Genome Sequencing & Assembly. Slides by Carl Kingsford
|
|
|
- Gwenda Carter
- 6 years ago
- Views:
Transcription
1 Genome Sequencing & Assembly Slides by Carl Kingsford
2 Genome Sequencing ACCGTCCAATTGG...! TGGCAGGTTAACC... E.g. human: 3 billion bases split into 23 chromosomes Main tool of traditional sequencing: DNA Synthesis DNA polymerase: enzyme that will grow a complementary DNA strand. gacgatcggtttatcc! ctgctagccaaataggctaatactacgga
3 Sanger Sequencing: Finding the As dxtp c c c c g g g g t t t t a a a a a * ddatp gacgatcgg ttta *! ctgctagcc aaataggctaatactacgga gacgatcgg tttatccgattatga *! ctgctagcc aaataggctaatactacgga gacgatcgg tttatccga *! ctgctagcc aaataggctaatactacgga gacgatcgg tttatccgatta *! ctgctagcc aaataggctaatactacgga gacgatcgg ttta *! ctgctagcc aaataggctaatactacgga gacgatcgg tttatccgattatga *! ctgctagcc aaataggctaatactacgga gacgatcgg tttatccgatta *! ctgctagcc aaataggctaatactacgga gacgatcgg tttatccga *! ctgctagcc aaataggctaatactacgga
4 Size Sequence gacgatcggttta * gacgatcggttta * gacgatcggtttatccga * gacgatcggtttatccga * gacgatcggtttatccgatta * gacgatcggtttatccgatta * gacgatcggtttatccgattatga * gacgatcggtttatccgattatga * Size gacgatcggtttatccgattatg * gacgatcggtttatccg * A C G T gacgatcggtttatc * gacgatcggtttatcc *
5 Size Sequence Single lane: ddxtp that fluoresce different colors gacgatcggttta * gacgatcggttta * gacgatcggtttatccga * gacgatcggtttatccga * gacgatcggtttatccgatta * gacgatcggtttatccgatta * gacgatcggtttatccgattatga * gacgatcggtttatccgattatga * Size gacgatcggtttatccgattatg * gacgatcggtttatccg * A C G T gacgatcggtttatc * gacgatcggtttatcc *
6 Size Sequence Single lane: ddxtp that fluoresce different colors gacgatcggttta * gacgatcggttta * gacgatcggtttatccga * gacgatcggtttatccga * gacgatcggtttatccgatta * gacgatcggtttatccgatta * gacgatcggtttatccgattatga * gacgatcggtttatccgattatga * Size gacgatcggtttatccgattatg * gacgatcggtttatccg * A C G T gacgatcggtttatc * gacgatcggtttatcc * Main problem: larger fragments take a long time to be sorted correctly (or don t sort correctly ever) letter maximum
7 Shotgun Sequencing Many copies of the DNA Shear it, randomly breaking them into many small pieces, read ends of each: Assemble into original genome:
8 We can only read ~ 1000 characters at a time from a random place: good-natured, she thought: still when it saw Alice. It looked ought to be treated good-natured, she thought, still Cat only a greet many It looked goodclaws and a great many teeth, so she The Cat only grinned when it saw Alice. be treated with respect. sti1l it had very long claws so she felt that it ought Algorithms are needed to piece the story together.
9 good-natured, she thought: still good-natured, she thought, still sti1l it had very long claws The Cat only grinned when it saw Alice. Cat only when it saw Alice. It looked It looked goodclaws and a great many teeth, so she a greet many so she felt that it ought ought to be treated be treated with respect. It s a jigsaw puzzle......except with 35 million pieces
10 Lander-Waterman Statistics How many reads to we need to be sure we cover the whole genome? g genome L N θ = fraction of L required to detect an overlap An island is a contiguous group of reads that are connected by overlaps of length θl. (Various colors above) Want: Expression for expected # of islands given N, g, L, θ.
11 Expected # of Islands λ := N/g = probability a read starts at a given position (assuming random sampling) Pr(k reads start in an interval of length x) x trials, want k successes, small probability λ of success Expected # of successes = λx Poisson approximation to binomial distribution: Pr(k reads in length x) =e x ( x)k Expected # of islands = N Pr(read is at rightmost end of island) (1-θ)L θl = N Pr(0 reads start in (1-θ)L) = Ne = Ne = Ne (1 )L 0 (1 )L 0! (1 )LN/g (from above) k! LN/g is called the coverage c.
12 Expected # of Islands, 2 Rewrite to depend more directly on the things we can control: c and θ Expected # of islands = Ne = Ne (1 )LN/g (1 )c Expected # islands θ = 0.15 L = 1000; g = θ = 0.35 = L/g L/g = g L Ne (1 )c ce (1 )c c
13 Shotgun Sequencing Summary Sanger sequencing widely used up through 2006 or 2007, including for the human genome project. Won Sanger his second Nobel prize. (Wikipedia) Lander-Waterman statistics estimate the number of islands you will get for a given coverage. Used as a way to guess how much sequencing you need to do for a given technology and genome size. Often hard in practice to guess the genome size g before you ve sequenced it.
14 Genome Assembly Paradigms
15 Shortest Common Superstring Def. Given strings s1,..., sn, find the shortest string T such that each si is a substring of T. NP-hard (contrast with case when requiring si to be subsequences of T) Approximation algorithms exist with factors: 4, 3, 2.89, 2.75, 2.67, 2.596, 2.5,... Basic greedy method: find pair of strings that overlap the best, merge them, repeat (4 approximation): Given match, mismatch, gap costs, how can we compute the score of the best overlap?
16 Overlap Alignment y C 9 0 Score of an optimal alignment between a suffix of Y and a prefix of X Initialize first column to 0s A G T T G C Answer is maximum score in top row (traceback starts from there until it falls off left side) A 2 0 A 1 0 y 0 0 1g 2g 3g 4g 5g 6g 7g 8g 9g 10g 11g 12g x A A G G T A T G A A T C x
17 Overlap Alignment y C 9 0 Score of an optimal alignment between a suffix of Y and a prefix of X Initialize first column to 0s A G T T G C Answer is maximum score in top row (traceback starts from there until it falls off left side) A 2 0 A 1 0 y 0 0 1g 2g 3g 4g 5g 6g 7g 8g 9g 10g 11g 12g x A A G G T A T G A A T C x
18 Only compute overlap alignment between reads that share a kmer: K-mer Hashing AAAA r1 r2 r10 r11 read AAAT AAAG r2 r3 AAAC AATA AATT AATG kmer AATC AAGA AAGT
19 The problem with Shortest Common Superstring (SCS): Repeats Truth: AAAAAAAAAAAAAAAAAAA AAAAA AAAAA AAAAA AAAAA AAAAA AAAAA SCS: AAAAA AAAAA AAAAA AAAAA AAAAA More complex example: 2 or 3 ACCGCCT ACCGCCT ACCGCCT copies?
20 Overlap Consensus Overlap graph: Nodes = reads Edges = suffix-prefix overlaps Given overlap graph, how can we find a good candidate assembly? Idea: Every read must be used in exactly one place in the genome.
21 Assembly by Traveling Salesman Traveling Salesman Problem: Find a path that visits every node in the graph exactly once. Optimal Traveling Salesman path of 24,978 cities in Sweden (Applegate et al, 2004, index.html).
22 Assembly via Eulerian Path
23 de Bruijn graph read k kmer k-1 de Bruijn graph: nodes represent kmers, edges connect k-mers that are known to follow each other based on an observed read.! Can have > 1 edge between nodes. k-mer k-mer
24 Examples agg tag aga gac acg cga gaa aac cgt cgg gta tac A directed graph has an Eulerian cycle if and only if: All nodes have the same number of edges entering and leaving ggt tagacgaacgtacggtagg acg cga gaa aac acc cca cac cgt gac gta acgaaccacgacgta
25 Example bacterial de Bruijn graph AAAGAAA AGTG..TAAA AATT..AACT GCTT..TTCA T TTAT..CTAA CAGC..CTAA TACC..CAGG ACAC..TTTA TAAC..TAGT CAAC..AGCC CACC..AATA TTCA..AACT GTTC..AATA ACCC..CATT ATAG..TCAC GTATCGC GTAT..GTTG GGGT..GAAA A AAAC..AAGT TGCT..TTAA GAAA..TTAACAAA..ACTC AAAG..GTTG TTAA..ACCA ATTG..CAAA GTTG..CAAA CATC..AACG CTACGCC AGTG..AACG ACCA..AGAA TGTG..CCCC GGTT..CAAT GACTTT CACC..CATT TTCAACTTC ACTAAAAA TAAT..AAGT AAAA..CAAC GAAG..CCCA GAAA..TTAC CACT..AGAA TCTT..AGCC GGGT..CCCC AGTT..AACA CATT..AACA TTCC..ATCC TACT..GGTT CGCT..ATCA ATGG..AAAC GATCA A TGTG..GGGG CCCAA TTGG..GTTG ATTTAAA GTAT..TTTT ATTA..GTTG GAAT..GGTT CCAA..ACTG GTTA..TTAC CAAC..TTCT ACAG..TCGG AGGT..AGTC A CAAG..CAAC TTTAT TGAG..AGTG TCAA..TCGG CATC..CCCA TATC..CAAC CTAG..CGGGAGGAT AAGA..TTTT TATC..TTTT AAGC..AAAT CAAT..GGAT AGTG..GGGG CGCA..AAAT ATCC..GGGA TGCG..AAAA CCAA..GGAT G CTTA..AGTGGCAG..AACC AATG..AGTC GCTA..CAGC AGCTTA CAAC..TAGT TCTT..AAAA AGTA..ACGG GATA..TATA TAGG..GGGG CAAA..TGGA GTAG..AACT GGGA..CAGC CTTG..TAGT CCAA..GGGA TCAA..ATCG CCAA..AACT TGAA..ATCG TTTG..AAAT AAGC..TAAA TTTG..AATC AAGA..CTTA CTCC..TCCC TTTT..AGTC AGAA..ACGG GGGG..GTTA CTGA..TTTA CCA TCAAC GTAGTA CTTC..GTTT TAGG..GTTG CGTG..GAGT GTTG..ACCA AACA..CAAG TCAA..GAAT AATT..GAAG TAGC..GAGT CCCA AGTC..ATGC GGGT..AGTA CAAG..CAAG GTAT..GTTA TGAT..GTTG GGTA..TTTC GATG..AATC ACTC..TGGG CTGC..ATCA AATC..TGTG TGGC AACA..TCCC GCTC..CTAC ATAT..AGCT ACAA..CAAC TTGC..TGTG TCAA..GTTT TGGT..TGCA ACAA..CATT CCAA..CAAC AGTA..GTTT TGAT..ACTT CTAG..GAAC TTCTAC CCAGC ATGT..TGCA CAAG..AGCT TTGT..TGGG TGCA..GTTT CCCA..CCAG GGCG..TAAA ATCT..CCAG G ACAG..CAAT TTCA..GTTG CTGG..ACCA TAGTT CAAC..ACCA CAGG..CCAA TCCATT CTTT..ATAA TTTT..ATAA GAGT..GAAT CAGG..CCAA TTTA..GAGG GGAA..AACT AGAA..AACT GTTG..TCAA TTAT..AAGG TTGG..GTTG Paths with no branches compressed into a single node Eulerian path = use every edge exactly once. With perfect data, the genome can be reconstructed by some Eulerian path through this graph
26 Assembly via Eulerian Path acg cga gaa aac acgaacgta cgt gta Let dg(s) be the de Bruijn graph of string s. Then s corresponds to some Eulerian path in dg(s). A directed graph has an Eulerian path if and only if: One node has one more edge leaving it than entering One node has one more edge entering than leaving All other nodes have the same number of edges entering and leaving How can we find such a path?
27 Connect node with out-degree < in-degree to node with out-degree < in-degree. So that we will have an Eulerian cycle.! Walk from some arbitrary node u until you return to u, creating a doubly liked list of the path you visit.! Repeat until all edges used: Eulerian Path Algorithm Why will you return to u? * How can find such a node quickly? Start from some node w on the current tour with unused edges *. Walk along unused edges until you return to w, inserting the visited nodes after w into the current tour list. u v w x y w u
28 Connect node with out-degree < in-degree to node with out-degree < in-degree. So that we will have an Eulerian cycle.! Walk from some arbitrary node u until you return to u, creating a doubly liked list of the path you visit.! Repeat until all edges used: Eulerian Path Algorithm Why will you return to u? * How can find such a node quickly? Start from some node w on the current tour with unused edges *. Walk along unused edges until you return to w, inserting the visited nodes after w into the current tour list. a b u v w x y duplicate this w node w u
29 The Problem with Eulerian Paths There are typically an astronomical number of possible Eulerian tours with perfect data.! Adding back constraints to limit # of tours leads to a NPhard problem.! With imperfect data, there are usually NO Eulerian tours! Estimating # of parallel edges is usually tricky. Aside: counting # of Eulerian tours in a directed graph is easy, but in an undirected graph is #Pcomplete (hard). (Kingsford, Schatz, Pop, 2010)
30 Mate Pairs chop up select for a given size mate pair: 2 reads, of opposite orientation, separated by an approximately known distance sequence 1000 bases from each end long range information
31 Islands = contigs Scaffolding
32 Islands = contigs Scaffolding
33 Islands = contigs Scaffolding
34 Comparative Assembly (Read Mapping) Align reads to known genome: known reference genome x xx x consistent differences = deviation from reference rare differences = sequencing errors Can use much lower coverage (e.g. 4X coverage instead of 20-30X for de novo assembly). Aligning a large # of short sequences to one large sequence is an important special case of sequence alignment.
35 Summary Sanger sequencing reads DNA via synthesis; bp. Assembly Paradigms: Shortest Common Superstring (NP-hard; sensitive to repeats) Hamiltonian cycle in overlap graph (NP-hard) Eulerian cycle in de Bruijn graph (polynomial in basic form, but large # of solutions) Overlap alignment can be computed with slight variant of sequence alignment DP. K-mer hashing technique avoids all pairs overlap alignment
36 Hard vs. Easy Eulerian path visit every edge once (easy) Hamiltonian path visit every node once (hard)! Shortest common supersequence (easy) Shortest common superstring (hard)! Counting Eulerian tours in directed graphs (easy) Counting Eulerian tours in undirected graphs (hard)! Aligning 2 sequences (easy) Aligning k > 2 sequences (hard)! Shortest path (easy) Longest path (hard)
37 Cufflinks Transcript Assembly Cole Trapnell, Brian A. Williams, Geo Pertea, Ali Mortazavi, Gordon Kwan, Marijke J. van Baren, Steven L. Salzberg, Barbara J. Wold, and Lior Pachter.. Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nat Biotechnol, 28(5): (2010)!
38 Partially Ordered Sets Def. A pair (S, ) is a partial order if, for all x, y S: (transitivity) x y and y z x z (reflexivity) x x (antisymmetry) x y and y x x = y y z x
39 Partially Ordered Sets Def. A pair (S, ) is a partial order if, for all x, y S: (transitivity) x y and y z x z (reflexivity) x x (antisymmetry) x y and y x x = y chain: every pair is comparable y z x
40 Partially Ordered Sets Def. A pair (S, ) is a partial order if, for all x, y S: (transitivity) x y and y z x z (reflexivity) x x (antisymmetry) x y and y x x = y chain: every pair is comparable antichain: every pair is incomparable z y x
41 Cufflink s Partial Order y x = sequenced fragment: = x aligns to the left of y and x and y have compatible intron structure y 1 x 1 incompatible b/c the right end of y 2 is split-mapped, implying an intron where there is no intron in x 2. x 3 and y 3 are nested, and so are merged into a single fragment. x 4 is uncertain because it could be compatible with either y 4 or y 5 ; x 4 is therefore thrown away. (Trapnell et al., 2010)
42 Cufflinks Assembly Algorithm Partitioning partial order into smallest # of chains parsimonious set of transcripts that explains the observed reads Smallest # of chains Largest antichain Vertex cover Maximum bipartite matching Dilworth s Theorem Dilworth König König s Theorem Solvable in O(E V)
43 Cufflinks Assembly Algorithm (covering) Partitioning partial order into smallest # of chains parsimonious set of transcripts that explains the observed reads Smallest # of chains Largest antichain Vertex cover Maximum bipartite matching Dilworth s Theorem Dilworth König König s Theorem Solvable in O(E V)
44 Dilworth s Theorem Thm (Dilworth). In a poset, the size of the largest antichain = the size of the minimum cover by chains. Proof intuition. The largest antichain must hit every chain (otherwise it could be made larger). It can t hit any chain twice, otherwise it would contain two comparable items.
45 König s Theorem Thm (König). In a bipartite graph, the # of edges in a maximum matching = # of vertices in the smalelst vertex cover. Proof intuition. In a maximum matching, every edge must be covered. Otherwise, if both endpoints are not matched, we could add that edge to the matching and increase its size.
46 Using Matching to Find a Minimal Chain Cover All items in poset Edge if x < y All items in poset Let M be the maximal matching. By König s theorem, there is a (minimal) vertex cover C of the same size as M. Let T be the elements of the poset that are not in C. T is an antichain. Why? Make a set W of chains by u v if (u,v) M. These equivalence classes are chains. Why?
47 Using Matching to Find a Minimal Chain Cover All items in poset Edge if x < y All items in poset Let M be the maximal matching. By König s theorem, there is a (minimal) vertex cover C of the same size as M. Let T be the elements of the poset that are not in C. If u and v were comparable, T is an antichain. Why? there would be an edge between them, and since neither u or v used in M, we could add that edge to M. Make a set W of chains by u v if (u,v) M. These equivalence classes are chains. Why?
48 Using Matching to Find a Minimal Chain Cover All items in poset Edge if x < y All items in poset Let M be the maximal matching. By König s theorem, there is a (minimal) vertex cover C of the same size as M. Let T be the elements of the poset that are not in C. If u and v were comparable, T is an antichain. Why? there would be an edge between them, and since neither u or v used in M, we could add that edge to M. Make a set W of chains by u v if (u,v) M. These equivalence classes are chains. Why? Every pair of items in each equivalence class had an edge between them, meaning they were comparable.
49 W = T M = maximal matching. C = vertex cover of the same size as M. n = # elements in poset m = # of edges in matching T = antichain elements of poset that are not in C. W = set of chains formed from edges of M. Size of T is n - m. Why? Size of W is n - m. Why?
50 W = T M = maximal matching. C = vertex cover of the same size as M. n = # elements in poset m = # of edges in matching T = antichain elements of poset that are not in C. W = set of chains formed from edges of M. Size of T is n - m. Why? Every edge uses up exactly one element on the LHS of the bipartite graph. Size of W is n - m. Why?
51 W = T M = maximal matching. C = vertex cover of the same size as M. n = # elements in poset m = # of edges in matching T = antichain elements of poset that are not in C. W = set of chains formed from edges of M. Size of T is n - m. Why? Every edge uses up exactly one element on the LHS of the bipartite graph. Size of W is n - m. Why? Consider set of n chains each consisting of a single element of poset. Each edge (u,v) that we use to put v into the same poset as u reduces the number of chains by 1. Number of equivalence-class chains = n - m
52 Why is W Minimum Size? chain covers All antichains must be of size all chain covers. Suppose not, and let A be an antichain bigger than cover Q. Then, by pigeonhole, A must contain at least 2 elements x, y from the same chain in Q. antichians But x, y are comparable because they are in the same chain. the pair (T,W) must be a largest antichain and a smallest W because they are the same size.
53 A Matching-Covering Example (Trapnell et al., 2010)
54 Selecting From Among Many Minimum Solutions v w x y z Correct Wrong Idea: exons included in same transcript should have similar expression Estimate Percent Spliced In (PSI, ψ): # of reads crossing exon x that are compatible with x divided by # of reads overlapping x (divided by length of x). weight(x, y) = log(1 x y )
55 Selecting From Among Many Minimum Solutions v w x y z Correct Wrong Idea: exons included in same transcript should have similar expression Estimate Percent measures Spliced how In similar (PSI, ψ): the # of exons reads PSI crossing values exon are x that are compatible with x divided by # of reads overlapping x (divided by length of x). weight(x, y) = log(1 x y )
56 Discovery of Novel Isoforms (Trapnell et al., 2010)
Read Mapping. de Novo Assembly. Genomics: Lecture #2 WS 2014/2015
 Mapping de Novo Assembly Institut für Medizinische Genetik und Humangenetik Charité Universitätsmedizin Berlin Genomics: Lecture #2 WS 2014/2015 Today Genome assembly: the basics Hamiltonian and Eulerian
Mapping de Novo Assembly Institut für Medizinische Genetik und Humangenetik Charité Universitätsmedizin Berlin Genomics: Lecture #2 WS 2014/2015 Today Genome assembly: the basics Hamiltonian and Eulerian
Read Mapping. Slides by Carl Kingsford
 Read Mapping Slides by Carl Kingsford Bowtie Ultrafast and memory-efficient alignment of short DNA sequences to the human genome Ben Langmead, Cole Trapnell, Mihai Pop and Steven L Salzberg, Genome Biology
Read Mapping Slides by Carl Kingsford Bowtie Ultrafast and memory-efficient alignment of short DNA sequences to the human genome Ben Langmead, Cole Trapnell, Mihai Pop and Steven L Salzberg, Genome Biology
de novo assembly Simon Rasmussen 36626: Next Generation Sequencing analysis DTU Bioinformatics Next Generation Sequencing Analysis
 de novo assembly Simon Rasmussen 36626: Next Generation Sequencing analysis DTU Bioinformatics 27626 - Next Generation Sequencing Analysis Generalized NGS analysis Data size Application Assembly: Compare
de novo assembly Simon Rasmussen 36626: Next Generation Sequencing analysis DTU Bioinformatics 27626 - Next Generation Sequencing Analysis Generalized NGS analysis Data size Application Assembly: Compare
Genome Reconstruction: A Puzzle with a Billion Pieces Phillip E. C. Compeau and Pavel A. Pevzner
 Genome Reconstruction: A Puzzle with a Billion Pieces Phillip E. C. Compeau and Pavel A. Pevzner Outline I. Problem II. Two Historical Detours III.Example IV.The Mathematics of DNA Sequencing V.Complications
Genome Reconstruction: A Puzzle with a Billion Pieces Phillip E. C. Compeau and Pavel A. Pevzner Outline I. Problem II. Two Historical Detours III.Example IV.The Mathematics of DNA Sequencing V.Complications
Sequencing. Computational Biology IST Ana Teresa Freitas 2011/2012. (BACs) Whole-genome shotgun sequencing Celera Genomics
 Computational Biology IST Ana Teresa Freitas 2011/2012 Sequencing Clone-by-clone shotgun sequencing Human Genome Project Whole-genome shotgun sequencing Celera Genomics (BACs) 1 Must take the fragments
Computational Biology IST Ana Teresa Freitas 2011/2012 Sequencing Clone-by-clone shotgun sequencing Human Genome Project Whole-genome shotgun sequencing Celera Genomics (BACs) 1 Must take the fragments
(for more info see:
 Genome assembly (for more info see: http://www.cbcb.umd.edu/research/assembly_primer.shtml) Introduction Sequencing technologies can only "read" short fragments from a genome. Reconstructing the entire
Genome assembly (for more info see: http://www.cbcb.umd.edu/research/assembly_primer.shtml) Introduction Sequencing technologies can only "read" short fragments from a genome. Reconstructing the entire
Algorithms for Bioinformatics
 Adapted from slides by Alexandru Tomescu, Leena Salmela and Veli Mäkinen, which are partly from http://bix.ucsd.edu/bioalgorithms/slides.php 582670 Algorithms for Bioinformatics Lecture 3: Graph Algorithms
Adapted from slides by Alexandru Tomescu, Leena Salmela and Veli Mäkinen, which are partly from http://bix.ucsd.edu/bioalgorithms/slides.php 582670 Algorithms for Bioinformatics Lecture 3: Graph Algorithms
Sequence Assembly. BMI/CS 576 Mark Craven Some sequencing successes
 Sequence Assembly BMI/CS 576 www.biostat.wisc.edu/bmi576/ Mark Craven craven@biostat.wisc.edu Some sequencing successes Yersinia pestis Cannabis sativa The sequencing problem We want to determine the identity
Sequence Assembly BMI/CS 576 www.biostat.wisc.edu/bmi576/ Mark Craven craven@biostat.wisc.edu Some sequencing successes Yersinia pestis Cannabis sativa The sequencing problem We want to determine the identity
Graph Algorithms in Bioinformatics
 Graph Algorithms in Bioinformatics Computational Biology IST Ana Teresa Freitas 2015/2016 Sequencing Clone-by-clone shotgun sequencing Human Genome Project Whole-genome shotgun sequencing Celera Genomics
Graph Algorithms in Bioinformatics Computational Biology IST Ana Teresa Freitas 2015/2016 Sequencing Clone-by-clone shotgun sequencing Human Genome Project Whole-genome shotgun sequencing Celera Genomics
10/15/2009 Comp 590/Comp Fall
 Lecture 13: Graph Algorithms Study Chapter 8.1 8.8 10/15/2009 Comp 590/Comp 790-90 Fall 2009 1 The Bridge Obsession Problem Find a tour crossing every bridge just once Leonhard Euler, 1735 Bridges of Königsberg
Lecture 13: Graph Algorithms Study Chapter 8.1 8.8 10/15/2009 Comp 590/Comp 790-90 Fall 2009 1 The Bridge Obsession Problem Find a tour crossing every bridge just once Leonhard Euler, 1735 Bridges of Königsberg
RESEARCH TOPIC IN BIOINFORMANTIC
 RESEARCH TOPIC IN BIOINFORMANTIC GENOME ASSEMBLY Instructor: Dr. Yufeng Wu Noted by: February 25, 2012 Genome Assembly is a kind of string sequencing problems. As we all know, the human genome is very
RESEARCH TOPIC IN BIOINFORMANTIC GENOME ASSEMBLY Instructor: Dr. Yufeng Wu Noted by: February 25, 2012 Genome Assembly is a kind of string sequencing problems. As we all know, the human genome is very
Algorithms for Bioinformatics
 Adapted from slides by Alexandru Tomescu, Leena Salmela and Veli Mäkinen, which are partly from http://bix.ucsd.edu/bioalgorithms/slides.php 58670 Algorithms for Bioinformatics Lecture 5: Graph Algorithms
Adapted from slides by Alexandru Tomescu, Leena Salmela and Veli Mäkinen, which are partly from http://bix.ucsd.edu/bioalgorithms/slides.php 58670 Algorithms for Bioinformatics Lecture 5: Graph Algorithms
BLAST & Genome assembly
 BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies May 15, 2014 1 BLAST What is BLAST? The algorithm 2 Genome assembly De novo assembly Mapping assembly 3
BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies May 15, 2014 1 BLAST What is BLAST? The algorithm 2 Genome assembly De novo assembly Mapping assembly 3
10/8/13 Comp 555 Fall
 10/8/13 Comp 555 Fall 2013 1 Find a tour crossing every bridge just once Leonhard Euler, 1735 Bridges of Königsberg 10/8/13 Comp 555 Fall 2013 2 Find a cycle that visits every edge exactly once Linear
10/8/13 Comp 555 Fall 2013 1 Find a tour crossing every bridge just once Leonhard Euler, 1735 Bridges of Königsberg 10/8/13 Comp 555 Fall 2013 2 Find a cycle that visits every edge exactly once Linear
Reducing Genome Assembly Complexity with Optical Maps
 Reducing Genome Assembly Complexity with Optical Maps Lee Mendelowitz LMendelo@math.umd.edu Advisor: Dr. Mihai Pop Computer Science Department Center for Bioinformatics and Computational Biology mpop@umiacs.umd.edu
Reducing Genome Assembly Complexity with Optical Maps Lee Mendelowitz LMendelo@math.umd.edu Advisor: Dr. Mihai Pop Computer Science Department Center for Bioinformatics and Computational Biology mpop@umiacs.umd.edu
DNA Sequencing The Shortest Superstring & Traveling Salesman Problems Sequencing by Hybridization
 Eulerian & Hamiltonian Cycle Problems DNA Sequencing The Shortest Superstring & Traveling Salesman Problems Sequencing by Hybridization The Bridge Obsession Problem Find a tour crossing every bridge just
Eulerian & Hamiltonian Cycle Problems DNA Sequencing The Shortest Superstring & Traveling Salesman Problems Sequencing by Hybridization The Bridge Obsession Problem Find a tour crossing every bridge just
Genome 373: Genome Assembly. Doug Fowler
 Genome 373: Genome Assembly Doug Fowler What are some of the things we ve seen we can do with HTS data? We ve seen that HTS can enable a wide variety of analyses ranging from ID ing variants to genome-
Genome 373: Genome Assembly Doug Fowler What are some of the things we ve seen we can do with HTS data? We ve seen that HTS can enable a wide variety of analyses ranging from ID ing variants to genome-
RNA-seq. Read mapping and Quantification. Genomics: Lecture #12. Institut für Medizinische Genetik und Humangenetik Charité Universitätsmedizin Berlin
 (1) Read and Quantification Institut für Medizinische Genetik und Humangenetik Charité Universitätsmedizin Berlin Genomics: Lecture #12 Today (1) Gene Expression Previous gold standard: Basic protocol
(1) Read and Quantification Institut für Medizinische Genetik und Humangenetik Charité Universitätsmedizin Berlin Genomics: Lecture #12 Today (1) Gene Expression Previous gold standard: Basic protocol
DNA Sequencing. Overview
 BINF 3350, Genomics and Bioinformatics DNA Sequencing Young-Rae Cho Associate Professor Department of Computer Science Baylor University Overview Backgrounds Eulerian Cycles Problem Hamiltonian Cycles
BINF 3350, Genomics and Bioinformatics DNA Sequencing Young-Rae Cho Associate Professor Department of Computer Science Baylor University Overview Backgrounds Eulerian Cycles Problem Hamiltonian Cycles
Sequencing. Short Read Alignment. Sequencing. Paired-End Sequencing 6/10/2010. Tobias Rausch 7 th June 2010 WGS. ChIP-Seq. Applied Biosystems.
 Sequencing Short Alignment Tobias Rausch 7 th June 2010 WGS RNA-Seq Exon Capture ChIP-Seq Sequencing Paired-End Sequencing Target genome Fragments Roche GS FLX Titanium Illumina Applied Biosystems SOLiD
Sequencing Short Alignment Tobias Rausch 7 th June 2010 WGS RNA-Seq Exon Capture ChIP-Seq Sequencing Paired-End Sequencing Target genome Fragments Roche GS FLX Titanium Illumina Applied Biosystems SOLiD
version /1/2011 Source code Linux x86_64 binary Mac OS X x86_64 binary
 Cufflinks RNA-Seq analysis tools - Getting Started 1 of 6 14.07.2011 09:42 Cufflinks Transcript assembly, differential expression, and differential regulation for RNA-Seq Site Map Home Getting started
Cufflinks RNA-Seq analysis tools - Getting Started 1 of 6 14.07.2011 09:42 Cufflinks Transcript assembly, differential expression, and differential regulation for RNA-Seq Site Map Home Getting started
CSCI2950-C Lecture 4 DNA Sequencing and Fragment Assembly
 CSCI2950-C Lecture 4 DNA Sequencing and Fragment Assembly Ben Raphael Sept. 22, 2009 http://cs.brown.edu/courses/csci2950-c/ l-mer composition Def: Given string s, the Spectrum ( s, l ) is unordered multiset
CSCI2950-C Lecture 4 DNA Sequencing and Fragment Assembly Ben Raphael Sept. 22, 2009 http://cs.brown.edu/courses/csci2950-c/ l-mer composition Def: Given string s, the Spectrum ( s, l ) is unordered multiset
Introduction to Genome Assembly. Tandy Warnow
 Introduction to Genome Assembly Tandy Warnow 2 Shotgun DNA Sequencing DNA target sample SHEAR & SIZE End Reads / Mate Pairs 550bp 10,000bp Not all sequencing technologies produce mate-pairs. Different
Introduction to Genome Assembly Tandy Warnow 2 Shotgun DNA Sequencing DNA target sample SHEAR & SIZE End Reads / Mate Pairs 550bp 10,000bp Not all sequencing technologies produce mate-pairs. Different
Reducing Genome Assembly Complexity with Optical Maps Mid-year Progress Report
 Reducing Genome Assembly Complexity with Optical Maps Mid-year Progress Report Lee Mendelowitz LMendelo@math.umd.edu Advisor: Dr. Mihai Pop Computer Science Department Center for Bioinformatics and Computational
Reducing Genome Assembly Complexity with Optical Maps Mid-year Progress Report Lee Mendelowitz LMendelo@math.umd.edu Advisor: Dr. Mihai Pop Computer Science Department Center for Bioinformatics and Computational
Sequence Assembly Required!
 Sequence Assembly Required! 1 October 3, ISMB 20172007 1 Sequence Assembly Genome Sequenced Fragments (reads) Assembled Contigs Finished Genome 2 Greedy solution is bounded 3 Typical assembly strategy
Sequence Assembly Required! 1 October 3, ISMB 20172007 1 Sequence Assembly Genome Sequenced Fragments (reads) Assembled Contigs Finished Genome 2 Greedy solution is bounded 3 Typical assembly strategy
BLAST & Genome assembly
 BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies November 17, 2012 1 Introduction Introduction 2 BLAST What is BLAST? The algorithm 3 Genome assembly De
BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies November 17, 2012 1 Introduction Introduction 2 BLAST What is BLAST? The algorithm 3 Genome assembly De
CSE 549: Genome Assembly Intro & OLC. All slides in this lecture not marked with * courtesy of Ben Langmead.
 CSE 9: Genome Assembly Intro & OLC All slides in this lecture not marked with * courtesy of Ben Langmead. Shotgun Sequencing Many copies of the DNA Shear it, randomly breaking them into many small pieces,
CSE 9: Genome Assembly Intro & OLC All slides in this lecture not marked with * courtesy of Ben Langmead. Shotgun Sequencing Many copies of the DNA Shear it, randomly breaking them into many small pieces,
Aligners. J Fass 21 June 2017
 Aligners J Fass 21 June 2017 Definitions Assembly: I ve found the shredded remains of an important document; put it back together! UC Davis Genome Center Bioinformatics Core J Fass Aligners 2017-06-21
Aligners J Fass 21 June 2017 Definitions Assembly: I ve found the shredded remains of an important document; put it back together! UC Davis Genome Center Bioinformatics Core J Fass Aligners 2017-06-21
Genome Assembly Using de Bruijn Graphs. Biostatistics 666
 Genome Assembly Using de Bruijn Graphs Biostatistics 666 Previously: Reference Based Analyses Individual short reads are aligned to reference Genotypes generated by examining reads overlapping each position
Genome Assembly Using de Bruijn Graphs Biostatistics 666 Previously: Reference Based Analyses Individual short reads are aligned to reference Genotypes generated by examining reads overlapping each position
NGS FASTQ file format
 NGS FASTQ file format Line1: Begins with @ and followed by a sequence idenefier and opeonal descripeon Line2: Raw sequence leiers Line3: + Line4: Encodes the quality values for the sequence in Line2 (see
NGS FASTQ file format Line1: Begins with @ and followed by a sequence idenefier and opeonal descripeon Line2: Raw sequence leiers Line3: + Line4: Encodes the quality values for the sequence in Line2 (see
Genome Reconstruction: A Puzzle with a Billion Pieces. Phillip Compeau Carnegie Mellon University Computational Biology Department
 http://cbd.cmu.edu Genome Reconstruction: A Puzzle with a Billion Pieces Phillip Compeau Carnegie Mellon University Computational Biology Department Eternity II: The Highest-Stakes Puzzle in History Courtesy:
http://cbd.cmu.edu Genome Reconstruction: A Puzzle with a Billion Pieces Phillip Compeau Carnegie Mellon University Computational Biology Department Eternity II: The Highest-Stakes Puzzle in History Courtesy:
Omega: an Overlap-graph de novo Assembler for Metagenomics
 Omega: an Overlap-graph de novo Assembler for Metagenomics B a h l e l H a i d e r, Ta e - H y u k A h n, B r i a n B u s h n e l l, J u a n j u a n C h a i, A l e x C o p e l a n d, C h o n g l e Pa n
Omega: an Overlap-graph de novo Assembler for Metagenomics B a h l e l H a i d e r, Ta e - H y u k A h n, B r i a n B u s h n e l l, J u a n j u a n C h a i, A l e x C o p e l a n d, C h o n g l e Pa n
I519 Introduction to Bioinformatics, Genome assembly. Yuzhen Ye School of Informatics & Computing, IUB
 I519 Introduction to Bioinformatics, 2014 Genome assembly Yuzhen Ye (yye@indiana.edu) School of Informatics & Computing, IUB Contents Genome assembly problem Approaches Comparative assembly The string
I519 Introduction to Bioinformatics, 2014 Genome assembly Yuzhen Ye (yye@indiana.edu) School of Informatics & Computing, IUB Contents Genome assembly problem Approaches Comparative assembly The string
Genome Assembly and De Novo RNAseq
 Genome Assembly and De Novo RNAseq BMI 7830 Kun Huang Department of Biomedical Informatics The Ohio State University Outline Problem formulation Hamiltonian path formulation Euler path and de Bruijin graph
Genome Assembly and De Novo RNAseq BMI 7830 Kun Huang Department of Biomedical Informatics The Ohio State University Outline Problem formulation Hamiltonian path formulation Euler path and de Bruijin graph
IDBA A Practical Iterative de Bruijn Graph De Novo Assembler
 IDBA A Practical Iterative de Bruijn Graph De Novo Assembler Yu Peng, Henry C.M. Leung, S.M. Yiu, and Francis Y.L. Chin Department of Computer Science, The University of Hong Kong Pokfulam Road, Hong Kong
IDBA A Practical Iterative de Bruijn Graph De Novo Assembler Yu Peng, Henry C.M. Leung, S.M. Yiu, and Francis Y.L. Chin Department of Computer Science, The University of Hong Kong Pokfulam Road, Hong Kong
Description of a genome assembler: CABOG
 Theo Zimmermann Description of a genome assembler: CABOG CABOG (Celera Assembler with the Best Overlap Graph) is an assembler built upon the Celera Assembler, which, at first, was designed for Sanger sequencing,
Theo Zimmermann Description of a genome assembler: CABOG CABOG (Celera Assembler with the Best Overlap Graph) is an assembler built upon the Celera Assembler, which, at first, was designed for Sanger sequencing,
7.36/7.91 recitation. DG Lectures 5 & 6 2/26/14
 7.36/7.91 recitation DG Lectures 5 & 6 2/26/14 1 Announcements project specific aims due in a little more than a week (March 7) Pset #2 due March 13, start early! Today: library complexity BWT and read
7.36/7.91 recitation DG Lectures 5 & 6 2/26/14 1 Announcements project specific aims due in a little more than a week (March 7) Pset #2 due March 13, start early! Today: library complexity BWT and read
David Crossman, Ph.D. UAB Heflin Center for Genomic Science. GCC2012 Wednesday, July 25, 2012
 David Crossman, Ph.D. UAB Heflin Center for Genomic Science GCC2012 Wednesday, July 25, 2012 Galaxy Splash Page Colors Random Galaxy icons/colors Queued Running Completed Download/Save Failed Icons Display
David Crossman, Ph.D. UAB Heflin Center for Genomic Science GCC2012 Wednesday, July 25, 2012 Galaxy Splash Page Colors Random Galaxy icons/colors Queued Running Completed Download/Save Failed Icons Display
IDBA - A Practical Iterative de Bruijn Graph De Novo Assembler
 IDBA - A Practical Iterative de Bruijn Graph De Novo Assembler Yu Peng, Henry Leung, S.M. Yiu, Francis Y.L. Chin Department of Computer Science, The University of Hong Kong Pokfulam Road, Hong Kong {ypeng,
IDBA - A Practical Iterative de Bruijn Graph De Novo Assembler Yu Peng, Henry Leung, S.M. Yiu, Francis Y.L. Chin Department of Computer Science, The University of Hong Kong Pokfulam Road, Hong Kong {ypeng,
DNA Fragment Assembly
 Algorithms in Bioinformatics Sami Khuri Department of Computer Science San José State University San José, California, USA khuri@cs.sjsu.edu www.cs.sjsu.edu/faculty/khuri DNA Fragment Assembly Overlap
Algorithms in Bioinformatics Sami Khuri Department of Computer Science San José State University San José, California, USA khuri@cs.sjsu.edu www.cs.sjsu.edu/faculty/khuri DNA Fragment Assembly Overlap
Read Mapping and Assembly
 Statistical Bioinformatics: Read Mapping and Assembly Stefan Seemann seemann@rth.dk University of Copenhagen April 9th 2019 Why sequencing? Why sequencing? Which organism does the sample comes from? Assembling
Statistical Bioinformatics: Read Mapping and Assembly Stefan Seemann seemann@rth.dk University of Copenhagen April 9th 2019 Why sequencing? Why sequencing? Which organism does the sample comes from? Assembling
Graph Algorithms in Bioinformatics
 Graph Algorithms in Bioinformatics Bioinformatics: Issues and Algorithms CSE 308-408 Fall 2007 Lecture 13 Lopresti Fall 2007 Lecture 13-1 - Outline Introduction to graph theory Eulerian & Hamiltonian Cycle
Graph Algorithms in Bioinformatics Bioinformatics: Issues and Algorithms CSE 308-408 Fall 2007 Lecture 13 Lopresti Fall 2007 Lecture 13-1 - Outline Introduction to graph theory Eulerian & Hamiltonian Cycle
Scalable Solutions for DNA Sequence Analysis
 Scalable Solutions for DNA Sequence Analysis Michael Schatz Dec 4, 2009 JHU/UMD Joint Sequencing Meeting The Evolution of DNA Sequencing Year Genome Technology Cost 2001 Venter et al. Sanger (ABI) $300,000,000
Scalable Solutions for DNA Sequence Analysis Michael Schatz Dec 4, 2009 JHU/UMD Joint Sequencing Meeting The Evolution of DNA Sequencing Year Genome Technology Cost 2001 Venter et al. Sanger (ABI) $300,000,000
Reducing Genome Assembly Complexity with Optical Maps
 Reducing Genome Assembly Complexity with Optical Maps AMSC 663 Mid-Year Progress Report 12/13/2011 Lee Mendelowitz Lmendelo@math.umd.edu Advisor: Mihai Pop mpop@umiacs.umd.edu Computer Science Department
Reducing Genome Assembly Complexity with Optical Maps AMSC 663 Mid-Year Progress Report 12/13/2011 Lee Mendelowitz Lmendelo@math.umd.edu Advisor: Mihai Pop mpop@umiacs.umd.edu Computer Science Department
Long Read RNA-seq Mapper
 UNIVERSITY OF ZAGREB FACULTY OF ELECTRICAL ENGENEERING AND COMPUTING MASTER THESIS no. 1005 Long Read RNA-seq Mapper Josip Marić Zagreb, February 2015. Table of Contents 1. Introduction... 1 2. RNA Sequencing...
UNIVERSITY OF ZAGREB FACULTY OF ELECTRICAL ENGENEERING AND COMPUTING MASTER THESIS no. 1005 Long Read RNA-seq Mapper Josip Marić Zagreb, February 2015. Table of Contents 1. Introduction... 1 2. RNA Sequencing...
Short Read Alignment. Mapping Reads to a Reference
 Short Read Alignment Mapping Reads to a Reference Brandi Cantarel, Ph.D. & Daehwan Kim, Ph.D. BICF 05/2018 Introduction to Mapping Short Read Aligners DNA vs RNA Alignment Quality Pitfalls and Improvements
Short Read Alignment Mapping Reads to a Reference Brandi Cantarel, Ph.D. & Daehwan Kim, Ph.D. BICF 05/2018 Introduction to Mapping Short Read Aligners DNA vs RNA Alignment Quality Pitfalls and Improvements
CS681: Advanced Topics in Computational Biology
 CS681: Advanced Topics in Computational Biology Can Alkan EA224 calkan@cs.bilkent.edu.tr Week 7 Lectures 2-3 http://www.cs.bilkent.edu.tr/~calkan/teaching/cs681/ Genome Assembly Test genome Random shearing
CS681: Advanced Topics in Computational Biology Can Alkan EA224 calkan@cs.bilkent.edu.tr Week 7 Lectures 2-3 http://www.cs.bilkent.edu.tr/~calkan/teaching/cs681/ Genome Assembly Test genome Random shearing
From the Schnable Lab:
 From the Schnable Lab: Yang Zhang and Daniel Ngu s Pipeline for Processing RNA-seq Data (As of November 17, 2016) yzhang91@unl.edu dngu2@huskers.unl.edu Pre-processing the reads: The alignment software
From the Schnable Lab: Yang Zhang and Daniel Ngu s Pipeline for Processing RNA-seq Data (As of November 17, 2016) yzhang91@unl.edu dngu2@huskers.unl.edu Pre-processing the reads: The alignment software
NP Completeness. Andreas Klappenecker [partially based on slides by Jennifer Welch]
![NP Completeness. Andreas Klappenecker [partially based on slides by Jennifer Welch] NP Completeness. Andreas Klappenecker [partially based on slides by Jennifer Welch]](/thumbs/96/128538415.jpg) NP Completeness Andreas Klappenecker [partially based on slides by Jennifer Welch] Dealing with NP-Complete Problems Dealing with NP-Completeness Suppose the problem you need to solve is NP-complete. What
NP Completeness Andreas Klappenecker [partially based on slides by Jennifer Welch] Dealing with NP-Complete Problems Dealing with NP-Completeness Suppose the problem you need to solve is NP-complete. What
From Smith-Waterman to BLAST
 From Smith-Waterman to BLAST Jeremy Buhler July 23, 2015 Smith-Waterman is the fundamental tool that we use to decide how similar two sequences are. Isn t that all that BLAST does? In principle, it is
From Smith-Waterman to BLAST Jeremy Buhler July 23, 2015 Smith-Waterman is the fundamental tool that we use to decide how similar two sequences are. Isn t that all that BLAST does? In principle, it is
Introduction and tutorial for SOAPdenovo. Xiaodong Fang Department of Science and BGI May, 2012
 Introduction and tutorial for SOAPdenovo Xiaodong Fang fangxd@genomics.org.cn Department of Science and Technology @ BGI May, 2012 Why de novo assembly? Genome is the genetic basis for different phenotypes
Introduction and tutorial for SOAPdenovo Xiaodong Fang fangxd@genomics.org.cn Department of Science and Technology @ BGI May, 2012 Why de novo assembly? Genome is the genetic basis for different phenotypes
Combinatorics Summary Sheet for Exam 1 Material 2019
 Combinatorics Summary Sheet for Exam 1 Material 2019 1 Graphs Graph An ordered three-tuple (V, E, F ) where V is a set representing the vertices, E is a set representing the edges, and F is a function
Combinatorics Summary Sheet for Exam 1 Material 2019 1 Graphs Graph An ordered three-tuple (V, E, F ) where V is a set representing the vertices, E is a set representing the edges, and F is a function
COMP260 Spring 2014 Notes: February 4th
 COMP260 Spring 2014 Notes: February 4th Andrew Winslow In these notes, all graphs are undirected. We consider matching, covering, and packing in bipartite graphs, general graphs, and hypergraphs. We also
COMP260 Spring 2014 Notes: February 4th Andrew Winslow In these notes, all graphs are undirected. We consider matching, covering, and packing in bipartite graphs, general graphs, and hypergraphs. We also
Purpose of sequence assembly
 Sequence Assembly Purpose of sequence assembly Reconstruct long DNA/RNA sequences from short sequence reads Genome sequencing RNA sequencing for gene discovery Amplicon sequencing But not for transcript
Sequence Assembly Purpose of sequence assembly Reconstruct long DNA/RNA sequences from short sequence reads Genome sequencing RNA sequencing for gene discovery Amplicon sequencing But not for transcript
Assembly in the Clouds
 Assembly in the Clouds Michael Schatz October 13, 2010 Beyond the Genome Shredded Book Reconstruction Dickens accidentally shreds the first printing of A Tale of Two Cities Text printed on 5 long spools
Assembly in the Clouds Michael Schatz October 13, 2010 Beyond the Genome Shredded Book Reconstruction Dickens accidentally shreds the first printing of A Tale of Two Cities Text printed on 5 long spools
How to apply de Bruijn graphs to genome assembly
 PRIMER How to apply de Bruijn graphs to genome assembly Phillip E C Compeau, Pavel A Pevzner & lenn Tesler A mathematical concept known as a de Bruijn graph turns the formidable challenge of assembling
PRIMER How to apply de Bruijn graphs to genome assembly Phillip E C Compeau, Pavel A Pevzner & lenn Tesler A mathematical concept known as a de Bruijn graph turns the formidable challenge of assembling
Discrete mathematics , Fall Instructor: prof. János Pach
 Discrete mathematics 2016-2017, Fall Instructor: prof. János Pach - covered material - Lecture 1. Counting problems To read: [Lov]: 1.2. Sets, 1.3. Number of subsets, 1.5. Sequences, 1.6. Permutations,
Discrete mathematics 2016-2017, Fall Instructor: prof. János Pach - covered material - Lecture 1. Counting problems To read: [Lov]: 1.2. Sets, 1.3. Number of subsets, 1.5. Sequences, 1.6. Permutations,
Read Naming Format Specification
 Read Naming Format Specification Karel Břinda Valentina Boeva Gregory Kucherov Version 0.1.3 (4 August 2015) Abstract This document provides a standard for naming simulated Next-Generation Sequencing (Ngs)
Read Naming Format Specification Karel Břinda Valentina Boeva Gregory Kucherov Version 0.1.3 (4 August 2015) Abstract This document provides a standard for naming simulated Next-Generation Sequencing (Ngs)
Manual of SOAPdenovo-Trans-v1.03. Yinlong Xie, Gengxiong Wu, Jingbo Tang,
 Manual of SOAPdenovo-Trans-v1.03 Yinlong Xie, 2013-07-19 Gengxiong Wu, 2013-07-19 Jingbo Tang, 2013-07-19 ********** Introduction SOAPdenovo-Trans is a de novo transcriptome assembler basing on the SOAPdenovo
Manual of SOAPdenovo-Trans-v1.03 Yinlong Xie, 2013-07-19 Gengxiong Wu, 2013-07-19 Jingbo Tang, 2013-07-19 ********** Introduction SOAPdenovo-Trans is a de novo transcriptome assembler basing on the SOAPdenovo
CSE 549: Genome Assembly De Bruijn Graph. All slides in this lecture not marked with * courtesy of Ben Langmead.
 CSE 549: Genome Assembly De Bruijn Graph All slides in this lecture not marked with * courtesy of Ben Langmead. Real-world assembly methods OLC: Overlap-Layout-Consensus assembly DBG: De Bruijn graph assembly
CSE 549: Genome Assembly De Bruijn Graph All slides in this lecture not marked with * courtesy of Ben Langmead. Real-world assembly methods OLC: Overlap-Layout-Consensus assembly DBG: De Bruijn graph assembly
T-IDBA: A de novo Iterative de Bruijn Graph Assembler for Transcriptome
 T-IDBA: A de novo Iterative de Bruin Graph Assembler for Transcriptome Yu Peng, Henry C.M. Leung, S.M. Yiu, Francis Y.L. Chin Department of Computer Science, The University of Hong Kong Pokfulam Road,
T-IDBA: A de novo Iterative de Bruin Graph Assembler for Transcriptome Yu Peng, Henry C.M. Leung, S.M. Yiu, Francis Y.L. Chin Department of Computer Science, The University of Hong Kong Pokfulam Road,
IDBA - A practical Iterative de Bruijn Graph De Novo Assembler
 IDBA - A practical Iterative de Bruijn Graph De Novo Assembler Speaker: Gabriele Capannini May 21, 2010 Introduction De Novo Assembly assembling reads together so that they form a new, previously unknown
IDBA - A practical Iterative de Bruijn Graph De Novo Assembler Speaker: Gabriele Capannini May 21, 2010 Introduction De Novo Assembly assembling reads together so that they form a new, previously unknown
Partha Sarathi Mandal
 MA 515: Introduction to Algorithms & MA353 : Design and Analysis of Algorithms [3-0-0-6] Lecture 39 http://www.iitg.ernet.in/psm/indexing_ma353/y09/index.html Partha Sarathi Mandal psm@iitg.ernet.in Dept.
MA 515: Introduction to Algorithms & MA353 : Design and Analysis of Algorithms [3-0-0-6] Lecture 39 http://www.iitg.ernet.in/psm/indexing_ma353/y09/index.html Partha Sarathi Mandal psm@iitg.ernet.in Dept.
Building approximate overlap graphs for DNA assembly using random-permutations-based search.
 An algorithm is presented for fast construction of graphs of reads, where an edge between two reads indicates an approximate overlap between the reads. Since the algorithm finds approximate overlaps directly,
An algorithm is presented for fast construction of graphs of reads, where an edge between two reads indicates an approximate overlap between the reads. Since the algorithm finds approximate overlaps directly,
Theorem 2.9: nearest addition algorithm
 There are severe limits on our ability to compute near-optimal tours It is NP-complete to decide whether a given undirected =(,)has a Hamiltonian cycle An approximation algorithm for the TSP can be used
There are severe limits on our ability to compute near-optimal tours It is NP-complete to decide whether a given undirected =(,)has a Hamiltonian cycle An approximation algorithm for the TSP can be used
Unit 8: Coping with NP-Completeness. Complexity classes Reducibility and NP-completeness proofs Coping with NP-complete problems. Y.-W.
 : Coping with NP-Completeness Course contents: Complexity classes Reducibility and NP-completeness proofs Coping with NP-complete problems Reading: Chapter 34 Chapter 35.1, 35.2 Y.-W. Chang 1 Complexity
: Coping with NP-Completeness Course contents: Complexity classes Reducibility and NP-completeness proofs Coping with NP-complete problems Reading: Chapter 34 Chapter 35.1, 35.2 Y.-W. Chang 1 Complexity
Reducing Genome Assembly Complexity with Optical Maps Final Report
 Reducing Genome Assembly Complexity with Optical Maps Final Report Lee Mendelowitz LMendelo@math.umd.edu Advisor: Dr. Mihai Pop Computer Science Department Center for Bioinformatics and Computational Biology
Reducing Genome Assembly Complexity with Optical Maps Final Report Lee Mendelowitz LMendelo@math.umd.edu Advisor: Dr. Mihai Pop Computer Science Department Center for Bioinformatics and Computational Biology
CS 68: BIOINFORMATICS. Prof. Sara Mathieson Swarthmore College Spring 2018
 CS 68: BIOINFORMATICS Prof. Sara Mathieson Swarthmore College Spring 2018 Outline: Jan 31 DBG assembly in practice Velvet assembler Evaluation of assemblies (if time) Start: string alignment Candidate
CS 68: BIOINFORMATICS Prof. Sara Mathieson Swarthmore College Spring 2018 Outline: Jan 31 DBG assembly in practice Velvet assembler Evaluation of assemblies (if time) Start: string alignment Candidate
02-711/ Computational Genomics and Molecular Biology Fall 2016
 Literature assignment 2 Due: Nov. 3 rd, 2016 at 4:00pm Your name: Article: Phillip E C Compeau, Pavel A. Pevzner, Glenn Tesler. How to apply de Bruijn graphs to genome assembly. Nature Biotechnology 29,
Literature assignment 2 Due: Nov. 3 rd, 2016 at 4:00pm Your name: Article: Phillip E C Compeau, Pavel A. Pevzner, Glenn Tesler. How to apply de Bruijn graphs to genome assembly. Nature Biotechnology 29,
RNA-seq. Manpreet S. Katari
 RNA-seq Manpreet S. Katari Evolution of Sequence Technology Normalizing the Data RPKM (Reads per Kilobase of exons per million reads) Score = R NT R = # of unique reads for the gene N = Size of the gene
RNA-seq Manpreet S. Katari Evolution of Sequence Technology Normalizing the Data RPKM (Reads per Kilobase of exons per million reads) Score = R NT R = # of unique reads for the gene N = Size of the gene
Sequence Analysis Pipeline
 Sequence Analysis Pipeline Transcript fragments 1. PREPROCESSING 2. ASSEMBLY (today) Removal of contaminants, vector, adaptors, etc Put overlapping sequence together and calculate bigger sequences 3. Analysis/Annotation
Sequence Analysis Pipeline Transcript fragments 1. PREPROCESSING 2. ASSEMBLY (today) Removal of contaminants, vector, adaptors, etc Put overlapping sequence together and calculate bigger sequences 3. Analysis/Annotation
MITOCW watch?v=zyw2aede6wu
 MITOCW watch?v=zyw2aede6wu The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high quality educational resources for free. To
MITOCW watch?v=zyw2aede6wu The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high quality educational resources for free. To
CS 173, Lecture B Introduction to Genome Assembly (using Eulerian Graphs) Tandy Warnow
 CS 173, Lecture B Introduction to Genome Assembly (using Eulerian Graphs) Tandy Warnow 2 Shotgun DNA Sequencing DNA target sample SHEAR & SIZE End Reads / Mate Pairs 550bp 10,000bp Not all sequencing technologies
CS 173, Lecture B Introduction to Genome Assembly (using Eulerian Graphs) Tandy Warnow 2 Shotgun DNA Sequencing DNA target sample SHEAR & SIZE End Reads / Mate Pairs 550bp 10,000bp Not all sequencing technologies
Module 6 NP-Complete Problems and Heuristics
 Module 6 NP-Complete Problems and Heuristics Dr. Natarajan Meghanathan Professor of Computer Science Jackson State University Jackson, MS 39217 E-mail: natarajan.meghanathan@jsums.edu P, NP-Problems Class
Module 6 NP-Complete Problems and Heuristics Dr. Natarajan Meghanathan Professor of Computer Science Jackson State University Jackson, MS 39217 E-mail: natarajan.meghanathan@jsums.edu P, NP-Problems Class
Design and Analysis of Algorithms
 CSE 101, Winter 018 D/Q Greed SP s DP LP, Flow B&B, Backtrack Metaheuristics P, NP Design and Analysis of Algorithms Lecture 8: Greed Class URL: http://vlsicad.ucsd.edu/courses/cse101-w18/ Optimization
CSE 101, Winter 018 D/Q Greed SP s DP LP, Flow B&B, Backtrack Metaheuristics P, NP Design and Analysis of Algorithms Lecture 8: Greed Class URL: http://vlsicad.ucsd.edu/courses/cse101-w18/ Optimization
CSCI 1820 Notes. Scribes: tl40. February 26 - March 02, Estimating size of graphs used to build the assembly.
 CSCI 1820 Notes Scribes: tl40 February 26 - March 02, 2018 Chapter 2. Genome Assembly Algorithms 2.1. Statistical Theory 2.2. Algorithmic Theory Idury-Waterman Algorithm Estimating size of graphs used
CSCI 1820 Notes Scribes: tl40 February 26 - March 02, 2018 Chapter 2. Genome Assembly Algorithms 2.1. Statistical Theory 2.2. Algorithmic Theory Idury-Waterman Algorithm Estimating size of graphs used
CS270 Combinatorial Algorithms & Data Structures Spring Lecture 19:
 CS270 Combinatorial Algorithms & Data Structures Spring 2003 Lecture 19: 4.1.03 Lecturer: Satish Rao Scribes: Kevin Lacker and Bill Kramer Disclaimer: These notes have not been subjected to the usual scrutiny
CS270 Combinatorial Algorithms & Data Structures Spring 2003 Lecture 19: 4.1.03 Lecturer: Satish Rao Scribes: Kevin Lacker and Bill Kramer Disclaimer: These notes have not been subjected to the usual scrutiny
Approximation Algorithms
 15-251: Great Ideas in Theoretical Computer Science Spring 2019, Lecture 14 March 5, 2019 Approximation Algorithms 1 2 SAT 3SAT Clique Hamiltonian- Cycle given a Boolean formula F, is it satisfiable? same,
15-251: Great Ideas in Theoretical Computer Science Spring 2019, Lecture 14 March 5, 2019 Approximation Algorithms 1 2 SAT 3SAT Clique Hamiltonian- Cycle given a Boolean formula F, is it satisfiable? same,
Questions? You are given the complete graph of Facebook. What questions would you ask? (What questions could we hope to answer?)
 P vs. NP What now? Attribution These slides were prepared for the New Jersey Governor s School course The Math Behind the Machine taught in the summer of 2011 by Grant Schoenebeck Large parts of these
P vs. NP What now? Attribution These slides were prepared for the New Jersey Governor s School course The Math Behind the Machine taught in the summer of 2011 by Grant Schoenebeck Large parts of these
More NP-complete Problems. CS255 Chris Pollett May 3, 2006.
 More NP-complete Problems CS255 Chris Pollett May 3, 2006. Outline More NP-Complete Problems Hamiltonian Cycle Recall a hamiltonian cycle is a permutation of the vertices v i_1,, v i_n of a graph G so
More NP-complete Problems CS255 Chris Pollett May 3, 2006. Outline More NP-Complete Problems Hamiltonian Cycle Recall a hamiltonian cycle is a permutation of the vertices v i_1,, v i_n of a graph G so
Eulerian Tours and Fleury s Algorithm
 Eulerian Tours and Fleury s Algorithm CSE21 Winter 2017, Day 12 (B00), Day 8 (A00) February 8, 2017 http://vlsicad.ucsd.edu/courses/cse21-w17 Vocabulary Path (or walk): describes a route from one vertex
Eulerian Tours and Fleury s Algorithm CSE21 Winter 2017, Day 12 (B00), Day 8 (A00) February 8, 2017 http://vlsicad.ucsd.edu/courses/cse21-w17 Vocabulary Path (or walk): describes a route from one vertex
KisSplice. Identifying and Quantifying SNPs, indels and Alternative Splicing Events from RNA-seq data. 29th may 2013
 Identifying and Quantifying SNPs, indels and Alternative Splicing Events from RNA-seq data 29th may 2013 Next Generation Sequencing A sequencing experiment now produces millions of short reads ( 100 nt)
Identifying and Quantifying SNPs, indels and Alternative Splicing Events from RNA-seq data 29th may 2013 Next Generation Sequencing A sequencing experiment now produces millions of short reads ( 100 nt)
Computational Architecture of Cloud Environments Michael Schatz. April 1, 2010 NHGRI Cloud Computing Workshop
 Computational Architecture of Cloud Environments Michael Schatz April 1, 2010 NHGRI Cloud Computing Workshop Cloud Architecture Computation Input Output Nebulous question: Cloud computing = Utility computing
Computational Architecture of Cloud Environments Michael Schatz April 1, 2010 NHGRI Cloud Computing Workshop Cloud Architecture Computation Input Output Nebulous question: Cloud computing = Utility computing
Discrete Mathematics and Probability Theory Fall 2013 Vazirani Note 7
 CS 70 Discrete Mathematics and Probability Theory Fall 2013 Vazirani Note 7 An Introduction to Graphs A few centuries ago, residents of the city of Königsberg, Prussia were interested in a certain problem.
CS 70 Discrete Mathematics and Probability Theory Fall 2013 Vazirani Note 7 An Introduction to Graphs A few centuries ago, residents of the city of Königsberg, Prussia were interested in a certain problem.
The Value of Mate-pairs for Repeat Resolution
 The Value of Mate-pairs for Repeat Resolution An Analysis on Graphs Created From Short Reads Joshua Wetzel Department of Computer Science Rutgers University Camden in conjunction with CBCB at University
The Value of Mate-pairs for Repeat Resolution An Analysis on Graphs Created From Short Reads Joshua Wetzel Department of Computer Science Rutgers University Camden in conjunction with CBCB at University
Alignment of Long Sequences
 Alignment of Long Sequences BMI/CS 776 www.biostat.wisc.edu/bmi776/ Spring 2009 Mark Craven craven@biostat.wisc.edu Pairwise Whole Genome Alignment: Task Definition Given a pair of genomes (or other large-scale
Alignment of Long Sequences BMI/CS 776 www.biostat.wisc.edu/bmi776/ Spring 2009 Mark Craven craven@biostat.wisc.edu Pairwise Whole Genome Alignment: Task Definition Given a pair of genomes (or other large-scale
Theory of Computing. Lecture 10 MAS 714 Hartmut Klauck
 Theory of Computing Lecture 10 MAS 714 Hartmut Klauck Seven Bridges of Königsberg Can one take a walk that crosses each bridge exactly once? Seven Bridges of Königsberg Model as a graph Is there a path
Theory of Computing Lecture 10 MAS 714 Hartmut Klauck Seven Bridges of Königsberg Can one take a walk that crosses each bridge exactly once? Seven Bridges of Königsberg Model as a graph Is there a path
14 More Graphs: Euler Tours and Hamilton Cycles
 14 More Graphs: Euler Tours and Hamilton Cycles 14.1 Degrees The degree of a vertex is the number of edges coming out of it. The following is sometimes called the First Theorem of Graph Theory : Lemma
14 More Graphs: Euler Tours and Hamilton Cycles 14.1 Degrees The degree of a vertex is the number of edges coming out of it. The following is sometimes called the First Theorem of Graph Theory : Lemma
Optimal tour along pubs in the UK
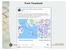 1 From Facebook Optimal tour along 24727 pubs in the UK Road distance (by google maps) see also http://www.math.uwaterloo.ca/tsp/pubs/index.html (part of TSP homepage http://www.math.uwaterloo.ca/tsp/
1 From Facebook Optimal tour along 24727 pubs in the UK Road distance (by google maps) see also http://www.math.uwaterloo.ca/tsp/pubs/index.html (part of TSP homepage http://www.math.uwaterloo.ca/tsp/
Walking with Euler through Ostpreußen and RNA
 Walking with Euler through Ostpreußen and RNA Mark Muldoon February 4, 2010 Königsberg (1652) Kaliningrad (2007)? The Königsberg Bridge problem asks whether it is possible to walk around the old city in
Walking with Euler through Ostpreußen and RNA Mark Muldoon February 4, 2010 Königsberg (1652) Kaliningrad (2007)? The Königsberg Bridge problem asks whether it is possible to walk around the old city in
Outline. CS38 Introduction to Algorithms. Approximation Algorithms. Optimization Problems. Set Cover. Set cover 5/29/2014. coping with intractibility
 Outline CS38 Introduction to Algorithms Lecture 18 May 29, 2014 coping with intractibility approximation algorithms set cover TSP center selection randomness in algorithms May 29, 2014 CS38 Lecture 18
Outline CS38 Introduction to Algorithms Lecture 18 May 29, 2014 coping with intractibility approximation algorithms set cover TSP center selection randomness in algorithms May 29, 2014 CS38 Lecture 18
CSE 100: B+ TREE, 2-3 TREE, NP- COMPLETNESS
 CSE 100: B+ TREE, 2-3 TREE, NP- COMPLETNESS Analyzing find in B-trees Since the B-tree is perfectly height-balanced, the worst case time cost for find is O(logN) Best case: If every internal node is completely
CSE 100: B+ TREE, 2-3 TREE, NP- COMPLETNESS Analyzing find in B-trees Since the B-tree is perfectly height-balanced, the worst case time cost for find is O(logN) Best case: If every internal node is completely
Instant Insanity Instructor s Guide Make-it and Take-it Kit for AMTNYS 2006
 Instant Insanity Instructor s Guide Make-it and Take-it Kit for AMTNYS 2006 THE KIT: This kit contains materials for two Instant Insanity games, a student activity sheet with answer key and this instructor
Instant Insanity Instructor s Guide Make-it and Take-it Kit for AMTNYS 2006 THE KIT: This kit contains materials for two Instant Insanity games, a student activity sheet with answer key and this instructor
Lecture 24: More Reductions (1997) Steven Skiena. skiena
 Lecture 24: More Reductions (1997) Steven Skiena Department of Computer Science State University of New York Stony Brook, NY 11794 4400 http://www.cs.sunysb.edu/ skiena Prove that subgraph isomorphism
Lecture 24: More Reductions (1997) Steven Skiena Department of Computer Science State University of New York Stony Brook, NY 11794 4400 http://www.cs.sunysb.edu/ skiena Prove that subgraph isomorphism
Sequence Design Problems in Discovery of Regulatory Elements
 Sequence Design Problems in Discovery of Regulatory Elements Yaron Orenstein, Bonnie Berger and Ron Shamir Regulatory Genomics workshop Simons Institute March 10th, 2016, Berkeley, CA Differentially methylated
Sequence Design Problems in Discovery of Regulatory Elements Yaron Orenstein, Bonnie Berger and Ron Shamir Regulatory Genomics workshop Simons Institute March 10th, 2016, Berkeley, CA Differentially methylated
Traveling Salesman Problem. Algorithms and Networks 2014/2015 Hans L. Bodlaender Johan M. M. van Rooij
 Traveling Salesman Problem Algorithms and Networks 2014/2015 Hans L. Bodlaender Johan M. M. van Rooij 1 Contents TSP and its applications Heuristics and approximation algorithms Construction heuristics,
Traveling Salesman Problem Algorithms and Networks 2014/2015 Hans L. Bodlaender Johan M. M. van Rooij 1 Contents TSP and its applications Heuristics and approximation algorithms Construction heuristics,
High-throughput sequencing: Alignment and related topic. Simon Anders EMBL Heidelberg
 High-throughput sequencing: Alignment and related topic Simon Anders EMBL Heidelberg Established platforms HTS Platforms Illumina HiSeq, ABI SOLiD, Roche 454 Newcomers: Benchtop machines 454 GS Junior,
High-throughput sequencing: Alignment and related topic Simon Anders EMBL Heidelberg Established platforms HTS Platforms Illumina HiSeq, ABI SOLiD, Roche 454 Newcomers: Benchtop machines 454 GS Junior,
CS161 - Final Exam Computer Science Department, Stanford University August 16, 2008
 CS161 - Final Exam Computer Science Department, Stanford University August 16, 2008 Name: Honor Code 1. The Honor Code is an undertaking of the students, individually and collectively: a) that they will
CS161 - Final Exam Computer Science Department, Stanford University August 16, 2008 Name: Honor Code 1. The Honor Code is an undertaking of the students, individually and collectively: a) that they will
Multiple sequence alignment accuracy estimation and its role in creating an automated bioinformatician
 Multiple sequence alignment accuracy estimation and its role in creating an automated bioinformatician Dan DeBlasio dandeblasio.com danfdeblasio StringBio 2018 Tunable parameters!2 Tunable parameters Quant
Multiple sequence alignment accuracy estimation and its role in creating an automated bioinformatician Dan DeBlasio dandeblasio.com danfdeblasio StringBio 2018 Tunable parameters!2 Tunable parameters Quant
Miniproject 1. Part 1 Due: 16 February. The coverage problem. Method. Why it is hard. Data. Task1
 Miniproject 1 Part 1 Due: 16 February The coverage problem given an assembled transcriptome (RNA) and a reference genome (DNA) 1. 2. what fraction (in bases) of the transcriptome sequences match to annotated
Miniproject 1 Part 1 Due: 16 February The coverage problem given an assembled transcriptome (RNA) and a reference genome (DNA) 1. 2. what fraction (in bases) of the transcriptome sequences match to annotated
