.1. Introduction into Multivariate Calibration Modelling... 1 1.1.1. Preparing Data... 1 1.2. Step 1: Calibration Wizard
Last Updated: 06.06.2007 Table Of Contents 1. HOW TO CREATE CALIBRATION MODELS...1 1.1. Introduction into Multivariate Calibration Modelling... 1 1.1.1. Preparing Data... 1 1.2. Step 1: Calibration Wizard
Olink NPX Manager. User Guide
 Olink NPX Manager User Guide Table of contents INTRODUCTION 3 Process 3 Limitations 3 Requirements for analysis 3 List of abbreviations 3 TO CREATE A HEAT MAP CT CSV FILE 4 OLINK NPX MANAGER STEP BY STEP
Olink NPX Manager User Guide Table of contents INTRODUCTION 3 Process 3 Limitations 3 Requirements for analysis 3 List of abbreviations 3 TO CREATE A HEAT MAP CT CSV FILE 4 OLINK NPX MANAGER STEP BY STEP
MA900 Multi-Application Cell Sorter
 MA9 Multi-Application Cell Sorter Sony Biotechnology Inc. MA9 Cell Sorter Sorting Made Simple The MA9 from Sony meets the needs of most sorting applications, supporting 12 fluorescence parameters and 4-way
MA9 Multi-Application Cell Sorter Sony Biotechnology Inc. MA9 Cell Sorter Sorting Made Simple The MA9 from Sony meets the needs of most sorting applications, supporting 12 fluorescence parameters and 4-way
C101-E137 TALK LETTER. Vol. 14
 C101-E137 TALK LETTER Vol. 14 Diffuse Reflectance Measurement of Powder Samples and Kubelka-Munk Transformation ------- 02 Application: Measuring Food Color ------- 08 Q&A: What effect does the scan speed
C101-E137 TALK LETTER Vol. 14 Diffuse Reflectance Measurement of Powder Samples and Kubelka-Munk Transformation ------- 02 Application: Measuring Food Color ------- 08 Q&A: What effect does the scan speed
Release Notes Life Technologies Attune NxT Software v2.3
 Release Notes Life Technologies Attune NxT Software v2.3 In the following pages you will find instructions describing: New software features Known software/system issues with troubleshooting guidance Software
Release Notes Life Technologies Attune NxT Software v2.3 In the following pages you will find instructions describing: New software features Known software/system issues with troubleshooting guidance Software
A Silicon Graphics CRT monitor was characterized so that multispectral images could be
 A Joint Research Program of The National Gallery of Art, Washington The Museum of Modern Art, New York Rochester Institute of Technology Technical Report April, 2002 Colorimetric Characterization of a
A Joint Research Program of The National Gallery of Art, Washington The Museum of Modern Art, New York Rochester Institute of Technology Technical Report April, 2002 Colorimetric Characterization of a
Tutorial 7: Automated Peak Picking in Skyline
 Tutorial 7: Automated Peak Picking in Skyline Skyline now supports the ability to create custom advanced peak picking and scoring models for both selected reaction monitoring (SRM) and data-independent
Tutorial 7: Automated Peak Picking in Skyline Skyline now supports the ability to create custom advanced peak picking and scoring models for both selected reaction monitoring (SRM) and data-independent
2014 Stat-Ease, Inc. All Rights Reserved.
 What s New in Design-Expert version 9 Factorial split plots (Two-Level, Multilevel, Optimal) Definitive Screening and Single Factor designs Journal Feature Design layout Graph Columns Design Evaluation
What s New in Design-Expert version 9 Factorial split plots (Two-Level, Multilevel, Optimal) Definitive Screening and Single Factor designs Journal Feature Design layout Graph Columns Design Evaluation
A Multiple-Line Fitting Algorithm Without Initialization Yan Guo
 A Multiple-Line Fitting Algorithm Without Initialization Yan Guo Abstract: The commonest way to fit multiple lines is to use methods incorporate the EM algorithm. However, the EM algorithm dose not guarantee
A Multiple-Line Fitting Algorithm Without Initialization Yan Guo Abstract: The commonest way to fit multiple lines is to use methods incorporate the EM algorithm. However, the EM algorithm dose not guarantee
Experiment 8 Wave Optics
 Physics 263 Experiment 8 Wave Optics In this laboratory, we will perform two experiments on wave optics. 1 Double Slit Interference In two-slit interference, light falls on an opaque screen with two closely
Physics 263 Experiment 8 Wave Optics In this laboratory, we will perform two experiments on wave optics. 1 Double Slit Interference In two-slit interference, light falls on an opaque screen with two closely
Curve Correction in Atomic Absorption
 Curve Correction in Atomic Absorption Application Note Atomic Absorption Authors B. E. Limbek C. J. Rowe Introduction The Atomic Absorption technique ultimately produces an output measured in optical units
Curve Correction in Atomic Absorption Application Note Atomic Absorption Authors B. E. Limbek C. J. Rowe Introduction The Atomic Absorption technique ultimately produces an output measured in optical units
Release Notes Life Technologies Attune NxT Software v2.2
 Release Notes Life Technologies Attune NxT Software v2.2 In the following pages you will find instructions describing: New software features Known software/system issues with troubleshooting guidance Software
Release Notes Life Technologies Attune NxT Software v2.2 In the following pages you will find instructions describing: New software features Known software/system issues with troubleshooting guidance Software
Cytometry Data Analysis in FlowJo V10. Timothy Quinn Crawford, PhD Application Scientist FlowJo, LLC
 Cytometry Data Analysis in FlowJo V10 Timothy Quinn Crawford, PhD Application Scientist FlowJo, LLC timc@flowjo.com Outline Part I Intro to FlowJo Navigating the V10 Workspace Customizing Ribbons Demo
Cytometry Data Analysis in FlowJo V10 Timothy Quinn Crawford, PhD Application Scientist FlowJo, LLC timc@flowjo.com Outline Part I Intro to FlowJo Navigating the V10 Workspace Customizing Ribbons Demo
LSRFortessa Guide. Cell Evaluation and Therapy Core HO324
 LSRFortessa Guide Cell Evaluation and Therapy Core HO324 Before Use: Prime the instrument twice with no tube and arm to the side Ensure that sheath is full and waste is empty To Begin: Log into ilabs tracker
LSRFortessa Guide Cell Evaluation and Therapy Core HO324 Before Use: Prime the instrument twice with no tube and arm to the side Ensure that sheath is full and waste is empty To Begin: Log into ilabs tracker
Thermo Xcalibur Getting Started (Quantitative Analysis)
 Thermo Xcalibur Getting Started (Quantitative Analysis) XCALI-97207 Revision B September 2010 2010 Thermo Fisher Scientific Inc. All rights reserved. Xcalibur, Surveyor, and Accela are registered trademarks
Thermo Xcalibur Getting Started (Quantitative Analysis) XCALI-97207 Revision B September 2010 2010 Thermo Fisher Scientific Inc. All rights reserved. Xcalibur, Surveyor, and Accela are registered trademarks
USER GUIDE TO BECKMAN COULTER FC500
 USER GUIDE TO BECKMAN COULTER FC500 Create a New Protocol Purpose: This procedure describes how to define the parameters and create rough analysis plots for a new experimental set-up. Some protocol templates
USER GUIDE TO BECKMAN COULTER FC500 Create a New Protocol Purpose: This procedure describes how to define the parameters and create rough analysis plots for a new experimental set-up. Some protocol templates
Standardized Tests: Best Practices for the TI-Nspire CX
 The role of TI technology in the classroom is intended to enhance student learning and deepen understanding. However, efficient and effective use of graphing calculator technology on high stakes tests
The role of TI technology in the classroom is intended to enhance student learning and deepen understanding. However, efficient and effective use of graphing calculator technology on high stakes tests
Hacking FlowJo VX. 42 Time-Saving FlowJo Shortcuts To Help You Get Your Data Published No Matter What Flow Cytometer It Came From
 Hacking FlowJo VX 42 Time-Saving FlowJo Shortcuts To Help You Get Your Data Published No Matter What Flow Cytometer It Came From Contents 1. Change the default name of your files. 2. Edit your workspace
Hacking FlowJo VX 42 Time-Saving FlowJo Shortcuts To Help You Get Your Data Published No Matter What Flow Cytometer It Came From Contents 1. Change the default name of your files. 2. Edit your workspace
Raster Image Correlation Spectroscopy and Number and Brightness Analysis
 Raster Image Correlation Spectroscopy and Number and Brightness Analysis 15 Principles of Fluorescence Course Paolo Annibale, PhD On behalf of Dr. Enrico Gratton lfd Raster Image Correlation Spectroscopy
Raster Image Correlation Spectroscopy and Number and Brightness Analysis 15 Principles of Fluorescence Course Paolo Annibale, PhD On behalf of Dr. Enrico Gratton lfd Raster Image Correlation Spectroscopy
Alternative Filters (all non-standard)
 Fluorochrome Excitation (nm) Emission (nm) Best Filter (available) LP Alternative Filters (all non-standard) Notes 7-AAD 488;546 647 710/50;670/30 685;635 670/30 BP with 635LP* NOTE: very high spectral
Fluorochrome Excitation (nm) Emission (nm) Best Filter (available) LP Alternative Filters (all non-standard) Notes 7-AAD 488;546 647 710/50;670/30 685;635 670/30 BP with 635LP* NOTE: very high spectral
Using Excel for Graphical Analysis of Data
 EXERCISE Using Excel for Graphical Analysis of Data Introduction In several upcoming experiments, a primary goal will be to determine the mathematical relationship between two variable physical parameters.
EXERCISE Using Excel for Graphical Analysis of Data Introduction In several upcoming experiments, a primary goal will be to determine the mathematical relationship between two variable physical parameters.
Merging Mixture Components for Cell Population Identification in Flow Cytometry Data The flowmerge package
 Merging Mixture Components for Cell Population Identification in Flow Cytometry Data The flowmerge package Greg Finak, Raphael Gottardo October 30, 2017 greg.finak@ircm.qc.ca, raphael.gottardo@ircm.qc.ca
Merging Mixture Components for Cell Population Identification in Flow Cytometry Data The flowmerge package Greg Finak, Raphael Gottardo October 30, 2017 greg.finak@ircm.qc.ca, raphael.gottardo@ircm.qc.ca
SPECTRAL IMAGING VIEWER SOFTWARE MANUAL
 103 Quality Circle, Suite 215 Huntsville, AL 35806 Phone: (256) 704-3332 Fax: (256) 971-2073 E-Mail: info@axometrics.com Website: http://www.axometrics.com SPECTRAL IMAGING VIEWER SOFTWARE MANUAL 2012
103 Quality Circle, Suite 215 Huntsville, AL 35806 Phone: (256) 704-3332 Fax: (256) 971-2073 E-Mail: info@axometrics.com Website: http://www.axometrics.com SPECTRAL IMAGING VIEWER SOFTWARE MANUAL 2012
Using LoggerPro. Nothing is more terrible than to see ignorance in action. J. W. Goethe ( )
 Using LoggerPro Nothing is more terrible than to see ignorance in action. J. W. Goethe (1749-1832) LoggerPro is a general-purpose program for acquiring, graphing and analyzing data. It can accept input
Using LoggerPro Nothing is more terrible than to see ignorance in action. J. W. Goethe (1749-1832) LoggerPro is a general-purpose program for acquiring, graphing and analyzing data. It can accept input
CyAn ADP Guide. Starting Up
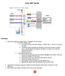 CyAn ADP Guide Starting Up 1. Check the sheath and waste fluids, if needed, fill and empty: 1. To fill the Sheath Tank: a. Unscrew the top of the tank labeled Sheath Tank, and set it on top of the tank.
CyAn ADP Guide Starting Up 1. Check the sheath and waste fluids, if needed, fill and empty: 1. To fill the Sheath Tank: a. Unscrew the top of the tank labeled Sheath Tank, and set it on top of the tank.
Engineered Diffusers Intensity vs Irradiance
 Engineered Diffusers Intensity vs Irradiance Engineered Diffusers are specified by their divergence angle and intensity profile. The divergence angle usually is given as the width of the intensity distribution
Engineered Diffusers Intensity vs Irradiance Engineered Diffusers are specified by their divergence angle and intensity profile. The divergence angle usually is given as the width of the intensity distribution
ChromQuest 5.0 Quick Reference Guide
 ChromQuest 5.0 Quick Reference Guide This guide contains an overview of the ChromQuest chromatography data system, with topics organized by workflow. For more information, refer to the ChromQuest User
ChromQuest 5.0 Quick Reference Guide This guide contains an overview of the ChromQuest chromatography data system, with topics organized by workflow. For more information, refer to the ChromQuest User
BD Multiwell AutoSampler Additional Features Tutorial
 BD Multiwell AutoSampler Additional Features Tutorial Introduction This tutorial provides step-by-step instructions on how to use the additional features available in BD Multiwell Plate Manager (MPM) software
BD Multiwell AutoSampler Additional Features Tutorial Introduction This tutorial provides step-by-step instructions on how to use the additional features available in BD Multiwell Plate Manager (MPM) software
Lab 9. Julia Janicki. Introduction
 Lab 9 Julia Janicki Introduction My goal for this project is to map a general land cover in the area of Alexandria in Egypt using supervised classification, specifically the Maximum Likelihood and Support
Lab 9 Julia Janicki Introduction My goal for this project is to map a general land cover in the area of Alexandria in Egypt using supervised classification, specifically the Maximum Likelihood and Support
Package GateFinder. March 17, 2019
 Type Package Package GateFinder March 17, 2019 Title Projection-based Gating Strategy Optimization for Flow and Mass Cytometry Version 1.3.2 Date 2018-03-28 Author Nima Aghaeepour ,
Type Package Package GateFinder March 17, 2019 Title Projection-based Gating Strategy Optimization for Flow and Mass Cytometry Version 1.3.2 Date 2018-03-28 Author Nima Aghaeepour ,
LIFA KEY FEATURES APPLICATIONS. Fluorescence Lifetime Attachment LIFA 15001A02 16/03/2015
 LIFA Fluorescence Lifetime Attachment LIFA 151A2 16/3/215 The LIFA is a dedicated system for Fluorescence Lifetime Imaging Microscopy (FLIM). It allows the generation of lifetime images on any widefield
LIFA Fluorescence Lifetime Attachment LIFA 151A2 16/3/215 The LIFA is a dedicated system for Fluorescence Lifetime Imaging Microscopy (FLIM). It allows the generation of lifetime images on any widefield
Chapter 2 Describing, Exploring, and Comparing Data
 Slide 1 Chapter 2 Describing, Exploring, and Comparing Data Slide 2 2-1 Overview 2-2 Frequency Distributions 2-3 Visualizing Data 2-4 Measures of Center 2-5 Measures of Variation 2-6 Measures of Relative
Slide 1 Chapter 2 Describing, Exploring, and Comparing Data Slide 2 2-1 Overview 2-2 Frequency Distributions 2-3 Visualizing Data 2-4 Measures of Center 2-5 Measures of Variation 2-6 Measures of Relative
NUMERICAL METHODS PERFORMANCE OPTIMIZATION IN ELECTROLYTES PROPERTIES MODELING
 NUMERICAL METHODS PERFORMANCE OPTIMIZATION IN ELECTROLYTES PROPERTIES MODELING Dmitry Potapov National Research Nuclear University MEPHI, Russia, Moscow, Kashirskoe Highway, The European Laboratory for
NUMERICAL METHODS PERFORMANCE OPTIMIZATION IN ELECTROLYTES PROPERTIES MODELING Dmitry Potapov National Research Nuclear University MEPHI, Russia, Moscow, Kashirskoe Highway, The European Laboratory for
LSM 5 MP, LSM 510 and LSM 510 META Laser Scanning Microscopes
 LSM 5 MP, LSM 510 and LSM 510 META Laser Scanning Microscopes Brief Operating Manual Release 4.2 January 2007 Contents Page Starting the System...3 Setting the microscope...6 Configuring the beam path
LSM 5 MP, LSM 510 and LSM 510 META Laser Scanning Microscopes Brief Operating Manual Release 4.2 January 2007 Contents Page Starting the System...3 Setting the microscope...6 Configuring the beam path
Operating instructions - Cary 100 Bio UV-visible Spectrophotometer
 Operating instructions - Cary 100 Bio UV-visible Spectrophotometer This document describes how to power up the spectrophotometer, set its measurement parameters, insert one or more of samples for analysis
Operating instructions - Cary 100 Bio UV-visible Spectrophotometer This document describes how to power up the spectrophotometer, set its measurement parameters, insert one or more of samples for analysis
Exercise: Graphing and Least Squares Fitting in Quattro Pro
 Chapter 5 Exercise: Graphing and Least Squares Fitting in Quattro Pro 5.1 Purpose The purpose of this experiment is to become familiar with using Quattro Pro to produce graphs and analyze graphical data.
Chapter 5 Exercise: Graphing and Least Squares Fitting in Quattro Pro 5.1 Purpose The purpose of this experiment is to become familiar with using Quattro Pro to produce graphs and analyze graphical data.
Package flowtype. R topics documented: November 27, Type Package Title Phenotyping Flow Cytometry Assays Version 2.16.
 Type Package Title Phenotyping Flow Cytometry Assays Version 2.16.0 Date 2014-11-26 Package flowtype November 27, 2017 Author Nima Aghaeepour, Kieran O'Neill, Adrin Jalali Maintainer Nima Aghaeepour
Type Package Title Phenotyping Flow Cytometry Assays Version 2.16.0 Date 2014-11-26 Package flowtype November 27, 2017 Author Nima Aghaeepour, Kieran O'Neill, Adrin Jalali Maintainer Nima Aghaeepour
Prepare a stem-and-leaf graph for the following data. In your final display, you should arrange the leaves for each stem in increasing order.
 Chapter 2 2.1 Descriptive Statistics A stem-and-leaf graph, also called a stemplot, allows for a nice overview of quantitative data without losing information on individual observations. It can be a good
Chapter 2 2.1 Descriptive Statistics A stem-and-leaf graph, also called a stemplot, allows for a nice overview of quantitative data without losing information on individual observations. It can be a good
/ Computational Genomics. Normalization
 10-810 /02-710 Computational Genomics Normalization Genes and Gene Expression Technology Display of Expression Information Yeast cell cycle expression Experiments (over time) baseline expression program
10-810 /02-710 Computational Genomics Normalization Genes and Gene Expression Technology Display of Expression Information Yeast cell cycle expression Experiments (over time) baseline expression program
