Voronoi-Based Segmentation of Cells on Image Manifolds
|
|
|
- Rosamund Brown
- 5 years ago
- Views:
Transcription
1 Voronoi-Based Segmentation of Cells on Image Manifolds Thouis R. Jones 1, Anne Carpenter 2, and Polina Golland 1 1 MIT CSAIL, Cambridge, MA, USA 2 Whitehead Institute for Biomedical Research, Cambridge, MA, USA Abstract. We present a method for finding the boundaries between adjacent regions in an image, where seed areas have already been identified in the individual regions to be segmented. This method was motivated by the problem of finding the borders of cells in microscopy images, given a labelling of the nuclei in the images. The method finds the Voronoi region of each seed on a manifold with a metric controlled by local image properties. We discuss similarities to other methods based on image-controlled metrics, such as Geodesic Active Contours, and give a fast algorithm for computing the Voronoi regions. We validate our method against hand-traced boundaries for cell images. 1 Introduction Image cytometry, the measurement of cell properties from microscopy images, has become an important tool for biological research. In particular, high-throughput experiments rely on automatic processing of images to deal with the large amount of data they produce [3]. A fundamental operation in cell-image analysis is identifying individual cell boundaries. This is often difficult because there are many different staining protocols, leading to dramatically different appearances for cells. Moreover, the difference between cell interior and cell border may not be very pronounced (see Figures 1 and 2). Identification of individual cells allows much more powerful analysis of the resulting data than methods that provide only mean measurements for cell populations. For example, expression data from a protein interaction chip cannot differentiate a bimodal population and a unimodal population if they have the same mean expression levels. Measurements of individual cells prevents conflation of subpopulations. It is almost always the case that the nuclei of cells are more easily identifiable, because they have a more uniform appearance and shape, are brighter relative to the background when stained, and do not abut one another, as cells do. They are also usually interior to the cells. This leads us to phrase the problem of segmenting cells as one of identifying boundaries between regions given seeds in individual regions from which to start the segmentation. Current methods for identifying cells for image cytometry sometimes use a fixed offset around the nuclei. However, this fixed offset requires tuning to different cell types, and does not provide information about phenotypes that cause changes in cell size or shape [2]. Another common approach is to use watershed segmentation to identify cell boundaries [1, 8], however, this is often fragile. Watershed segmentation treats the image as a
2 height field, and segments pixels according to which minimum a drop of water would flow to if placed on that pixel in the height field. Morphological operations are used to impose a limited set of minima, equivalent to our seed regions. Watershed is quite unstable, because a single noisy pixel can allow large groups of pixels to change segmentation, by creating a gap leading to a different minimum. We avoid this fragility in two ways: first, by comparing neighborhoods of pixels rather than individual pixels, and second, by including a regularization factor to provide reasonable behavior when the image data does not contain a strong enough edge between two seed regions. In the limit, our regularization approaches a 2D Voronoi segmentation, i.e., pixels are assigned to the nearest seed region measured in the image plane, without reference to image features. Our approach is to define a metric in the image plane and to calculate distances from seed regions according to this metric. Pixels are then assigned to cells according to their distance from the corresponding nucleus under that metric. The metric uses information about image edges, both their strength and their orientation, as well as a regularization term corresponding to inter-pixel distance within the image. 2 Method Our method operates by computing a discretized approximation of the Voronoi regions of each seed on a manifold with a metric controlled by local image features. The metric defines the incremental distance in a particular direction in the image/manifold. Its behavior is such that adjacent pixels with similar surrounds are close to one another, while pixels whose surrounds differ are treated as farther apart. We introduce a Riemannian metric defined in terms of the image I and a regularization parameter λ, as G = g(i) gt (I) + λi, (1) 1 + λ where I is the 2 2 identity matrix. The function g maps images to images, and in our application is generally a small-radius blur. The effect of this blur is to combine a weighted neighborhood in the gradient computation, to avoid relying too much on single (noisy) pixel values. Infinitesimal distances under G are measured by dx 2 G dx T Gdx = (dxt g(i)) 2 + λ(dx T dx) 2. (2) λ + 1 The first term in the numerator of (2), dx T g(i) 2, increases distances measured parallel to large gradients in g(i). The regularization effect of λ can be seen by lim λ dx 2 G = dx T dx = dx 2 2, (3) i.e., G becomes more Euclidean as λ increases. Given (2), we can compute the distance between any two points in an image as the shortest path between those points. We use a discretized approximation, i.e., a chamfer distance, applied to an 8-connected neighborhood. This approximation to the distance
3 is generally no worse than 10% of the correct value [7], which for our application is generally not more than a single pixel. Pixels overlapping seed regions are initialized to be distance 0 from their seed, and the distances of the remaining pixels are computed by Dijkstra s algorithm [6]. Each pixel is labelled with the seed it is closest to, i.e., the Voronoi region of that seed in the manifold defined by (2). Computing the seed-to-pixel distances in this manner also makes it trivial to limit the segmentation to a predetermined foreground region (for example, the result from a global thresholding step). The computation of Voronoi regions places the boundary between two adjacent regions at pixels that are equidistant from each seed, as measured on the manifold from (2). The inter-pixel distance is larger where the image is changing more according to g(i), so boundaries tend to align with image differences. The regularizing parameter λ allows the user to make the manifold more flat according to prior knowledge about region shapes, and the choice of g (e.g., the radius of smoothing) controls how sensitive distances are to small features. In this work, we use a narrow (3- or 5-pixel radius) Gaussian blur for g. 2.1 Connection to Geodesic Active Contours Our algorithm is related to Geodesic Active Contours [5]. The full details of that work are not given here, but we discuss the connection briefly. Active contours can be seen as finding a shortest path in a Riemannian space, where distances between pixels are defined by an edge stopping function g : R + R +. 3 Examining equation (8) from Caselles et al. [5] helps establish the similarity: Min 1 0 g( I(C(q) ) C (q) dq (4) where I is the image, C(q) is the curve on image that we are minimizing over, and q is the parameter along the curve. The edge stopping function g is strictly decreasing and positive, with g( ) = 0. The effect of g s interaction with I is such that the minimum curve follows larger gradients in the image. The minimization can also be written as (equation (12) of [5]) Min L(C) 0 g( I(C(s) )ds (5) where s is the arclength parameter for C, and L(C) is the length of C. Therefore, active contours can be seen as seeking a minimum length curve where the length depends on image characteristics [5]. Our goal is different from active contours, since we hope to find boundaries between regions corresponding to different seeds. However, we do seek shortest paths with a distance metric controlled by image characteristics. We do not wish to follow boundaries, as in active contours, but rather to avoid them (equivalent to making boundaries far 3 In this subsection, g is an edge stopping function, not the same as g in the previous subsection.
4 from the seed regions in our formulation). However, just as Voronoi regions can be defined in terms of shortest paths, our segmentation algorithm can be defined in terms of shortest paths defined by equation (1) and image properties. The metrics in the two approaches differ, specifically in their treatment of edges in the image: edges in the active contour setting have a metric that makes points along the edge closer, while our metric makes the points across the edge more separated. Moreover, active contours use a directionally uniform metric, while ours is not, since it is larger across edges rather than along them. However, the goal is the same: allow the computation of inter-pixel distances that simplify the problem at hand and map it to a simpler framework. In both cases, the problem reduces to that of finding shortest paths (though this is simply a jumping-off point for active contours to more powerful and efficient methods such as level-sets). 3 Experiments and Evaluation In order to provide insight into the behavior of the metric defined in (1), we experiment with synthetic data and adjust λ, trading off the effect of image features on the region segmentation versus the Euclidean distance from the seed. As can be seen in Figures 3 and 4, the segmentation approaches the Voronoi diagram of the seed regions (i.e., ignoring image features) as λ increases. Also, note that in the noisy example less regularization (i.e., a lower value for λ) is needed to achieve similar segmentations because the noise makes the edges in the image less pronounced than in the noise-free example. In effect, increasing noise provides a form of regularization. Our algorithm is currently part of an automatic image cytometry program [4], and has been used in a variety of experiments with several cell types of varying morphologies. To evaluate our algorithm on real world data, we compare it to the manual segmentation of cell images. Sixteen images taken from Drosophila cells stained for DNA (to label nuclei) and Actin (a cytoskeletal protein, to show the cell body) were outlined. First, nuclei were outlined by hand. The nuclear outlines were overlaid on the cell images, and one cell per nucleus was outlined. Our algorithm was then applied to the same cell images and nuclear-outlines data, and the results compared by computing the signed distance between boundary pixels. It effectively computes the distance for each pixel in the automatic segmentation boundary to the corresponding manual segmentation boundary. Hand-outlined nuclei were used, rather than automatically segmented nuclei, since our algorithm does not address nuclei segmentation, and we want the comparison to be as meaningful as possible. In general, automatic segmentation of nuclei is fairly simple (See section 1.) Our algorithm does not compute a foreground/background separation, but instead relies on a such a label for each pixel to be given as input. For comparing our algorithm to the manual segmentation, we compute the foreground pixels as the union of cells identified in the manual segmentation. Methods exist for automatically choosing a foreground/background labelling, but distinguishing foreground from background is not part of our algorithm. For the purposes of this comparison, we also use seed regions from hand-outlined nuclei. In general, nuclei are more compact, separated, and brightly stained than cells,
5 Fig. 1. Typical results from our algorithm. Internal shapes are seed regions, taken from manual segmentation of nuclei, and are the same in both rows. Bold lines show cell/cell boundaries that are compared. Top row: Cell images, with nuclei outlined. Middle row: Automatic segmentation with our method. Bottom row: Manual segmentation. Fig. 2. Five worst outliers from evaluation set, as measured by distance between manual and automatic segmentation boundaries. Internal shapes are seed regions, taken from manual segmentation of nuclei, and are the same in both rows. Bold lines show cell/cell boundaries that are compared. Top row: Cell images, with nuclei outlined. Middle row: Automatic segmentation with our method. Bottom row: Manual segmentation.
6 Fig. 3. Synthetic example. The input image is in the upper left, with seed sites marked with dots. From left to right across the two rows, the resulting distances calculated with our metric are shown, with the resulting segmentation overlaid white lines, for λ equal to 0.2, 0.3, 0.4, 0.5, 0.6, 0.8, and 3.0. The segmentation lines follow the ridges in the distances function. As can be seen, as λ increases, the segmentation approaches the Voronoi diagram of the seed regions. Fig. 4. Synthetic example with noise. The same input as in Figure 3 is used, with zero-mean Gaussian noise with standard deviation 0.5 added to each pixel. Edges were 1.0 and background was 0.0 before noise was added. The layout is the same as in Figure 3, but with λ equal to 0.025, 0.05, 0.075, 0.1, 0.125, 0.2, and 0.75.
7 Fig. 5. Combined histogram for the signed distances and cumulative distribution of absolute distances from automatic segmentation to manual segmentation for the sixteen images in our test set. and so are usually easily segmented by simple thresholding. We chose to use the nuclei from manual segmentation in order to keep the conditions for manual segmentation and our algorithm as similar as possible. Under these conditions, our algorithm is only responsible for computing cell-cell boundaries, rather than cell-background boundaries, which are fixed. We therefore only consider the distance between boundary pixels in the two images that have a common cell-label on either border. When evaluating the algorithm, we use the one-sided signed distance (negative inside cells) from the automatic segmentation to the manual segmentation. We set λ in (1) to 0.05 times the distance between the average foreground and background pixel intensities on a per-image basis. This value for λ was found to be close to optimal in our experiments, with fairly stable behavior for a reasonably large range (within a factor of two). Our test set includes a wide variety of cell types, with different sizes and morphologies. In general, most applications would have more homogeneous data, to which λ should be tuned. Sixteen images made up the test set. Each image was roughly 512x512 pixels on a side, with cells roughly 25 pixels in diameter, and 80 cells per image on average. Across the entire set, there were 21.6k pixels on a cell-cell boundary in the automatic segmentation. A histogram of their signed distances with respect to the manual segmentation is shown in Figure 5. Sixty-four percent (14.0k) of the boundary pixels in the automatic segmentation are within 2 pixels in distance from the corresponding manual boundary. Ninety-two percent (19.8k) of the pixels are within 5 pixels. The accuracy of the hand-labelling is around 3 pixels, based on the width of the marker used to outline the cells. Typical results on the test set are shown in Figure 1. We show the top 5 worst outliers from the data set, based on maximum boundary distance, in Figure 2. In some cases, the automatic method has chosen a different edge in the image to use as the cell-cell boundary. In others, the close proximity of the nucleus to a cell boundary has caused the
8 automatic segmentation to move past the boundary chosen in the manual segmentation, causing large deviations between the two. 4 Discussion In some segmentation tasks seed regions for segmentation are easily identified. Nuclei act as such seed regions in our application, the segmentation of cells in microscopy images. The more difficult task is finding the boundaries between cells that share a common border. Cell appearance in the images is not uniform, varying depending on the type of cells and the protocol used to stain them, and simplistic approaches are not sufficiently accurate or robust. We have demonstrated an algorithm for segmentation of image regions based on seed areas that respects image boundaries as defined by a difference operator. The algorithm computes an approximation of the Voronoi region of a seed on a manifold, implicitly defined by a difference operator that operates on image neighborhoods. The only assumption that the algorithm relies on is that images change more near the borders of regions. This is similar to the behavior of Geodesic Active Contours, with an inversion in the behavior of the metrics near edges. We approximate distances using a chamfer-like difference operator, and use Dijkstra s algorithm for computing individual regions quickly. Our algorithm is currently implemented in an automatic image cytometry package [4], and has been used successfully in several experiments with a variety of cell types and morphologies. We have compared our algorithm to manual segmentation by an expert of microscopy cell images. Our algorithm performs well. There are a few failure cases, perhaps due to the fairly simple prior on cell shape and size that our metric implies. A more complex prior could incorporate measures of, e.g., cell roundness or cell area. Although our method is more accurate for segmenting cells than others, such as using fixed offsets from nuclei or the watershed transform, the true test of its usefulness is in whether it produces more accurate measurements of cellular phenotypes. In order to understand how segmentation accuracy affects measurement accuracy, we plan to validate against data from flow cytometry or another method not based on image processing. In the future, we would like to explore other choices for g in (1). It might also be possible to use the distance defined by our metric to compute foreground/background labellings and to detect poorly-segmented cells. Acknowledgements The authors would like to thank In-Han Kang for his work on validation of the algorithm, and Chris Gang for the hand-outlined nucleus and cell images, and David Sabatini for his support. References 1. S. Beucher The Watershed Transformation Applied to Image Segmentation, Scanning Microscopy International, Vol. 6, pp , 1992.
9 2. Boutros M., A. Kiger, S. Armknecht, K. Kerr, M. Hild, B. Koch, S. Haas, Heidelberg Fly Array Consortium, R. Paro and N. Perrimon Genome-wide RNAi Analysis of Cell Growth and Viability in Drosophila, Science, Vol. 303, pp , A.E. Carpenter and D.M. Sabatini Systematic Genome-wide Screens of Gene Function, Nature Reviews Genetics, Vol. 5, pp , CellProfiler: versatile software for high throughput image analysis, A.E. Carpenter, T.R. Jones, D.B. Wheeler, D.A. Guertin, I.H. Kang, P. Golland, and D.M. Sabatini, in preparation. 5. V. Caselles, R. Kimmel, and G. Sapiro Geodesic Active Contours, Int. J. Comput. Vision, Vol. 22, pp , E. W. Dijkstra A Note on Two Problems in Connection with Graphs, Numerische Math., Vol. 1, pp , U. Montanari A Method for Obtaining Skeletons Using a Quasi-Euclidean Distance, Journal of the ACM, Vol. 15, pp , C. Wählby Algorithms for Applied Digital Image Cytometry, PhD Thesis, 2003
Introduction. Loading Images
 Introduction CellProfiler is a free Open Source software for automated image analysis. Versions for Mac, Windows and Linux are available and can be downloaded at: http://www.cellprofiler.org/. CellProfiler
Introduction CellProfiler is a free Open Source software for automated image analysis. Versions for Mac, Windows and Linux are available and can be downloaded at: http://www.cellprofiler.org/. CellProfiler
Lecture: Segmentation I FMAN30: Medical Image Analysis. Anders Heyden
 Lecture: Segmentation I FMAN30: Medical Image Analysis Anders Heyden 2017-11-13 Content What is segmentation? Motivation Segmentation methods Contour-based Voxel/pixel-based Discussion What is segmentation?
Lecture: Segmentation I FMAN30: Medical Image Analysis Anders Heyden 2017-11-13 Content What is segmentation? Motivation Segmentation methods Contour-based Voxel/pixel-based Discussion What is segmentation?
EECS490: Digital Image Processing. Lecture #22
 Lecture #22 Gold Standard project images Otsu thresholding Local thresholding Region segmentation Watershed segmentation Frequency-domain techniques Project Images 1 Project Images 2 Project Images 3 Project
Lecture #22 Gold Standard project images Otsu thresholding Local thresholding Region segmentation Watershed segmentation Frequency-domain techniques Project Images 1 Project Images 2 Project Images 3 Project
C E N T E R A T H O U S T O N S C H O O L of H E A L T H I N F O R M A T I O N S C I E N C E S. Image Operations II
 T H E U N I V E R S I T Y of T E X A S H E A L T H S C I E N C E C E N T E R A T H O U S T O N S C H O O L of H E A L T H I N F O R M A T I O N S C I E N C E S Image Operations II For students of HI 5323
T H E U N I V E R S I T Y of T E X A S H E A L T H S C I E N C E C E N T E R A T H O U S T O N S C H O O L of H E A L T H I N F O R M A T I O N S C I E N C E S Image Operations II For students of HI 5323
Coarse-to-fine image registration
 Today we will look at a few important topics in scale space in computer vision, in particular, coarseto-fine approaches, and the SIFT feature descriptor. I will present only the main ideas here to give
Today we will look at a few important topics in scale space in computer vision, in particular, coarseto-fine approaches, and the SIFT feature descriptor. I will present only the main ideas here to give
Mathematical Morphology and Distance Transforms. Robin Strand
 Mathematical Morphology and Distance Transforms Robin Strand robin.strand@it.uu.se Morphology Form and structure Mathematical framework used for: Pre-processing Noise filtering, shape simplification,...
Mathematical Morphology and Distance Transforms Robin Strand robin.strand@it.uu.se Morphology Form and structure Mathematical framework used for: Pre-processing Noise filtering, shape simplification,...
Image Processing
 Image Processing 159.731 Canny Edge Detection Report Syed Irfanullah, Azeezullah 00297844 Danh Anh Huynh 02136047 1 Canny Edge Detection INTRODUCTION Edges Edges characterize boundaries and are therefore
Image Processing 159.731 Canny Edge Detection Report Syed Irfanullah, Azeezullah 00297844 Danh Anh Huynh 02136047 1 Canny Edge Detection INTRODUCTION Edges Edges characterize boundaries and are therefore
EE795: Computer Vision and Intelligent Systems
 EE795: Computer Vision and Intelligent Systems Spring 2012 TTh 17:30-18:45 WRI C225 Lecture 04 130131 http://www.ee.unlv.edu/~b1morris/ecg795/ 2 Outline Review Histogram Equalization Image Filtering Linear
EE795: Computer Vision and Intelligent Systems Spring 2012 TTh 17:30-18:45 WRI C225 Lecture 04 130131 http://www.ee.unlv.edu/~b1morris/ecg795/ 2 Outline Review Histogram Equalization Image Filtering Linear
Segmentation of Three Dimensional Cell Culture Models from a Single Focal Plane
 Segmentation of Three Dimensional Cell Culture Models from a Single Focal Plane Hang Chang 1,2 and Bahram Parvin 1 1 Lawrence Berkeley National Laboratory, Berkeley, CA 94720 2 Institute of Automation,
Segmentation of Three Dimensional Cell Culture Models from a Single Focal Plane Hang Chang 1,2 and Bahram Parvin 1 1 Lawrence Berkeley National Laboratory, Berkeley, CA 94720 2 Institute of Automation,
ECG782: Multidimensional Digital Signal Processing
 Professor Brendan Morris, SEB 3216, brendan.morris@unlv.edu ECG782: Multidimensional Digital Signal Processing Spring 2014 TTh 14:30-15:45 CBC C313 Lecture 10 Segmentation 14/02/27 http://www.ee.unlv.edu/~b1morris/ecg782/
Professor Brendan Morris, SEB 3216, brendan.morris@unlv.edu ECG782: Multidimensional Digital Signal Processing Spring 2014 TTh 14:30-15:45 CBC C313 Lecture 10 Segmentation 14/02/27 http://www.ee.unlv.edu/~b1morris/ecg782/
doi: /
 Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
Biomedical Image Analysis. Point, Edge and Line Detection
 Biomedical Image Analysis Point, Edge and Line Detection Contents: Point and line detection Advanced edge detection: Canny Local/regional edge processing Global processing: Hough transform BMIA 15 V. Roth
Biomedical Image Analysis Point, Edge and Line Detection Contents: Point and line detection Advanced edge detection: Canny Local/regional edge processing Global processing: Hough transform BMIA 15 V. Roth
EE368 Project Report CD Cover Recognition Using Modified SIFT Algorithm
 EE368 Project Report CD Cover Recognition Using Modified SIFT Algorithm Group 1: Mina A. Makar Stanford University mamakar@stanford.edu Abstract In this report, we investigate the application of the Scale-Invariant
EE368 Project Report CD Cover Recognition Using Modified SIFT Algorithm Group 1: Mina A. Makar Stanford University mamakar@stanford.edu Abstract In this report, we investigate the application of the Scale-Invariant
Introduction to Medical Imaging (5XSA0) Module 5
 Introduction to Medical Imaging (5XSA0) Module 5 Segmentation Jungong Han, Dirk Farin, Sveta Zinger ( s.zinger@tue.nl ) 1 Outline Introduction Color Segmentation region-growing region-merging watershed
Introduction to Medical Imaging (5XSA0) Module 5 Segmentation Jungong Han, Dirk Farin, Sveta Zinger ( s.zinger@tue.nl ) 1 Outline Introduction Color Segmentation region-growing region-merging watershed
SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS.
 SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS. 1. 3D AIRWAY TUBE RECONSTRUCTION. RELATED TO FIGURE 1 AND STAR METHODS
SUPPLEMENTARY FILE S1: 3D AIRWAY TUBE RECONSTRUCTION AND CELL-BASED MECHANICAL MODEL. RELATED TO FIGURE 1, FIGURE 7, AND STAR METHODS. 1. 3D AIRWAY TUBE RECONSTRUCTION. RELATED TO FIGURE 1 AND STAR METHODS
Perception. Autonomous Mobile Robots. Sensors Vision Uncertainties, Line extraction from laser scans. Autonomous Systems Lab. Zürich.
 Autonomous Mobile Robots Localization "Position" Global Map Cognition Environment Model Local Map Path Perception Real World Environment Motion Control Perception Sensors Vision Uncertainties, Line extraction
Autonomous Mobile Robots Localization "Position" Global Map Cognition Environment Model Local Map Path Perception Real World Environment Motion Control Perception Sensors Vision Uncertainties, Line extraction
Computer Vision I - Basics of Image Processing Part 1
 Computer Vision I - Basics of Image Processing Part 1 Carsten Rother 28/10/2014 Computer Vision I: Basics of Image Processing Link to lectures Computer Vision I: Basics of Image Processing 28/10/2014 2
Computer Vision I - Basics of Image Processing Part 1 Carsten Rother 28/10/2014 Computer Vision I: Basics of Image Processing Link to lectures Computer Vision I: Basics of Image Processing 28/10/2014 2
Edge and local feature detection - 2. Importance of edge detection in computer vision
 Edge and local feature detection Gradient based edge detection Edge detection by function fitting Second derivative edge detectors Edge linking and the construction of the chain graph Edge and local feature
Edge and local feature detection Gradient based edge detection Edge detection by function fitting Second derivative edge detectors Edge linking and the construction of the chain graph Edge and local feature
6. Object Identification L AK S H M O U. E D U
 6. Object Identification L AK S H M AN @ O U. E D U Objects Information extracted from spatial grids often need to be associated with objects not just an individual pixel Group of pixels that form a real-world
6. Object Identification L AK S H M AN @ O U. E D U Objects Information extracted from spatial grids often need to be associated with objects not just an individual pixel Group of pixels that form a real-world
technique: seam carving Image and Video Processing Chapter 9
 Chapter 9 Seam Carving for Images and Videos Distributed Algorithms for 2 Introduction Goals Enhance the visual content of images Adapted images should look natural Most relevant content should be clearly
Chapter 9 Seam Carving for Images and Videos Distributed Algorithms for 2 Introduction Goals Enhance the visual content of images Adapted images should look natural Most relevant content should be clearly
Filters. Advanced and Special Topics: Filters. Filters
 Filters Advanced and Special Topics: Filters Dr. Edmund Lam Department of Electrical and Electronic Engineering The University of Hong Kong ELEC4245: Digital Image Processing (Second Semester, 2016 17)
Filters Advanced and Special Topics: Filters Dr. Edmund Lam Department of Electrical and Electronic Engineering The University of Hong Kong ELEC4245: Digital Image Processing (Second Semester, 2016 17)
Optimization. Intelligent Scissors (see also Snakes)
 Optimization We can define a cost for possible solutions Number of solutions is large (eg., exponential) Efficient search is needed Global methods: cleverly find best solution without considering all.
Optimization We can define a cost for possible solutions Number of solutions is large (eg., exponential) Efficient search is needed Global methods: cleverly find best solution without considering all.
SUMMARY: DISTINCTIVE IMAGE FEATURES FROM SCALE- INVARIANT KEYPOINTS
 SUMMARY: DISTINCTIVE IMAGE FEATURES FROM SCALE- INVARIANT KEYPOINTS Cognitive Robotics Original: David G. Lowe, 004 Summary: Coen van Leeuwen, s1460919 Abstract: This article presents a method to extract
SUMMARY: DISTINCTIVE IMAGE FEATURES FROM SCALE- INVARIANT KEYPOINTS Cognitive Robotics Original: David G. Lowe, 004 Summary: Coen van Leeuwen, s1460919 Abstract: This article presents a method to extract
Biometrics Technology: Image Processing & Pattern Recognition (by Dr. Dickson Tong)
 Biometrics Technology: Image Processing & Pattern Recognition (by Dr. Dickson Tong) References: [1] http://homepages.inf.ed.ac.uk/rbf/hipr2/index.htm [2] http://www.cs.wisc.edu/~dyer/cs540/notes/vision.html
Biometrics Technology: Image Processing & Pattern Recognition (by Dr. Dickson Tong) References: [1] http://homepages.inf.ed.ac.uk/rbf/hipr2/index.htm [2] http://www.cs.wisc.edu/~dyer/cs540/notes/vision.html
Chapter 3 Image Registration. Chapter 3 Image Registration
 Chapter 3 Image Registration Distributed Algorithms for Introduction (1) Definition: Image Registration Input: 2 images of the same scene but taken from different perspectives Goal: Identify transformation
Chapter 3 Image Registration Distributed Algorithms for Introduction (1) Definition: Image Registration Input: 2 images of the same scene but taken from different perspectives Goal: Identify transformation
Fast and accurate automated cell boundary determination for fluorescence microscopy
 Fast and accurate automated cell boundary determination for fluorescence microscopy Stephen Hugo Arce, Pei-Hsun Wu &, and Yiider Tseng Department of Chemical Engineering, University of Florida and National
Fast and accurate automated cell boundary determination for fluorescence microscopy Stephen Hugo Arce, Pei-Hsun Wu &, and Yiider Tseng Department of Chemical Engineering, University of Florida and National
Image Segmentation. Selim Aksoy. Bilkent University
 Image Segmentation Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr Examples of grouping in vision [http://poseidon.csd.auth.gr/lab_research/latest/imgs/s peakdepvidindex_img2.jpg]
Image Segmentation Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr Examples of grouping in vision [http://poseidon.csd.auth.gr/lab_research/latest/imgs/s peakdepvidindex_img2.jpg]
Image Segmentation. Selim Aksoy. Bilkent University
 Image Segmentation Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr Examples of grouping in vision [http://poseidon.csd.auth.gr/lab_research/latest/imgs/s peakdepvidindex_img2.jpg]
Image Segmentation Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr Examples of grouping in vision [http://poseidon.csd.auth.gr/lab_research/latest/imgs/s peakdepvidindex_img2.jpg]
Image Segmentation Based on Watershed and Edge Detection Techniques
 0 The International Arab Journal of Information Technology, Vol., No., April 00 Image Segmentation Based on Watershed and Edge Detection Techniques Nassir Salman Computer Science Department, Zarqa Private
0 The International Arab Journal of Information Technology, Vol., No., April 00 Image Segmentation Based on Watershed and Edge Detection Techniques Nassir Salman Computer Science Department, Zarqa Private
Report: Reducing the error rate of a Cat classifier
 Report: Reducing the error rate of a Cat classifier Raphael Sznitman 6 August, 2007 Abstract The following report discusses my work at the IDIAP from 06.2007 to 08.2007. This work had for objective to
Report: Reducing the error rate of a Cat classifier Raphael Sznitman 6 August, 2007 Abstract The following report discusses my work at the IDIAP from 06.2007 to 08.2007. This work had for objective to
Image Analysis Image Segmentation (Basic Methods)
 Image Analysis Image Segmentation (Basic Methods) Christophoros Nikou cnikou@cs.uoi.gr Images taken from: R. Gonzalez and R. Woods. Digital Image Processing, Prentice Hall, 2008. Computer Vision course
Image Analysis Image Segmentation (Basic Methods) Christophoros Nikou cnikou@cs.uoi.gr Images taken from: R. Gonzalez and R. Woods. Digital Image Processing, Prentice Hall, 2008. Computer Vision course
Biomedical Image Analysis. Mathematical Morphology
 Biomedical Image Analysis Mathematical Morphology Contents: Foundation of Mathematical Morphology Structuring Elements Applications BMIA 15 V. Roth & P. Cattin 265 Foundations of Mathematical Morphology
Biomedical Image Analysis Mathematical Morphology Contents: Foundation of Mathematical Morphology Structuring Elements Applications BMIA 15 V. Roth & P. Cattin 265 Foundations of Mathematical Morphology
Using CellProfiler for Biological Image Analysis Quantitative Analysis of Large-Scale Biological Image Data
 Using CellProfiler for Biological Image Analysis Quantitative Analysis of Large-Scale Biological Image Data Mark-Anthony Bray, Ph.D Imaging Platform, Broad Institute Cambridge, Massachusetts, USA mbray@broadinstitute.org
Using CellProfiler for Biological Image Analysis Quantitative Analysis of Large-Scale Biological Image Data Mark-Anthony Bray, Ph.D Imaging Platform, Broad Institute Cambridge, Massachusetts, USA mbray@broadinstitute.org
Ulrik Söderström 16 Feb Image Processing. Segmentation
 Ulrik Söderström ulrik.soderstrom@tfe.umu.se 16 Feb 2011 Image Processing Segmentation What is Image Segmentation? To be able to extract information from an image it is common to subdivide it into background
Ulrik Söderström ulrik.soderstrom@tfe.umu.se 16 Feb 2011 Image Processing Segmentation What is Image Segmentation? To be able to extract information from an image it is common to subdivide it into background
CS4733 Class Notes, Computer Vision
 CS4733 Class Notes, Computer Vision Sources for online computer vision tutorials and demos - http://www.dai.ed.ac.uk/hipr and Computer Vision resources online - http://www.dai.ed.ac.uk/cvonline Vision
CS4733 Class Notes, Computer Vision Sources for online computer vision tutorials and demos - http://www.dai.ed.ac.uk/hipr and Computer Vision resources online - http://www.dai.ed.ac.uk/cvonline Vision
Basic Algorithms for Digital Image Analysis: a course
 Institute of Informatics Eötvös Loránd University Budapest, Hungary Basic Algorithms for Digital Image Analysis: a course Dmitrij Csetverikov with help of Attila Lerch, Judit Verestóy, Zoltán Megyesi,
Institute of Informatics Eötvös Loránd University Budapest, Hungary Basic Algorithms for Digital Image Analysis: a course Dmitrij Csetverikov with help of Attila Lerch, Judit Verestóy, Zoltán Megyesi,
Normalized cuts and image segmentation
 Normalized cuts and image segmentation Department of EE University of Washington Yeping Su Xiaodan Song Normalized Cuts and Image Segmentation, IEEE Trans. PAMI, August 2000 5/20/2003 1 Outline 1. Image
Normalized cuts and image segmentation Department of EE University of Washington Yeping Su Xiaodan Song Normalized Cuts and Image Segmentation, IEEE Trans. PAMI, August 2000 5/20/2003 1 Outline 1. Image
Operators-Based on Second Derivative double derivative Laplacian operator Laplacian Operator Laplacian Of Gaussian (LOG) Operator LOG
 Operators-Based on Second Derivative The principle of edge detection based on double derivative is to detect only those points as edge points which possess local maxima in the gradient values. Laplacian
Operators-Based on Second Derivative The principle of edge detection based on double derivative is to detect only those points as edge points which possess local maxima in the gradient values. Laplacian
Computer Vision I. Announcements. Fourier Tansform. Efficient Implementation. Edge and Corner Detection. CSE252A Lecture 13.
 Announcements Edge and Corner Detection HW3 assigned CSE252A Lecture 13 Efficient Implementation Both, the Box filter and the Gaussian filter are separable: First convolve each row of input image I with
Announcements Edge and Corner Detection HW3 assigned CSE252A Lecture 13 Efficient Implementation Both, the Box filter and the Gaussian filter are separable: First convolve each row of input image I with
SYDE Winter 2011 Introduction to Pattern Recognition. Clustering
 SYDE 372 - Winter 2011 Introduction to Pattern Recognition Clustering Alexander Wong Department of Systems Design Engineering University of Waterloo Outline 1 2 3 4 5 All the approaches we have learned
SYDE 372 - Winter 2011 Introduction to Pattern Recognition Clustering Alexander Wong Department of Systems Design Engineering University of Waterloo Outline 1 2 3 4 5 All the approaches we have learned
CS 5540 Spring 2013 Assignment 3, v1.0 Due: Apr. 24th 11:59PM
 1 Introduction In this programming project, we are going to do a simple image segmentation task. Given a grayscale image with a bright object against a dark background and we are going to do a binary decision
1 Introduction In this programming project, we are going to do a simple image segmentation task. Given a grayscale image with a bright object against a dark background and we are going to do a binary decision
Segmentation and descriptors for cellular immunofluoresence images
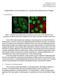 Christopher Probert cprobert@stanford.edu CS231A Project Report Segmentation and descriptors for cellular immunofluoresence images 1: Introduction Figure 1: Example immunoflouresence images of human cancer
Christopher Probert cprobert@stanford.edu CS231A Project Report Segmentation and descriptors for cellular immunofluoresence images 1: Introduction Figure 1: Example immunoflouresence images of human cancer
Lecture 6: Edge Detection
 #1 Lecture 6: Edge Detection Saad J Bedros sbedros@umn.edu Review From Last Lecture Options for Image Representation Introduced the concept of different representation or transformation Fourier Transform
#1 Lecture 6: Edge Detection Saad J Bedros sbedros@umn.edu Review From Last Lecture Options for Image Representation Introduced the concept of different representation or transformation Fourier Transform
Non-linear dimension reduction
 Sta306b May 23, 2011 Dimension Reduction: 1 Non-linear dimension reduction ISOMAP: Tenenbaum, de Silva & Langford (2000) Local linear embedding: Roweis & Saul (2000) Local MDS: Chen (2006) all three methods
Sta306b May 23, 2011 Dimension Reduction: 1 Non-linear dimension reduction ISOMAP: Tenenbaum, de Silva & Langford (2000) Local linear embedding: Roweis & Saul (2000) Local MDS: Chen (2006) all three methods
Segmentation
 Lecture 6: Segmentation 215-13-11 Filip Malmberg Centre for Image Analysis Uppsala University 2 Today What is image segmentation? A smörgåsbord of methods for image segmentation: Thresholding Edge-based
Lecture 6: Segmentation 215-13-11 Filip Malmberg Centre for Image Analysis Uppsala University 2 Today What is image segmentation? A smörgåsbord of methods for image segmentation: Thresholding Edge-based
Segmentation
 Lecture 6: Segmentation 24--4 Robin Strand Centre for Image Analysis Dept. of IT Uppsala University Today What is image segmentation? A smörgåsbord of methods for image segmentation: Thresholding Edge-based
Lecture 6: Segmentation 24--4 Robin Strand Centre for Image Analysis Dept. of IT Uppsala University Today What is image segmentation? A smörgåsbord of methods for image segmentation: Thresholding Edge-based
Local Features: Detection, Description & Matching
 Local Features: Detection, Description & Matching Lecture 08 Computer Vision Material Citations Dr George Stockman Professor Emeritus, Michigan State University Dr David Lowe Professor, University of British
Local Features: Detection, Description & Matching Lecture 08 Computer Vision Material Citations Dr George Stockman Professor Emeritus, Michigan State University Dr David Lowe Professor, University of British
Globally Stabilized 3L Curve Fitting
 Globally Stabilized 3L Curve Fitting Turker Sahin and Mustafa Unel Department of Computer Engineering, Gebze Institute of Technology Cayirova Campus 44 Gebze/Kocaeli Turkey {htsahin,munel}@bilmuh.gyte.edu.tr
Globally Stabilized 3L Curve Fitting Turker Sahin and Mustafa Unel Department of Computer Engineering, Gebze Institute of Technology Cayirova Campus 44 Gebze/Kocaeli Turkey {htsahin,munel}@bilmuh.gyte.edu.tr
Fast marching methods
 1 Fast marching methods Lecture 3 Alexander & Michael Bronstein tosca.cs.technion.ac.il/book Numerical geometry of non-rigid shapes Stanford University, Winter 2009 Metric discretization 2 Approach I:
1 Fast marching methods Lecture 3 Alexander & Michael Bronstein tosca.cs.technion.ac.il/book Numerical geometry of non-rigid shapes Stanford University, Winter 2009 Metric discretization 2 Approach I:
Region-based Segmentation
 Region-based Segmentation Image Segmentation Group similar components (such as, pixels in an image, image frames in a video) to obtain a compact representation. Applications: Finding tumors, veins, etc.
Region-based Segmentation Image Segmentation Group similar components (such as, pixels in an image, image frames in a video) to obtain a compact representation. Applications: Finding tumors, veins, etc.
Edge Detection CSC 767
 Edge Detection CSC 767 Edge detection Goal: Identify sudden changes (discontinuities) in an image Most semantic and shape information from the image can be encoded in the edges More compact than pixels
Edge Detection CSC 767 Edge detection Goal: Identify sudden changes (discontinuities) in an image Most semantic and shape information from the image can be encoded in the edges More compact than pixels
University of Florida CISE department Gator Engineering. Clustering Part 4
 Clustering Part 4 Dr. Sanjay Ranka Professor Computer and Information Science and Engineering University of Florida, Gainesville DBSCAN DBSCAN is a density based clustering algorithm Density = number of
Clustering Part 4 Dr. Sanjay Ranka Professor Computer and Information Science and Engineering University of Florida, Gainesville DBSCAN DBSCAN is a density based clustering algorithm Density = number of
Robotics Programming Laboratory
 Chair of Software Engineering Robotics Programming Laboratory Bertrand Meyer Jiwon Shin Lecture 8: Robot Perception Perception http://pascallin.ecs.soton.ac.uk/challenges/voc/databases.html#caltech car
Chair of Software Engineering Robotics Programming Laboratory Bertrand Meyer Jiwon Shin Lecture 8: Robot Perception Perception http://pascallin.ecs.soton.ac.uk/challenges/voc/databases.html#caltech car
SIFT: SCALE INVARIANT FEATURE TRANSFORM SURF: SPEEDED UP ROBUST FEATURES BASHAR ALSADIK EOS DEPT. TOPMAP M13 3D GEOINFORMATION FROM IMAGES 2014
 SIFT: SCALE INVARIANT FEATURE TRANSFORM SURF: SPEEDED UP ROBUST FEATURES BASHAR ALSADIK EOS DEPT. TOPMAP M13 3D GEOINFORMATION FROM IMAGES 2014 SIFT SIFT: Scale Invariant Feature Transform; transform image
SIFT: SCALE INVARIANT FEATURE TRANSFORM SURF: SPEEDED UP ROBUST FEATURES BASHAR ALSADIK EOS DEPT. TOPMAP M13 3D GEOINFORMATION FROM IMAGES 2014 SIFT SIFT: Scale Invariant Feature Transform; transform image
CS 664 Segmentation. Daniel Huttenlocher
 CS 664 Segmentation Daniel Huttenlocher Grouping Perceptual Organization Structural relationships between tokens Parallelism, symmetry, alignment Similarity of token properties Often strong psychophysical
CS 664 Segmentation Daniel Huttenlocher Grouping Perceptual Organization Structural relationships between tokens Parallelism, symmetry, alignment Similarity of token properties Often strong psychophysical
Lecture 7: Most Common Edge Detectors
 #1 Lecture 7: Most Common Edge Detectors Saad Bedros sbedros@umn.edu Edge Detection Goal: Identify sudden changes (discontinuities) in an image Intuitively, most semantic and shape information from the
#1 Lecture 7: Most Common Edge Detectors Saad Bedros sbedros@umn.edu Edge Detection Goal: Identify sudden changes (discontinuities) in an image Intuitively, most semantic and shape information from the
CITS 4402 Computer Vision
 CITS 4402 Computer Vision A/Prof Ajmal Mian Adj/A/Prof Mehdi Ravanbakhsh, CEO at Mapizy (www.mapizy.com) and InFarm (www.infarm.io) Lecture 02 Binary Image Analysis Objectives Revision of image formation
CITS 4402 Computer Vision A/Prof Ajmal Mian Adj/A/Prof Mehdi Ravanbakhsh, CEO at Mapizy (www.mapizy.com) and InFarm (www.infarm.io) Lecture 02 Binary Image Analysis Objectives Revision of image formation
Anno accademico 2006/2007. Davide Migliore
 Robotica Anno accademico 6/7 Davide Migliore migliore@elet.polimi.it Today What is a feature? Some useful information The world of features: Detectors Edges detection Corners/Points detection Descriptors?!?!?
Robotica Anno accademico 6/7 Davide Migliore migliore@elet.polimi.it Today What is a feature? Some useful information The world of features: Detectors Edges detection Corners/Points detection Descriptors?!?!?
Network Snakes for the Segmentation of Adjacent Cells in Confocal Images
 Network Snakes for the Segmentation of Adjacent Cells in Confocal Images Matthias Butenuth 1 and Fritz Jetzek 2 1 Institut für Photogrammetrie und GeoInformation, Leibniz Universität Hannover, 30167 Hannover
Network Snakes for the Segmentation of Adjacent Cells in Confocal Images Matthias Butenuth 1 and Fritz Jetzek 2 1 Institut für Photogrammetrie und GeoInformation, Leibniz Universität Hannover, 30167 Hannover
Multiple Contour Finding and Perceptual Grouping as a set of Energy Minimizing Paths
 Multiple Contour Finding and Perceptual Grouping as a set of Energy Minimizing Paths Laurent D. COHEN and Thomas DESCHAMPS CEREMADE, UMR 7534, Université Paris-Dauphine 75775 Paris cedex 16, France cohen@ceremade.dauphine.fr
Multiple Contour Finding and Perceptual Grouping as a set of Energy Minimizing Paths Laurent D. COHEN and Thomas DESCHAMPS CEREMADE, UMR 7534, Université Paris-Dauphine 75775 Paris cedex 16, France cohen@ceremade.dauphine.fr
Edge detection. Winter in Kraków photographed by Marcin Ryczek
 Edge detection Winter in Kraków photographed by Marcin Ryczek Edge detection Goal: Identify sudden changes (discontinuities) in an image Intuitively, most semantic and shape information from the image
Edge detection Winter in Kraków photographed by Marcin Ryczek Edge detection Goal: Identify sudden changes (discontinuities) in an image Intuitively, most semantic and shape information from the image
ISSN: X International Journal of Advanced Research in Electronics and Communication Engineering (IJARECE) Volume 6, Issue 8, August 2017
 ENTROPY BASED CONSTRAINT METHOD FOR IMAGE SEGMENTATION USING ACTIVE CONTOUR MODEL M.Nirmala Department of ECE JNTUA college of engineering, Anantapuramu Andhra Pradesh,India Abstract: Over the past existing
ENTROPY BASED CONSTRAINT METHOD FOR IMAGE SEGMENTATION USING ACTIVE CONTOUR MODEL M.Nirmala Department of ECE JNTUA college of engineering, Anantapuramu Andhra Pradesh,India Abstract: Over the past existing
Clustering Part 4 DBSCAN
 Clustering Part 4 Dr. Sanjay Ranka Professor Computer and Information Science and Engineering University of Florida, Gainesville DBSCAN DBSCAN is a density based clustering algorithm Density = number of
Clustering Part 4 Dr. Sanjay Ranka Professor Computer and Information Science and Engineering University of Florida, Gainesville DBSCAN DBSCAN is a density based clustering algorithm Density = number of
Part 3: Image Processing
 Part 3: Image Processing Image Filtering and Segmentation Georgy Gimel farb COMPSCI 373 Computer Graphics and Image Processing 1 / 60 1 Image filtering 2 Median filtering 3 Mean filtering 4 Image segmentation
Part 3: Image Processing Image Filtering and Segmentation Georgy Gimel farb COMPSCI 373 Computer Graphics and Image Processing 1 / 60 1 Image filtering 2 Median filtering 3 Mean filtering 4 Image segmentation
Cell Segmentation from Cellular Image
 Global Journal of Computer Science and Technology Vol. 10 Issue 13 (Ver. 1.0 ) October 2010 P a g e 55 Cell Segmentation from Cellular Image Mohammad Ibrahim Khan, Muhammad Kamal Hossen, Md.Sabbir Ali
Global Journal of Computer Science and Technology Vol. 10 Issue 13 (Ver. 1.0 ) October 2010 P a g e 55 Cell Segmentation from Cellular Image Mohammad Ibrahim Khan, Muhammad Kamal Hossen, Md.Sabbir Ali
(Refer Slide Time: 0:32)
 Digital Image Processing. Professor P. K. Biswas. Department of Electronics and Electrical Communication Engineering. Indian Institute of Technology, Kharagpur. Lecture-57. Image Segmentation: Global Processing
Digital Image Processing. Professor P. K. Biswas. Department of Electronics and Electrical Communication Engineering. Indian Institute of Technology, Kharagpur. Lecture-57. Image Segmentation: Global Processing
CellProfiler Analyst data exploration software
 CellProfiler Analyst data exploration software TM Created in the laboratories of Anne E. Carpenter, David M. Sabatini and Polina Golland at: Computer Sciences/ Artificial Intelligence Laboratory at the
CellProfiler Analyst data exploration software TM Created in the laboratories of Anne E. Carpenter, David M. Sabatini and Polina Golland at: Computer Sciences/ Artificial Intelligence Laboratory at the
Locally Weighted Learning for Control. Alexander Skoglund Machine Learning Course AASS, June 2005
 Locally Weighted Learning for Control Alexander Skoglund Machine Learning Course AASS, June 2005 Outline Locally Weighted Learning, Christopher G. Atkeson et. al. in Artificial Intelligence Review, 11:11-73,1997
Locally Weighted Learning for Control Alexander Skoglund Machine Learning Course AASS, June 2005 Outline Locally Weighted Learning, Christopher G. Atkeson et. al. in Artificial Intelligence Review, 11:11-73,1997
Identify Curvilinear Structure Based on Oriented Phase Congruency in Live Cell Images
 International Journal of Information and Computation Technology. ISSN 0974-2239 Volume 3, Number 4 (2013), pp. 335-340 International Research Publications House http://www. irphouse.com /ijict.htm Identify
International Journal of Information and Computation Technology. ISSN 0974-2239 Volume 3, Number 4 (2013), pp. 335-340 International Research Publications House http://www. irphouse.com /ijict.htm Identify
Let s start with occluding contours (or interior and exterior silhouettes), and look at image-space algorithms. A very simple technique is to render
 1 There are two major classes of algorithms for extracting most kinds of lines from 3D meshes. First, there are image-space algorithms that render something (such as a depth map or cosine-shaded model),
1 There are two major classes of algorithms for extracting most kinds of lines from 3D meshes. First, there are image-space algorithms that render something (such as a depth map or cosine-shaded model),
convolution shift invariant linear system Fourier Transform Aliasing and sampling scale representation edge detection corner detection
 COS 429: COMPUTER VISON Linear Filters and Edge Detection convolution shift invariant linear system Fourier Transform Aliasing and sampling scale representation edge detection corner detection Reading:
COS 429: COMPUTER VISON Linear Filters and Edge Detection convolution shift invariant linear system Fourier Transform Aliasing and sampling scale representation edge detection corner detection Reading:
Supervised texture detection in images
 Supervised texture detection in images Branislav Mičušík and Allan Hanbury Pattern Recognition and Image Processing Group, Institute of Computer Aided Automation, Vienna University of Technology Favoritenstraße
Supervised texture detection in images Branislav Mičušík and Allan Hanbury Pattern Recognition and Image Processing Group, Institute of Computer Aided Automation, Vienna University of Technology Favoritenstraße
CS443: Digital Imaging and Multimedia Binary Image Analysis. Spring 2008 Ahmed Elgammal Dept. of Computer Science Rutgers University
 CS443: Digital Imaging and Multimedia Binary Image Analysis Spring 2008 Ahmed Elgammal Dept. of Computer Science Rutgers University Outlines A Simple Machine Vision System Image segmentation by thresholding
CS443: Digital Imaging and Multimedia Binary Image Analysis Spring 2008 Ahmed Elgammal Dept. of Computer Science Rutgers University Outlines A Simple Machine Vision System Image segmentation by thresholding
Chapter 11 Representation & Description
 Chain Codes Chain codes are used to represent a boundary by a connected sequence of straight-line segments of specified length and direction. The direction of each segment is coded by using a numbering
Chain Codes Chain codes are used to represent a boundary by a connected sequence of straight-line segments of specified length and direction. The direction of each segment is coded by using a numbering
Segmentation tools and workflows in PerGeos
 Segmentation tools and workflows in PerGeos 1. Introduction Segmentation typically consists of a complex workflow involving multiple algorithms at multiple steps. Smart denoising and morphological filters
Segmentation tools and workflows in PerGeos 1. Introduction Segmentation typically consists of a complex workflow involving multiple algorithms at multiple steps. Smart denoising and morphological filters
CS664 Lecture #21: SIFT, object recognition, dynamic programming
 CS664 Lecture #21: SIFT, object recognition, dynamic programming Some material taken from: Sebastian Thrun, Stanford http://cs223b.stanford.edu/ Yuri Boykov, Western Ontario David Lowe, UBC http://www.cs.ubc.ca/~lowe/keypoints/
CS664 Lecture #21: SIFT, object recognition, dynamic programming Some material taken from: Sebastian Thrun, Stanford http://cs223b.stanford.edu/ Yuri Boykov, Western Ontario David Lowe, UBC http://www.cs.ubc.ca/~lowe/keypoints/
Bioimage Informatics
 Bioimage Informatics Lecture 14, Spring 2012 Bioimage Data Analysis (IV) Image Segmentation (part 3) Lecture 14 March 07, 2012 1 Outline Review: intensity thresholding based image segmentation Morphological
Bioimage Informatics Lecture 14, Spring 2012 Bioimage Data Analysis (IV) Image Segmentation (part 3) Lecture 14 March 07, 2012 1 Outline Review: intensity thresholding based image segmentation Morphological
LEVEL SET ALGORITHMS COMPARISON FOR MULTI-SLICE CT LEFT VENTRICLE SEGMENTATION
 LEVEL SET ALGORITHMS COMPARISON FOR MULTI-SLICE CT LEFT VENTRICLE SEGMENTATION 1 Investigador Prometeo, Universidad de Cuenca, Departamento de Electrónica y Telecomunicaciones, Cuenca, Ecuador 2 Departamento
LEVEL SET ALGORITHMS COMPARISON FOR MULTI-SLICE CT LEFT VENTRICLE SEGMENTATION 1 Investigador Prometeo, Universidad de Cuenca, Departamento de Electrónica y Telecomunicaciones, Cuenca, Ecuador 2 Departamento
Structured Light II. Thanks to Ronen Gvili, Szymon Rusinkiewicz and Maks Ovsjanikov
 Structured Light II Johannes Köhler Johannes.koehler@dfki.de Thanks to Ronen Gvili, Szymon Rusinkiewicz and Maks Ovsjanikov Introduction Previous lecture: Structured Light I Active Scanning Camera/emitter
Structured Light II Johannes Köhler Johannes.koehler@dfki.de Thanks to Ronen Gvili, Szymon Rusinkiewicz and Maks Ovsjanikov Introduction Previous lecture: Structured Light I Active Scanning Camera/emitter
Clustering k-mean clustering
 Clustering k-mean clustering Genome 373 Genomic Informatics Elhanan Borenstein The clustering problem: partition genes into distinct sets with high homogeneity and high separation Clustering (unsupervised)
Clustering k-mean clustering Genome 373 Genomic Informatics Elhanan Borenstein The clustering problem: partition genes into distinct sets with high homogeneity and high separation Clustering (unsupervised)
GENERAL AUTOMATED FLAW DETECTION SCHEME FOR NDE X-RAY IMAGES
 GENERAL AUTOMATED FLAW DETECTION SCHEME FOR NDE X-RAY IMAGES Karl W. Ulmer and John P. Basart Center for Nondestructive Evaluation Department of Electrical and Computer Engineering Iowa State University
GENERAL AUTOMATED FLAW DETECTION SCHEME FOR NDE X-RAY IMAGES Karl W. Ulmer and John P. Basart Center for Nondestructive Evaluation Department of Electrical and Computer Engineering Iowa State University
The SIFT (Scale Invariant Feature
 The SIFT (Scale Invariant Feature Transform) Detector and Descriptor developed by David Lowe University of British Columbia Initial paper ICCV 1999 Newer journal paper IJCV 2004 Review: Matt Brown s Canonical
The SIFT (Scale Invariant Feature Transform) Detector and Descriptor developed by David Lowe University of British Columbia Initial paper ICCV 1999 Newer journal paper IJCV 2004 Review: Matt Brown s Canonical
Segmentation and Grouping
 Segmentation and Grouping How and what do we see? Fundamental Problems ' Focus of attention, or grouping ' What subsets of pixels do we consider as possible objects? ' All connected subsets? ' Representation
Segmentation and Grouping How and what do we see? Fundamental Problems ' Focus of attention, or grouping ' What subsets of pixels do we consider as possible objects? ' All connected subsets? ' Representation
EECS150 - Digital Design Lecture 14 FIFO 2 and SIFT. Recap and Outline
 EECS150 - Digital Design Lecture 14 FIFO 2 and SIFT Oct. 15, 2013 Prof. Ronald Fearing Electrical Engineering and Computer Sciences University of California, Berkeley (slides courtesy of Prof. John Wawrzynek)
EECS150 - Digital Design Lecture 14 FIFO 2 and SIFT Oct. 15, 2013 Prof. Ronald Fearing Electrical Engineering and Computer Sciences University of California, Berkeley (slides courtesy of Prof. John Wawrzynek)
Data Mining Chapter 3: Visualizing and Exploring Data Fall 2011 Ming Li Department of Computer Science and Technology Nanjing University
 Data Mining Chapter 3: Visualizing and Exploring Data Fall 2011 Ming Li Department of Computer Science and Technology Nanjing University Exploratory data analysis tasks Examine the data, in search of structures
Data Mining Chapter 3: Visualizing and Exploring Data Fall 2011 Ming Li Department of Computer Science and Technology Nanjing University Exploratory data analysis tasks Examine the data, in search of structures
Automatic Segmentation of Semantic Classes in Raster Map Images
 Automatic Segmentation of Semantic Classes in Raster Map Images Thomas C. Henderson, Trevor Linton, Sergey Potupchik and Andrei Ostanin School of Computing, University of Utah, Salt Lake City, UT 84112
Automatic Segmentation of Semantic Classes in Raster Map Images Thomas C. Henderson, Trevor Linton, Sergey Potupchik and Andrei Ostanin School of Computing, University of Utah, Salt Lake City, UT 84112
Morphometric Analysis of Biomedical Images. Sara Rolfe 10/9/17
 Morphometric Analysis of Biomedical Images Sara Rolfe 10/9/17 Morphometric Analysis of Biomedical Images Object surface contours Image difference features Compact representation of feature differences
Morphometric Analysis of Biomedical Images Sara Rolfe 10/9/17 Morphometric Analysis of Biomedical Images Object surface contours Image difference features Compact representation of feature differences
Detecting Salient Contours Using Orientation Energy Distribution. Part I: Thresholding Based on. Response Distribution
 Detecting Salient Contours Using Orientation Energy Distribution The Problem: How Does the Visual System Detect Salient Contours? CPSC 636 Slide12, Spring 212 Yoonsuck Choe Co-work with S. Sarma and H.-C.
Detecting Salient Contours Using Orientation Energy Distribution The Problem: How Does the Visual System Detect Salient Contours? CPSC 636 Slide12, Spring 212 Yoonsuck Choe Co-work with S. Sarma and H.-C.
Image Segmentation Via Iterative Geodesic Averaging
 Image Segmentation Via Iterative Geodesic Averaging Asmaa Hosni, Michael Bleyer and Margrit Gelautz Institute for Software Technology and Interactive Systems, Vienna University of Technology Favoritenstr.
Image Segmentation Via Iterative Geodesic Averaging Asmaa Hosni, Michael Bleyer and Margrit Gelautz Institute for Software Technology and Interactive Systems, Vienna University of Technology Favoritenstr.
Modern Medical Image Analysis 8DC00 Exam
 Parts of answers are inside square brackets [... ]. These parts are optional. Answers can be written in Dutch or in English, as you prefer. You can use drawings and diagrams to support your textual answers.
Parts of answers are inside square brackets [... ]. These parts are optional. Answers can be written in Dutch or in English, as you prefer. You can use drawings and diagrams to support your textual answers.
Algorithm User Guide:
 Algorithm User Guide: Membrane Quantification Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to
Algorithm User Guide: Membrane Quantification Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to
Binary Image Processing. Introduction to Computer Vision CSE 152 Lecture 5
 Binary Image Processing CSE 152 Lecture 5 Announcements Homework 2 is due Apr 25, 11:59 PM Reading: Szeliski, Chapter 3 Image processing, Section 3.3 More neighborhood operators Binary System Summary 1.
Binary Image Processing CSE 152 Lecture 5 Announcements Homework 2 is due Apr 25, 11:59 PM Reading: Szeliski, Chapter 3 Image processing, Section 3.3 More neighborhood operators Binary System Summary 1.
Maximum flows and minimal cuts. Filip Malmberg
 Maximum flows and minimal cuts Filip Malmberg MSF cuts I A Minimum Spanning Forest with respect to a set of seed-points divides a graph into a number of connected components. Thus, it defines a cut on
Maximum flows and minimal cuts Filip Malmberg MSF cuts I A Minimum Spanning Forest with respect to a set of seed-points divides a graph into a number of connected components. Thus, it defines a cut on
Binary Shape Characterization using Morphological Boundary Class Distribution Functions
 Binary Shape Characterization using Morphological Boundary Class Distribution Functions Marcin Iwanowski Institute of Control and Industrial Electronics, Warsaw University of Technology, ul.koszykowa 75,
Binary Shape Characterization using Morphological Boundary Class Distribution Functions Marcin Iwanowski Institute of Control and Industrial Electronics, Warsaw University of Technology, ul.koszykowa 75,
arxiv: v1 [cs.cv] 28 Sep 2018
![arxiv: v1 [cs.cv] 28 Sep 2018 arxiv: v1 [cs.cv] 28 Sep 2018](/thumbs/93/113542646.jpg) Camera Pose Estimation from Sequence of Calibrated Images arxiv:1809.11066v1 [cs.cv] 28 Sep 2018 Jacek Komorowski 1 and Przemyslaw Rokita 2 1 Maria Curie-Sklodowska University, Institute of Computer Science,
Camera Pose Estimation from Sequence of Calibrated Images arxiv:1809.11066v1 [cs.cv] 28 Sep 2018 Jacek Komorowski 1 and Przemyslaw Rokita 2 1 Maria Curie-Sklodowska University, Institute of Computer Science,
Introduction to Digital Image Processing
 Introduction to Digital Image Processing Ranga Rodrigo June 9, 29 Outline Contents Introduction 2 Point Operations 2 Histogram Processing 5 Introduction We can process images either in spatial domain or
Introduction to Digital Image Processing Ranga Rodrigo June 9, 29 Outline Contents Introduction 2 Point Operations 2 Histogram Processing 5 Introduction We can process images either in spatial domain or
Medial Scaffolds for 3D data modelling: status and challenges. Frederic Fol Leymarie
 Medial Scaffolds for 3D data modelling: status and challenges Frederic Fol Leymarie Outline Background Method and some algorithmic details Applications Shape representation: From the Medial Axis to the
Medial Scaffolds for 3D data modelling: status and challenges Frederic Fol Leymarie Outline Background Method and some algorithmic details Applications Shape representation: From the Medial Axis to the
SCALE INVARIANT FEATURE TRANSFORM (SIFT)
 1 SCALE INVARIANT FEATURE TRANSFORM (SIFT) OUTLINE SIFT Background SIFT Extraction Application in Content Based Image Search Conclusion 2 SIFT BACKGROUND Scale-invariant feature transform SIFT: to detect
1 SCALE INVARIANT FEATURE TRANSFORM (SIFT) OUTLINE SIFT Background SIFT Extraction Application in Content Based Image Search Conclusion 2 SIFT BACKGROUND Scale-invariant feature transform SIFT: to detect
SECTION 5 IMAGE PROCESSING 2
 SECTION 5 IMAGE PROCESSING 2 5.1 Resampling 3 5.1.1 Image Interpolation Comparison 3 5.2 Convolution 3 5.3 Smoothing Filters 3 5.3.1 Mean Filter 3 5.3.2 Median Filter 4 5.3.3 Pseudomedian Filter 6 5.3.4
SECTION 5 IMAGE PROCESSING 2 5.1 Resampling 3 5.1.1 Image Interpolation Comparison 3 5.2 Convolution 3 5.3 Smoothing Filters 3 5.3.1 Mean Filter 3 5.3.2 Median Filter 4 5.3.3 Pseudomedian Filter 6 5.3.4
Towards Automated Cellular Image Segmentation for RNAi Genome-Wide Screening
 Towards Automated Cellular Image Segmentation for RNAi Genome-Wide Screening Xiaobo Zhou 1,2, K.-Y. Liu 1,2, P. Bradley 3, N. Perrimon 3, and Stephen TC Wong 1,2, * 1 Center for Bioinformatics, Harvard
Towards Automated Cellular Image Segmentation for RNAi Genome-Wide Screening Xiaobo Zhou 1,2, K.-Y. Liu 1,2, P. Bradley 3, N. Perrimon 3, and Stephen TC Wong 1,2, * 1 Center for Bioinformatics, Harvard
