Predictive Coding of Aligned Next-Generation Sequencing Data
|
|
|
- Francine Blair
- 5 years ago
- Views:
Transcription
1 Predictive Coding of Aligned Next-Generation Sequencing Data Jan Voges +, Marco Munderloh, Jörn Ostermann Institut für Informationsverarbeitung (TNT) Leibniz Universität Hannover Appelstr. 9A Hannover, Germany +To whom correspondence should be addressed. Abstract: Due to novel high-throughput next-generation sequencing technologies, the sequencing of huge amounts of genetic information has become affordable. On account of this flood of data, IT costs have become a major obstacle compared to sequencing costs. High-performance compression of genomic data is required to reduce the storage size and transmission costs. The high coverage inherent in next-generation sequencing technologies produces highly redundant data. This paper describes a compression algorithm for aligned sequence reads. The proposed algorithm combines alignment information to implicitly assemble local parts of the donor genome in order to compress the sequence reads. In contrast to other algorithms, the proposed compressor does not need a reference to encode sequence reads. Compression is performed on-the-fly using solely a sliding window (i.e. a permanently updated short-time memory) as context for the prediction of sequence reads. The algorithm yields compression results on par or better than the state-of-theart, compressing the data down to 1.9% of the original size at speeds of up to 60 MB/s and with a minute memory consumption of only several kilobytes, fitting in today s level 1 CPU caches. 1. Introduction 2016 Data Compression Conference Next-generation sequencing (NGS) machines do not read out an entire genomic template. Instead, they produce short random fragments of nucleotide sequences known as sequence reads. A quality score, expressing the confidence that a particular nucleotide has actually been found at a specific location, is associated with each nucleotide in a sequence read. This raw sequencing data generated by NGS machines is commonly stored in FASTQ files [1]. Recently, several tools for FASTQ compression have been developed, which without performing any kind of internal alignment or assembly are approaching the Kolmogorov complexity [1] [4]. FASTQ data can be aligned with tools such as BWA [5], SAMtools [6] or Bowtie2 [7]. Subsequently, the data is stored in Sequence Alignment/Map (SAM) format files which, compared to FASTQ files, contain the alignment/mapping information as well as additional meta data [6], [8]. Mapped sequencing data represented in SAM files contains more redundancy, as multiple sequence reads are typically mapped to the same location on the donor genome. The average amount of reads mapping to the same location is referred to as coverage. We propose a novel nucleotide sequence compression algorithm called tsc that exploits the redundancy introduced by the coverage as well as redundant information within sequence reads. In contrast to other tools such as Scramble [9] or DeeZ [10], our /16 $ IEEE DOI /DCC
2 algorithm does not rely on a reference. Compression is solely performed on the aligned sequence reads. Furthermore, the algorithm only requires partially sorted reads. The prediction context for sequence reads is derived by tracking previously encountered reads in a sliding window (i.e. a short-time memory). In order to encode a new sequence read, the best matching read is selected from the sliding window according to a predefined policy. Before new reads are pushed to the sliding window, they are preprocessed using the alignment information; this can be interpreted as a local assembly of the donor genome. This self-referential sliding window-technique ensures a minute memory footprint and prevents redundant encoding of allelic regions. A major feature of the algorithm is that aligned sequence reads are coded in independent blocks. Random access (RA) to the compressed data is thereby ensured. The algorithm was furthermore designed to conform to requirements for genome compression issued by ISO/IEC JTC 1 SC 29/WG 11, also known as Moving Picture Experts Group (MPEG) [11]. 2. Compression of Aligned Nucleotide Sequences To introduce this topic, we will briefly examine the SAM file format [8]. Each sequence read combined with mapping/alignment information is stored in a single line of a SAM file. This is referred to as a SAM record in the scope of this paper to avoid ambiguities. A SAM record consists of 11 mandatory fields. Among these are the mapping positions ( ), the so-called CIGAR strings ( ), and the actual nucleotide sequences ( ). The CIGAR strings contain information about inserts and/or deletions (indels). This information is generated during the alignment process. The proposed algorithm jointly encodes these SAM fields, producing a new representation called tsc record Related Work In the scope of this paper, three tools have been selected for the evaluation and comparison of the compression performance of the proposed algorithm: Scramble [9] (implementing the CRAM file format [12]), DeeZ [10], and Quip [4]. Table 1 Selected compression tools Tool Version Reference-based RA tsc 1.0 N Y Quip (normal mode) N N Quip (de-novo assembly mode) N N DeeZ 1.1 Y Y Scramble (CRAM 3.0) Y Y Scramble performs a reference-based compression of sequence reads. DeeZ performs a reference-based local assembly internally to obtain the consensus for a given position. Sequence reads are then encoded with reference to the obtained consensus. The third tool, Quip, mainly focuses on FASTQ compression, but also supports SAM file compression. Quip was used in two modes. The first mode make use of high-order Markov chains for the prediction of nucleotides followed by an arithmetic coder. The second mode is employing a de-novo assembly algorithm. Table 1 shows an overview of the mentioned tools. Tsc is the only tool which does not rely on a reference and at the same time ensures RA to the compressed data. 242
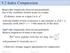
3 2.2. Proposed Algorithm The proposed algorithm solely requires SAM files which contain regions that are sorted by their mapping positions. Figure 1 visualizes a snapshot of subsequent nucleotide sequences from the data sample DH10B (see Table 2). Figure 2 shows the same sequences which now have been shifted according to their mapping positions. The algorithm is based on the unwinding of the sequence reads by using the mapping positions (as shown in Figure 2) and the additional alignment information provided by the CIGAR strings. The general mechanism is shown in Listing 1. The sample SAM records in rows 1-3 are taken from an example in the SAM file format specification [8]. First, we discuss the SAM record in line 1. The nucleotide sequence has been positioned at mapping position 7. The CIGAR string (8M2I4M1D3M) indicates that the first eight bases match to the reference which has been used for alignment (8M). According to the CIGAR string, the next two bases have been inserted into the read (2I), and so on. Figure 1 Sorted reads from data sample DH10B. Different gray-scale values represent the different nucleotides. Figure 2 The same nucleotide sequences, shifted to their mapping positions Line 5 in Listing 1 shows the positions of the reference genome used for alignment. Finally, line 6 shows the expanded nucleotide sequence from line 1. The sequence has been shifted to its mapping position. The insertions have been skipped from the expanded sequence. The deletion is represented by a question mark. The process of expanding the nucleotide sequence yields a modified CIGAR string, which we call, and an expanded nucleotide sequence, called. For the SAM record from line 1, and are shown in line 7. Additionally, an i, indicating the first record in a block (i.e. an I-record), and the mapping position have been prepended to line 7 in Listing 1. The tilde is used to indicate the end of the new sequence read representation. Analogous to a SAM record, we call this joint representation a tsc record. After encoding the first read, the mapping position and the expanded read are pushed to a circular buffer of size which implements the sliding window. The following SAM records are also expanded and pushed to the circular buffer, again obtaining and. Subsequently, the best matching read ( and ) is selected from the sliding window. The position offset, as well as the expanded reads and are then used to compute the modifications from to and/or the trailing sequence part from with respect to. In the next step, the tsc records are composed using,,, and, as shown in lines 10 and 13 in Listing
4 1 7 8M2I4M1D3M TTAGATAAAGGATACTG 2 9 3S6M1P1I9M AAAAGATAAGGATAACTGG 3 9 5S6M GCCTAAGCTAA T T A G A T A A G A T A? C T G POS STOGY EXS 7 i7:8m2iag4m1d3m:ttagataagata?ctg~ 8 9 A A A A G A T A A G A T A A C T G G POSOFF STOGY TRAIL 10 2:3SAAA6M1P1IG9M:G~ G C C T A A G C T A A POSOFF STOGY MOD 13 0:5SGCCTA6M:2C~ Listing 1 Exemplary operating principle of the proposed algorithm In addition to the first record in a block and to a normally predicted record, there might be incomplete SAM records. This yields in total three different types of tsc records (square brackets indicate optional parts): 1. I-records: 2. If,, or is missing: 3. (Normally) predicted record: Tsc records of type 2 are not pushed to the sliding window. Furthermore, new I-records are also produced if the position offset is larger than the length of the expanded reference sequence. Upon constructing a new I-record, the sliding window is cleared and a new block might be started. New I-records are also inserted every records to ensure block-wise RA to the compressed data. The ability to add new I-records ensures that the algorithm also works on only partially sorted input data. The sliding window is tracking previous encountered sequence reads. In order to select the best matching read from the sliding window, a metric called is computed for each tsc record and also pushed to the circular buffer. Let denote the length of the argument. With this notation, the parameter can be used to adjust the selection of reference reads from the sliding window, as shown in Equation 1. (1) The best matching record is then the one with as shown in Equation 2. (2) The I-records have a value calculated according to Equation 3. (3) The tsc records as a new sequence read representation are ultimately passed to a dictionary-based codec in order to exploit the redundancy along the tsc record stream. Specifically, error-free and properly aligned sequence reads produce a tsc record stream which is highly suitable for dictionary-based compression because of repetitions in the 244
5 data stream. In this paper, zlib [13], [14] has been used. It is based on the DEFLATE algorithm [15], which is in turn a combination of the LZ77 algorithm [16] and Huffman coding [17]. 3. Evaluation and Results The following sections describe the selection of test data, the test conditions, and compression results. In addition to the overall compression results, we evaluated the block-wise CRs of the proposed algorithm to verify its operating principle Test Data MPEG issued a reference collection of NGS data which was compiled according to specific criteria to ensure a wide variety of challenges for existing and future compression tools [18]. Therefore, the algorithms are evaluated using SAM/BAM files from the MPEG database. The selected data is shown in Table 2. Table 2 Selected test data from the MPEG database ID BAM size (GB) Record count Coverage cancermixed/hccmix ,395,158 26x DH10B ,175, x LID8465/K ,476,391 16x ERR317482WGS/9827# ,463, x ERP002490/NA ,286,194,550 26x 3.2. Implementation The proposed compression algorithm was implemented in a software also called tsc, which has been developed at Institut für Informationsverarbeitung. Tsc is a highly modular software: after parsing a SAM file and splitting it into the twelve distinct streams (one stream for each SAM field), each stream or a combination of streams can be passed to several so-called sub-block codecs. One of these sub-block codecs is dedicated to implementing the proposed algorithm. It jointly encodes the mapping positions, the CIGAR strings, and the nucleotide sequences. Zlib version is used to obtain the binary data. In the scope of this paper, the parameters for the proposed algorithm were empirically derived by finding a good trade-off between compression ratio and runtime. The sliding window size of the nucleotide sequence compressor was set to, and the selection of context reads from it was done using Equations 1-3 with. Figure 3 shows the average tsc CR for the first 100,000 reads from all data samples in Table 2 over the block size. As the CR converges toward larger block sizes, a block size of records has been chosen for the experiments. Since the size of the sliding window is limited to context reads, the tsc algorithm requires only tiny amounts of memory. If we, for example, assume an average read length of 200 base pairs, then the sliding window consumes about (8 bytes are used for, and 4 bytes are used for ). This amount of data easily fits into level 1 caches of modern CPUs. In contrast, for the compression of the data samples from Table 2, the memory consumption of Scramble ranges from about 0.2 GB up to 1 GB. Quip uses about 1 GB, and DeeZ uses roughly GB. 245
6 Average CR 120% 100% 80% 60% 40% 20% 0% % 20.59% 10.53% 9.06% 8.83% Nb 3.3. Compression Results Figure 3 Average tsc CR versus block size In this section, the compression ratio (CR) is defined as, where and denote the compressed and uncompressed size, respectively. Figure 4 shows the CRs obtained using tsc, Scramble, DeeZ, and Quip (in both the statistical and the de-novo assembly mode) on the data from Table 2. It is worth mentioning that the tools operate with slightly different input and output signals. The uncompressed sizes for each data sample were computed by accumulating the sizes of the SAM fields RNAME, POS, CIGAR, and SEQ. The compressed sizes were computed by accumulating those parts of the individual compressed bit streams that are associated with one of the mentioned SAM fields. 25% 23.36% 23.13% 23.73% 23.40% 21.32% 21.18% 20% 17.38% 15% 15.90% 15.89% 15.48% 15.44% 6.49% CR 10% 5% 4.56% 3.77% 1.90% 0.88% 0.79% 2.89% 1.19% 0.64% 2.50% 2.12% 5.77% 6.45% 7.03% 0% cancermixed/hccmix DH10B LID8465/K562 ERR317492WGS/9827#49 ERP002490/NA12878 tsc-1.0 quip quip a deez-1.1 scramble / CRAM 3.0 Figure 4 Comparison of the sequence read compression results Tsc outperforms all competing tools for the data sample ERP002490/NA12878, as shown in Figure 4. Furthermore, tsc performs better than Quip on all data samples and is only outperformed by the reference-based tools. Data sample ERR317492WGS/9827#49 has a very low coverage of only 2.3x as shown in Table 2. Hence, the proposed algorithm does not perform well compared to the reference-based 246
7 compressors. However, on average, tsc is able to compress the data down to 6.88% of the original size. The compression speed of the tsc software was measured on an Intel Core TM i7-3770k CPU with GHz and 32 GB RAM. Tsc achieves compression speeds ranging from about 10 MB/s for LID8465/K562 up to 60 MB/s for DH10B. The decompression is roughly twice as fast. This indicates that the tsc compression and decompression speeds are only bounded by the employed dictionary-based codec. A table with file pointer offsets to the compressed blocks is stored in the tsc file header. Therefore, RA speed is only bounded by the speed of file seek operations if the desired block has been found, it can be decompressed at full speed Block-wise Evaluation of Compression Ratios To evaluate the block-wise compression performance of the proposed algorithm, several values have been monitored for each block. Among these are: The mean length of the string per block The mean length of the string per block The compression ratio per block Each indel coded in the string and each modification (stored in ) increase the size of a tsc record. Therefore, additional indels or modifications should impose an increase in CR. Nevertheless, due to the self-referential algorithm, mutations and/or allelic regions in the donor genome should not increase the CR. Thereby, low-error alignments should be ideally suited for compression with the proposed algorithm. To show this behavior, we define the average number of edits per block as shown in Equation 4. The distribution of block-wise CRs over the average number of edits per block for the data samples from Table 2 is shown in Figure 5 to Figure 8. Furthermore, the Pearson correlation coefficient and the Spearman correlation coefficient were added to the figures. The Spearman correlation coefficient is less sensitive to outliers than the Pearson correlation coefficient and is thus more suitable for the data. With ranging from 0.20 up to 0.99, it is clearly shown that the obtainable CRs are correlated with the errors introduced by the sequencing technology and poor alignments. Emerging sequencing technologies yielding fewer errors or alignment tools finding better alignments should produce data that can be compressed at much lower bit rates using the proposed algorithm. 4. Conclusion It has been shown that the tsc algorithm for the compression of aligned nucleotide sequences is able to reduce the storage size of the combined mapping positions, CIGAR strings, and nucleotide sequences down to 1.9% for the dataset DH10B. On average, the algorithm is able to compress the data to about 6.88% of the original size and is only outperformed by the reference-based tools. Furthermore, tsc performs better than Quip on all data samples and outperforms all competing tools on the dataset ERP002490/NA (4) 247
8 In addition to its competitive compression ratios, a major advantage of the algorithm is its minute memory consumption and its low complexity. In contrast to the other tools which consume up to several gigabytes of memory, the context data used by the proposed algorithm solely consumes several kilobytes. This amount of data easily fits into level 1 caches of modern CPUs. Furthermore, the context for the prediction of aligned sequence reads is only derived from previously encountered reads. This naturally prevents the redundant encoding of allelic regions in the donor genome. Moreover, no reference is required for compression respectively decompression. Figure 5 cancermixed/hccmix Figure 6 DH10B Figure 7 LID8465/K562 Figure 8 ERR317492WGS/9827#49 Figure 9 ERP002490/NA12878 In addition, the algorithm is suitable for only partially sorted input data. Competitive compression ratios can be obtained, while at the same time fast and fine-grained random access to the compressed data is ensured. Due to these features and a novel compressed record structure, computation on the compressed data is encouraged. Furthermore, it has been shown that the obtained compression ratios depend on the error rates of the employed sequencing technologies and alignment tools. Low-error data produced by emerging sequencing technologies will have an immediate positive impact on the compression ratios obtained by the proposed algorithm. 248
9 References [1] P. J. A. Cock, C. J. Fields, N. Goto, M. L. Heuer, and P. M. Rice, The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants, Nucleic Acids Res., vol. 38, no. 6, pp , [2] J. K. Bonfield and M. V. Mahoney, Compression of FASTQ and SAM Format Sequencing Data, PLoS One, vol. 8, no. 3, p. e59190, [3] F. Campagne, K. C. Dorff, N. Chambwe, J. T. Robinson, and J. P. Mesirov, Compression of structured high-throughput sequencing data, PLoS One, vol. 8, no. 11, p. e79871, Jan [4] D. C. Jones, W. L. Ruzzo, X. Peng, and M. G. Katze, Compression of nextgeneration sequencing reads aided by highly efficient de novo assembly, Nucleic Acids Res., vol. 40, no. 22, pp. e171 e171, [5] H. Li and R. Durbin, Fast and accurate short read alignment with Burrows- Wheeler transform., Bioinformatics, vol. 25, no. 14, pp , [6] H. Li, B. Handsaker, A. Wysoker, T. Fennell, J. Ruan, N. Homer, G. Marth, G. Abecasis, and R. Durbin, The Sequence Alignment/Map format and SAMtools, Bioinformatics, vol. 25, no. 16, pp , [7] B. Langmead and S. L. Salzberg, Fast gapped-read alignment with Bowtie 2., Nat. Methods, vol. 9, no. 4, pp , [8] The SAM/BAM Format Specification Working Group, Sequence Alignment/Map Format Specification, [9] J. K. Bonfield, The Scramble conversion tool., Bioinformatics, vol. 30, no. 19, pp , Oct [10] F. Hach, I. Numanagi, and S. C. Sahinalp, DeeZ: reference-based compression by local assembly., Nat. Methods, vol. 11, no. 11, pp , Nov [11] C. Alberti, M. Mattavelli, L. Chiariglione, I. Xenarios, N. Guex, H. Stockinger, T. Schuepbach, P. Kahlem, C. Iseli, D. Zerzion, D. Kuznetsov, Y. Thoma, E. Petraglio, C. Sahinalp, I. Numanagi, J. Bonfield, V. Zalunin, J. Delgado, W. Allasia, S. Cheng, and J. Voges, Requirements on Genome Compression and Storage. ISO/IEC JTC 1/SC 29/WG 11 (MPEG), Document number N15345, Warsaw, [12] M. Hsi-Yang Fritz, R. Leinonen, G. Cochrane, and E. Birney, Efficient storage of high throughput DNA sequencing data using reference-based compression, Genome Res., vol. 21, no. 5, pp , Jan [13] J.-L. Gailly and P. Deutsch, ZLIB Compressed Data Format Specification version 3.3. Network Working Group,
10 [14] P. Deutsch, GZIP file format specification version 4.3. Network Working Group, [15] P. Deutsch, DEFLATE Compressed Data Format Specification version 1.3. Network Working Group, [16] J. Ziv and a. Lempel, A universal algorithm for sequential data compression, IEEE Trans. Inf. Theory, vol. 23, no. 3, pp , [17] D. A. Huffman, A Method for the Construction of Minimum-Redundancy Codes, Proc. IRE, vol. 40, no. 9, pp , [18] C. Alberti, M. Mattavelli, L. Chiariglione, I. Xenarios, N. Guex, H. Stockinger, T. Schuepbach, P. Kahlem, C. Iseli, D. Zerzion, D. Kuznetsov, Y. Thoma, E. Petraglio, C. Sahinalp, I. Numanagi, and J. Delgado, Database for Evaluation of Genome Compression and Storage. ISO/IEC JTC 1/SC 29/WG 11 (MPEG), Document number N15092, Geneva,
Bioinformatics in next generation sequencing projects
 Bioinformatics in next generation sequencing projects Rickard Sandberg Assistant Professor Department of Cell and Molecular Biology Karolinska Institutet March 2011 Once sequenced the problem becomes computational
Bioinformatics in next generation sequencing projects Rickard Sandberg Assistant Professor Department of Cell and Molecular Biology Karolinska Institutet March 2011 Once sequenced the problem becomes computational
SAM / BAM Tutorial. EMBL Heidelberg. Course Materials. Tobias Rausch September 2012
 SAM / BAM Tutorial EMBL Heidelberg Course Materials Tobias Rausch September 2012 Contents 1 SAM / BAM 3 1.1 Introduction................................... 3 1.2 Tasks.......................................
SAM / BAM Tutorial EMBL Heidelberg Course Materials Tobias Rausch September 2012 Contents 1 SAM / BAM 3 1.1 Introduction................................... 3 1.2 Tasks.......................................
Accurate Long-Read Alignment using Similarity Based Multiple Pattern Alignment and Prefix Tree Indexing
 Proposal for diploma thesis Accurate Long-Read Alignment using Similarity Based Multiple Pattern Alignment and Prefix Tree Indexing Astrid Rheinländer 01-09-2010 Supervisor: Prof. Dr. Ulf Leser Motivation
Proposal for diploma thesis Accurate Long-Read Alignment using Similarity Based Multiple Pattern Alignment and Prefix Tree Indexing Astrid Rheinländer 01-09-2010 Supervisor: Prof. Dr. Ulf Leser Motivation
Aligned genomic data compression via improved modeling
 Journal of Bioinformatics and Computational Biology Vol. 12, No. 6 (2014) 1442002 (17 pages) #.c Imperial College Press DOI: 10.1142/S0219720014420025 Aligned genomic data compression via improved modeling
Journal of Bioinformatics and Computational Biology Vol. 12, No. 6 (2014) 1442002 (17 pages) #.c Imperial College Press DOI: 10.1142/S0219720014420025 Aligned genomic data compression via improved modeling
File Formats: SAM, BAM, and CRAM. UCD Genome Center Bioinformatics Core Tuesday 15 September 2015
 File Formats: SAM, BAM, and CRAM UCD Genome Center Bioinformatics Core Tuesday 15 September 2015 / BAM / CRAM NEW! http://samtools.sourceforge.net/ - deprecated! http://www.htslib.org/ - SAMtools 1.0 and
File Formats: SAM, BAM, and CRAM UCD Genome Center Bioinformatics Core Tuesday 15 September 2015 / BAM / CRAM NEW! http://samtools.sourceforge.net/ - deprecated! http://www.htslib.org/ - SAMtools 1.0 and
Welcome to MAPHiTS (Mapping Analysis Pipeline for High-Throughput Sequences) tutorial page.
 Welcome to MAPHiTS (Mapping Analysis Pipeline for High-Throughput Sequences) tutorial page. In this page you will learn to use the tools of the MAPHiTS suite. A little advice before starting : rename your
Welcome to MAPHiTS (Mapping Analysis Pipeline for High-Throughput Sequences) tutorial page. In this page you will learn to use the tools of the MAPHiTS suite. A little advice before starting : rename your
So, what is data compression, and why do we need it?
 In the last decade we have been witnessing a revolution in the way we communicate 2 The major contributors in this revolution are: Internet; The explosive development of mobile communications; and The
In the last decade we have been witnessing a revolution in the way we communicate 2 The major contributors in this revolution are: Internet; The explosive development of mobile communications; and The
Lecture 12. Short read aligners
 Lecture 12 Short read aligners Ebola reference genome We will align ebola sequencing data against the 1976 Mayinga reference genome. We will hold the reference gnome and all indices: mkdir -p ~/reference/ebola
Lecture 12 Short read aligners Ebola reference genome We will align ebola sequencing data against the 1976 Mayinga reference genome. We will hold the reference gnome and all indices: mkdir -p ~/reference/ebola
High-throughput sequencing: Alignment and related topic. Simon Anders EMBL Heidelberg
 High-throughput sequencing: Alignment and related topic Simon Anders EMBL Heidelberg Established platforms HTS Platforms Illumina HiSeq, ABI SOLiD, Roche 454 Newcomers: Benchtop machines 454 GS Junior,
High-throughput sequencing: Alignment and related topic Simon Anders EMBL Heidelberg Established platforms HTS Platforms Illumina HiSeq, ABI SOLiD, Roche 454 Newcomers: Benchtop machines 454 GS Junior,
INTRODUCTION AUX FORMATS DE FICHIERS
 INTRODUCTION AUX FORMATS DE FICHIERS Plan. Formats de séquences brutes.. Format fasta.2. Format fastq 2. Formats d alignements 2.. Format SAM 2.2. Format BAM 4. Format «Variant Calling» 4.. Format Varscan
INTRODUCTION AUX FORMATS DE FICHIERS Plan. Formats de séquences brutes.. Format fasta.2. Format fastq 2. Formats d alignements 2.. Format SAM 2.2. Format BAM 4. Format «Variant Calling» 4.. Format Varscan
High-throughput sequencing: Alignment and related topic. Simon Anders EMBL Heidelberg
 High-throughput sequencing: Alignment and related topic Simon Anders EMBL Heidelberg Established platforms HTS Platforms Illumina HiSeq, ABI SOLiD, Roche 454 Newcomers: Benchtop machines: Illumina MiSeq,
High-throughput sequencing: Alignment and related topic Simon Anders EMBL Heidelberg Established platforms HTS Platforms Illumina HiSeq, ABI SOLiD, Roche 454 Newcomers: Benchtop machines: Illumina MiSeq,
Compression of structured high-throughput sequencing data
 Compression of structured high-throughput sequencing data Fabien Campagne 1,2 *, Kevin C. Dorff 1, Nyasha Chambwe 1,2, James T. Robinson, Jill P. Mesirov, Thomas D. Wu. 1 The HRH Prince Alwaleed Bin Talal
Compression of structured high-throughput sequencing data Fabien Campagne 1,2 *, Kevin C. Dorff 1, Nyasha Chambwe 1,2, James T. Robinson, Jill P. Mesirov, Thomas D. Wu. 1 The HRH Prince Alwaleed Bin Talal
Compressing and Decoding Term Statistics Time Series
 Compressing and Decoding Term Statistics Time Series Jinfeng Rao 1,XingNiu 1,andJimmyLin 2(B) 1 University of Maryland, College Park, USA {jinfeng,xingniu}@cs.umd.edu 2 University of Waterloo, Waterloo,
Compressing and Decoding Term Statistics Time Series Jinfeng Rao 1,XingNiu 1,andJimmyLin 2(B) 1 University of Maryland, College Park, USA {jinfeng,xingniu}@cs.umd.edu 2 University of Waterloo, Waterloo,
Genomic Files. University of Massachusetts Medical School. October, 2014
 .. Genomic Files University of Massachusetts Medical School October, 2014 2 / 39. A Typical Deep-Sequencing Workflow Samples Fastq Files Fastq Files Sam / Bam Files Various files Deep Sequencing Further
.. Genomic Files University of Massachusetts Medical School October, 2014 2 / 39. A Typical Deep-Sequencing Workflow Samples Fastq Files Fastq Files Sam / Bam Files Various files Deep Sequencing Further
Mapping NGS reads for genomics studies
 Mapping NGS reads for genomics studies Valencia, 28-30 Sep 2015 BIER Alejandro Alemán aaleman@cipf.es Genomics Data Analysis CIBERER Where are we? Fastq Sequence preprocessing Fastq Alignment BAM Visualization
Mapping NGS reads for genomics studies Valencia, 28-30 Sep 2015 BIER Alejandro Alemán aaleman@cipf.es Genomics Data Analysis CIBERER Where are we? Fastq Sequence preprocessing Fastq Alignment BAM Visualization
Galaxy Platform For NGS Data Analyses
 Galaxy Platform For NGS Data Analyses Weihong Yan wyan@chem.ucla.edu Collaboratory Web Site http://qcb.ucla.edu/collaboratory Collaboratory Workshops Workshop Outline ü Day 1 UCLA galaxy and user account
Galaxy Platform For NGS Data Analyses Weihong Yan wyan@chem.ucla.edu Collaboratory Web Site http://qcb.ucla.edu/collaboratory Collaboratory Workshops Workshop Outline ü Day 1 UCLA galaxy and user account
Optimized architectures of CABAC codec for IA-32-, DSP- and FPGAbased
 Optimized architectures of CABAC codec for IA-32-, DSP- and FPGAbased platforms Damian Karwowski, Marek Domański Poznan University of Technology, Chair of Multimedia Telecommunications and Microelectronics
Optimized architectures of CABAC codec for IA-32-, DSP- and FPGAbased platforms Damian Karwowski, Marek Domański Poznan University of Technology, Chair of Multimedia Telecommunications and Microelectronics
Image Compression for Mobile Devices using Prediction and Direct Coding Approach
 Image Compression for Mobile Devices using Prediction and Direct Coding Approach Joshua Rajah Devadason M.E. scholar, CIT Coimbatore, India Mr. T. Ramraj Assistant Professor, CIT Coimbatore, India Abstract
Image Compression for Mobile Devices using Prediction and Direct Coding Approach Joshua Rajah Devadason M.E. scholar, CIT Coimbatore, India Mr. T. Ramraj Assistant Professor, CIT Coimbatore, India Abstract
Dindel User Guide, version 1.0
 Dindel User Guide, version 1.0 Kees Albers University of Cambridge, Wellcome Trust Sanger Institute caa@sanger.ac.uk October 26, 2010 Contents 1 Introduction 2 2 Requirements 2 3 Optional input 3 4 Dindel
Dindel User Guide, version 1.0 Kees Albers University of Cambridge, Wellcome Trust Sanger Institute caa@sanger.ac.uk October 26, 2010 Contents 1 Introduction 2 2 Requirements 2 3 Optional input 3 4 Dindel
SAM : Sequence Alignment/Map format. A TAB-delimited text format storing the alignment information. A header section is optional.
 Alignment of NGS reads, samtools and visualization Hands-on Software used in this practical BWA MEM : Burrows-Wheeler Aligner. A software package for mapping low-divergent sequences against a large reference
Alignment of NGS reads, samtools and visualization Hands-on Software used in this practical BWA MEM : Burrows-Wheeler Aligner. A software package for mapping low-divergent sequences against a large reference
Compression of Stereo Images using a Huffman-Zip Scheme
 Compression of Stereo Images using a Huffman-Zip Scheme John Hamann, Vickey Yeh Department of Electrical Engineering, Stanford University Stanford, CA 94304 jhamann@stanford.edu, vickey@stanford.edu Abstract
Compression of Stereo Images using a Huffman-Zip Scheme John Hamann, Vickey Yeh Department of Electrical Engineering, Stanford University Stanford, CA 94304 jhamann@stanford.edu, vickey@stanford.edu Abstract
ECE 533 Digital Image Processing- Fall Group Project Embedded Image coding using zero-trees of Wavelet Transform
 ECE 533 Digital Image Processing- Fall 2003 Group Project Embedded Image coding using zero-trees of Wavelet Transform Harish Rajagopal Brett Buehl 12/11/03 Contributions Tasks Harish Rajagopal (%) Brett
ECE 533 Digital Image Processing- Fall 2003 Group Project Embedded Image coding using zero-trees of Wavelet Transform Harish Rajagopal Brett Buehl 12/11/03 Contributions Tasks Harish Rajagopal (%) Brett
GPUBwa -Parallelization of Burrows Wheeler Aligner using Graphical Processing Units
 GPUBwa -Parallelization of Burrows Wheeler Aligner using Graphical Processing Units Abstract A very popular discipline in bioinformatics is Next-Generation Sequencing (NGS) or DNA sequencing. It specifies
GPUBwa -Parallelization of Burrows Wheeler Aligner using Graphical Processing Units Abstract A very popular discipline in bioinformatics is Next-Generation Sequencing (NGS) or DNA sequencing. It specifies
NGS Data Analysis. Roberto Preste
 NGS Data Analysis Roberto Preste 1 Useful info http://bit.ly/2r1y2dr Contacts: roberto.preste@gmail.com Slides: http://bit.ly/ngs-data 2 NGS data analysis Overview 3 NGS Data Analysis: the basic idea http://bit.ly/2r1y2dr
NGS Data Analysis Roberto Preste 1 Useful info http://bit.ly/2r1y2dr Contacts: roberto.preste@gmail.com Slides: http://bit.ly/ngs-data 2 NGS data analysis Overview 3 NGS Data Analysis: the basic idea http://bit.ly/2r1y2dr
A High-Performance FPGA-Based Implementation of the LZSS Compression Algorithm
 2012 IEEE 2012 26th IEEE International 26th International Parallel Parallel and Distributed and Distributed Processing Processing Symposium Symposium Workshops Workshops & PhD Forum A High-Performance
2012 IEEE 2012 26th IEEE International 26th International Parallel Parallel and Distributed and Distributed Processing Processing Symposium Symposium Workshops Workshops & PhD Forum A High-Performance
CS 493: Algorithms for Massive Data Sets Dictionary-based compression February 14, 2002 Scribe: Tony Wirth LZ77
 CS 493: Algorithms for Massive Data Sets February 14, 2002 Dictionary-based compression Scribe: Tony Wirth This lecture will explore two adaptive dictionary compression schemes: LZ77 and LZ78. We use the
CS 493: Algorithms for Massive Data Sets February 14, 2002 Dictionary-based compression Scribe: Tony Wirth This lecture will explore two adaptive dictionary compression schemes: LZ77 and LZ78. We use the
Image compression. Stefano Ferrari. Università degli Studi di Milano Methods for Image Processing. academic year
 Image compression Stefano Ferrari Università degli Studi di Milano stefano.ferrari@unimi.it Methods for Image Processing academic year 2017 2018 Data and information The representation of images in a raw
Image compression Stefano Ferrari Università degli Studi di Milano stefano.ferrari@unimi.it Methods for Image Processing academic year 2017 2018 Data and information The representation of images in a raw
To Zip or not to Zip. Effective Resource Usage for Real-Time Compression
 To Zip or not to Zip Effective Resource Usage for Real-Time Compression Danny Harnik, Oded Margalit, Ronen Kat, Dmitry Sotnikov, Avishay Traeger IBM Research - Haifa Our scope Real-Time Compression Compression
To Zip or not to Zip Effective Resource Usage for Real-Time Compression Danny Harnik, Oded Margalit, Ronen Kat, Dmitry Sotnikov, Avishay Traeger IBM Research - Haifa Our scope Real-Time Compression Compression
CS 335 Graphics and Multimedia. Image Compression
 CS 335 Graphics and Multimedia Image Compression CCITT Image Storage and Compression Group 3: Huffman-type encoding for binary (bilevel) data: FAX Group 4: Entropy encoding without error checks of group
CS 335 Graphics and Multimedia Image Compression CCITT Image Storage and Compression Group 3: Huffman-type encoding for binary (bilevel) data: FAX Group 4: Entropy encoding without error checks of group
NGS Data and Sequence Alignment
 Applications and Servers SERVER/REMOTE Compute DB WEB Data files NGS Data and Sequence Alignment SSH WEB SCP Manpreet S. Katari App Aug 11, 2016 Service Terminal IGV Data files Window Personal Computer/Local
Applications and Servers SERVER/REMOTE Compute DB WEB Data files NGS Data and Sequence Alignment SSH WEB SCP Manpreet S. Katari App Aug 11, 2016 Service Terminal IGV Data files Window Personal Computer/Local
International Journal of Emerging Technology and Advanced Engineering Website: (ISSN , Volume 2, Issue 4, April 2012)
 A Technical Analysis Towards Digital Video Compression Rutika Joshi 1, Rajesh Rai 2, Rajesh Nema 3 1 Student, Electronics and Communication Department, NIIST College, Bhopal, 2,3 Prof., Electronics and
A Technical Analysis Towards Digital Video Compression Rutika Joshi 1, Rajesh Rai 2, Rajesh Nema 3 1 Student, Electronics and Communication Department, NIIST College, Bhopal, 2,3 Prof., Electronics and
NA12878 Platinum Genome GENALICE MAP Analysis Report
 NA12878 Platinum Genome GENALICE MAP Analysis Report Bas Tolhuis, PhD Jan-Jaap Wesselink, PhD GENALICE B.V. INDEX EXECUTIVE SUMMARY...4 1. MATERIALS & METHODS...5 1.1 SEQUENCE DATA...5 1.2 WORKFLOWS......5
NA12878 Platinum Genome GENALICE MAP Analysis Report Bas Tolhuis, PhD Jan-Jaap Wesselink, PhD GENALICE B.V. INDEX EXECUTIVE SUMMARY...4 1. MATERIALS & METHODS...5 1.1 SEQUENCE DATA...5 1.2 WORKFLOWS......5
Analysis of Parallelization Effects on Textual Data Compression
 Analysis of Parallelization Effects on Textual Data GORAN MARTINOVIC, CASLAV LIVADA, DRAGO ZAGAR Faculty of Electrical Engineering Josip Juraj Strossmayer University of Osijek Kneza Trpimira 2b, 31000
Analysis of Parallelization Effects on Textual Data GORAN MARTINOVIC, CASLAV LIVADA, DRAGO ZAGAR Faculty of Electrical Engineering Josip Juraj Strossmayer University of Osijek Kneza Trpimira 2b, 31000
A Nearest Neighbors Algorithm for Strings. R. Lederman Technical Report YALEU/DCS/TR-1453 April 5, 2012
 A randomized algorithm is presented for fast nearest neighbors search in libraries of strings. The algorithm is discussed in the context of one of the practical applications: aligning DNA reads to a reference
A randomized algorithm is presented for fast nearest neighbors search in libraries of strings. The algorithm is discussed in the context of one of the practical applications: aligning DNA reads to a reference
A Novel Image Compression Technique using Simple Arithmetic Addition
 Proc. of Int. Conf. on Recent Trends in Information, Telecommunication and Computing, ITC A Novel Image Compression Technique using Simple Arithmetic Addition Nadeem Akhtar, Gufran Siddiqui and Salman
Proc. of Int. Conf. on Recent Trends in Information, Telecommunication and Computing, ITC A Novel Image Compression Technique using Simple Arithmetic Addition Nadeem Akhtar, Gufran Siddiqui and Salman
Accelrys Pipeline Pilot and HP ProLiant servers
 Accelrys Pipeline Pilot and HP ProLiant servers A performance overview Technical white paper Table of contents Introduction... 2 Accelrys Pipeline Pilot benchmarks on HP ProLiant servers... 2 NGS Collection
Accelrys Pipeline Pilot and HP ProLiant servers A performance overview Technical white paper Table of contents Introduction... 2 Accelrys Pipeline Pilot benchmarks on HP ProLiant servers... 2 NGS Collection
SAM and VCF formats. UCD Genome Center Bioinformatics Core Tuesday 14 June 2016
 SAM and VCF formats UCD Genome Center Bioinformatics Core Tuesday 14 June 2016 File Format: SAM / BAM / CRAM! NEW http://samtools.sourceforge.net/ - deprecated! http://www.htslib.org/ - SAMtools 1.0 and
SAM and VCF formats UCD Genome Center Bioinformatics Core Tuesday 14 June 2016 File Format: SAM / BAM / CRAM! NEW http://samtools.sourceforge.net/ - deprecated! http://www.htslib.org/ - SAMtools 1.0 and
Genomic Files. University of Massachusetts Medical School. October, 2015
 .. Genomic Files University of Massachusetts Medical School October, 2015 2 / 55. A Typical Deep-Sequencing Workflow Samples Fastq Files Fastq Files Sam / Bam Files Various files Deep Sequencing Further
.. Genomic Files University of Massachusetts Medical School October, 2015 2 / 55. A Typical Deep-Sequencing Workflow Samples Fastq Files Fastq Files Sam / Bam Files Various files Deep Sequencing Further
MRT based Fixed Block size Transform Coding
 3 MRT based Fixed Block size Transform Coding Contents 3.1 Transform Coding..64 3.1.1 Transform Selection...65 3.1.2 Sub-image size selection... 66 3.1.3 Bit Allocation.....67 3.2 Transform coding using
3 MRT based Fixed Block size Transform Coding Contents 3.1 Transform Coding..64 3.1.1 Transform Selection...65 3.1.2 Sub-image size selection... 66 3.1.3 Bit Allocation.....67 3.2 Transform coding using
Quark enables semi-reference-based compression of RNA-seq data
 Bioinformatics, 33(21), 2017, 3380 3386 doi: 10.1093/bioinformatics/btx428 Advance Access Publication Date: 3 July 2017 Original Paper Sequence analysis Quark enables semi-reference-based compression of
Bioinformatics, 33(21), 2017, 3380 3386 doi: 10.1093/bioinformatics/btx428 Advance Access Publication Date: 3 July 2017 Original Paper Sequence analysis Quark enables semi-reference-based compression of
Sequence mapping and assembly. Alistair Ward - Boston College
 Sequence mapping and assembly Alistair Ward - Boston College Sequenced a genome? Fragmented a genome -> DNA library PCR amplification Sequence reads (ends of DNA fragment for mate pairs) We no longer have
Sequence mapping and assembly Alistair Ward - Boston College Sequenced a genome? Fragmented a genome -> DNA library PCR amplification Sequence reads (ends of DNA fragment for mate pairs) We no longer have
Academy of Sciences Via Accademia delle Scienze, Torino
 Academy of Sciences Via Accademia delle Scienze, 6 10123 Torino July 19, 2017 25 Years of MPEG Digital Media 25 Years of MPEG Digital Media 3 25 Years of MPEG Digital Media Just a few clicks 4 25 Years
Academy of Sciences Via Accademia delle Scienze, 6 10123 Torino July 19, 2017 25 Years of MPEG Digital Media 25 Years of MPEG Digital Media 3 25 Years of MPEG Digital Media Just a few clicks 4 25 Years
Scalable RNA Sequencing on Clusters of Multicore Processors
 JOAQUÍN DOPAZO JOAQUÍN TARRAGA SERGIO BARRACHINA MARÍA ISABEL CASTILLO HÉCTOR MARTÍNEZ ENRIQUE S. QUINTANA ORTÍ IGNACIO MEDINA INTRODUCTION DNA Exon 0 Exon 1 Exon 2 Intron 0 Intron 1 Reads Sequencing RNA
JOAQUÍN DOPAZO JOAQUÍN TARRAGA SERGIO BARRACHINA MARÍA ISABEL CASTILLO HÉCTOR MARTÍNEZ ENRIQUE S. QUINTANA ORTÍ IGNACIO MEDINA INTRODUCTION DNA Exon 0 Exon 1 Exon 2 Intron 0 Intron 1 Reads Sequencing RNA
Introduction to Read Alignment. UCD Genome Center Bioinformatics Core Tuesday 15 September 2015
 Introduction to Read Alignment UCD Genome Center Bioinformatics Core Tuesday 15 September 2015 From reads to molecules Why align? Individual A Individual B ATGATAGCATCGTCGGGTGTCTGCTCAATAATAGTGCCGTATCATGCTGGTGTTATAATCGCCGCATGACATGATCAATGG
Introduction to Read Alignment UCD Genome Center Bioinformatics Core Tuesday 15 September 2015 From reads to molecules Why align? Individual A Individual B ATGATAGCATCGTCGGGTGTCTGCTCAATAATAGTGCCGTATCATGCTGGTGTTATAATCGCCGCATGACATGATCAATGG
Multimedia Networking ECE 599
 Multimedia Networking ECE 599 Prof. Thinh Nguyen School of Electrical Engineering and Computer Science Based on B. Lee s lecture notes. 1 Outline Compression basics Entropy and information theory basics
Multimedia Networking ECE 599 Prof. Thinh Nguyen School of Electrical Engineering and Computer Science Based on B. Lee s lecture notes. 1 Outline Compression basics Entropy and information theory basics
NGS Data Visualization and Exploration Using IGV
 1 What is Galaxy Galaxy for Bioinformaticians Galaxy for Experimental Biologists Using Galaxy for NGS Analysis NGS Data Visualization and Exploration Using IGV 2 What is Galaxy Galaxy for Bioinformaticians
1 What is Galaxy Galaxy for Bioinformaticians Galaxy for Experimental Biologists Using Galaxy for NGS Analysis NGS Data Visualization and Exploration Using IGV 2 What is Galaxy Galaxy for Bioinformaticians
De Novo NGS Data Compression
 De Novo NGS Data Compression Gaëtan Benoit, Claire Lemaitre, Guillaume Rizk, Erwan Drezen, Dominique Lavenier To cite this version: Gaëtan Benoit, Claire Lemaitre, Guillaume Rizk, Erwan Drezen, Dominique
De Novo NGS Data Compression Gaëtan Benoit, Claire Lemaitre, Guillaume Rizk, Erwan Drezen, Dominique Lavenier To cite this version: Gaëtan Benoit, Claire Lemaitre, Guillaume Rizk, Erwan Drezen, Dominique
Basic Compression Library
 Basic Compression Library Manual API version 1.2 July 22, 2006 c 2003-2006 Marcus Geelnard Summary This document describes the algorithms used in the Basic Compression Library, and how to use the library
Basic Compression Library Manual API version 1.2 July 22, 2006 c 2003-2006 Marcus Geelnard Summary This document describes the algorithms used in the Basic Compression Library, and how to use the library
Binary Encoded Attribute-Pairing Technique for Database Compression
 Binary Encoded Attribute-Pairing Technique for Database Compression Akanksha Baid and Swetha Krishnan Computer Sciences Department University of Wisconsin, Madison baid,swetha@cs.wisc.edu Abstract Data
Binary Encoded Attribute-Pairing Technique for Database Compression Akanksha Baid and Swetha Krishnan Computer Sciences Department University of Wisconsin, Madison baid,swetha@cs.wisc.edu Abstract Data
REPORT. NA12878 Platinum Genome. GENALICE MAP Analysis Report. Bas Tolhuis, PhD GENALICE B.V.
 REPORT NA12878 Platinum Genome GENALICE MAP Analysis Report Bas Tolhuis, PhD GENALICE B.V. INDEX EXECUTIVE SUMMARY...4 1. MATERIALS & METHODS...5 1.1 SEQUENCE DATA...5 1.2 WORKFLOWS......5 1.3 ACCURACY
REPORT NA12878 Platinum Genome GENALICE MAP Analysis Report Bas Tolhuis, PhD GENALICE B.V. INDEX EXECUTIVE SUMMARY...4 1. MATERIALS & METHODS...5 1.1 SEQUENCE DATA...5 1.2 WORKFLOWS......5 1.3 ACCURACY
GSNAP: Fast and SNP-tolerant detection of complex variants and splicing in short reads by Thomas D. Wu and Serban Nacu
 GSNAP: Fast and SNP-tolerant detection of complex variants and splicing in short reads by Thomas D. Wu and Serban Nacu Matt Huska Freie Universität Berlin Computational Methods for High-Throughput Omics
GSNAP: Fast and SNP-tolerant detection of complex variants and splicing in short reads by Thomas D. Wu and Serban Nacu Matt Huska Freie Universität Berlin Computational Methods for High-Throughput Omics
RNA-Seq in Galaxy: Tuxedo protocol. Igor Makunin, UQ RCC, QCIF
 RNA-Seq in Galaxy: Tuxedo protocol Igor Makunin, UQ RCC, QCIF Acknowledgments Genomics Virtual Lab: gvl.org.au Galaxy for tutorials: galaxy-tut.genome.edu.au Galaxy Australia: galaxy-aust.genome.edu.au
RNA-Seq in Galaxy: Tuxedo protocol Igor Makunin, UQ RCC, QCIF Acknowledgments Genomics Virtual Lab: gvl.org.au Galaxy for tutorials: galaxy-tut.genome.edu.au Galaxy Australia: galaxy-aust.genome.edu.au
arxiv: v2 [q-bio.qm] 17 Nov 2013
![arxiv: v2 [q-bio.qm] 17 Nov 2013 arxiv: v2 [q-bio.qm] 17 Nov 2013](/thumbs/90/101417919.jpg) arxiv:1308.2150v2 [q-bio.qm] 17 Nov 2013 GeneZip: A software package for storage-efficient processing of genotype data Palmer, Cameron 1 and Pe er, Itsik 1 1 Center for Computational Biology and Bioinformatics,
arxiv:1308.2150v2 [q-bio.qm] 17 Nov 2013 GeneZip: A software package for storage-efficient processing of genotype data Palmer, Cameron 1 and Pe er, Itsik 1 1 Center for Computational Biology and Bioinformatics,
SlopMap: a software application tool for quick and flexible identification of similar sequences using exact k-mer matching
 SlopMap: a software application tool for quick and flexible identification of similar sequences using exact k-mer matching Ilya Y. Zhbannikov 1, Samuel S. Hunter 1,2, Matthew L. Settles 1,2, and James
SlopMap: a software application tool for quick and flexible identification of similar sequences using exact k-mer matching Ilya Y. Zhbannikov 1, Samuel S. Hunter 1,2, Matthew L. Settles 1,2, and James
ChIP-seq (NGS) Data Formats
 ChIP-seq (NGS) Data Formats Biological samples Sequence reads SRA/SRF, FASTQ Quality control SAM/BAM/Pileup?? Mapping Assembly... DE Analysis Variant Detection Peak Calling...? Counts, RPKM VCF BED/narrowPeak/
ChIP-seq (NGS) Data Formats Biological samples Sequence reads SRA/SRF, FASTQ Quality control SAM/BAM/Pileup?? Mapping Assembly... DE Analysis Variant Detection Peak Calling...? Counts, RPKM VCF BED/narrowPeak/
ROOT I/O compression algorithms. Oksana Shadura, Brian Bockelman University of Nebraska-Lincoln
 ROOT I/O compression algorithms Oksana Shadura, Brian Bockelman University of Nebraska-Lincoln Introduction Compression Algorithms 2 Compression algorithms Los Reduces size by permanently eliminating certain
ROOT I/O compression algorithms Oksana Shadura, Brian Bockelman University of Nebraska-Lincoln Introduction Compression Algorithms 2 Compression algorithms Los Reduces size by permanently eliminating certain
MIGRATORY COMPRESSION Coarse-grained Data Reordering to Improve Compressibility
 MIGRATORY COMPRESSION Coarse-grained Data Reordering to Improve Compressibility Xing Lin *, Guanlin Lu, Fred Douglis, Philip Shilane, Grant Wallace * University of Utah EMC Corporation Data Protection
MIGRATORY COMPRESSION Coarse-grained Data Reordering to Improve Compressibility Xing Lin *, Guanlin Lu, Fred Douglis, Philip Shilane, Grant Wallace * University of Utah EMC Corporation Data Protection
V.2 Index Compression
 V.2 Index Compression Heap s law (empirically observed and postulated): Size of the vocabulary (distinct terms) in a corpus E[ distinct terms in corpus] n with total number of term occurrences n, and constants,
V.2 Index Compression Heap s law (empirically observed and postulated): Size of the vocabulary (distinct terms) in a corpus E[ distinct terms in corpus] n with total number of term occurrences n, and constants,
Data Preprocessing. Next Generation Sequencing analysis DTU Bioinformatics Next Generation Sequencing Analysis
 Data Preprocessing Next Generation Sequencing analysis DTU Bioinformatics Generalized NGS analysis Data size Application Assembly: Compare Raw Pre- specific: Question Alignment / samples / Answer? reads
Data Preprocessing Next Generation Sequencing analysis DTU Bioinformatics Generalized NGS analysis Data size Application Assembly: Compare Raw Pre- specific: Question Alignment / samples / Answer? reads
Toward High Utilization of Heterogeneous Computing Resources in SNP Detection
 Toward High Utilization of Heterogeneous Computing Resources in SNP Detection Myungeun Lim, Minho Kim, Ho-Youl Jung, Dae-Hee Kim, Jae-Hun Choi, Wan Choi, and Kyu-Chul Lee As the amount of re-sequencing
Toward High Utilization of Heterogeneous Computing Resources in SNP Detection Myungeun Lim, Minho Kim, Ho-Youl Jung, Dae-Hee Kim, Jae-Hun Choi, Wan Choi, and Kyu-Chul Lee As the amount of re-sequencing
Short Read Alignment. Mapping Reads to a Reference
 Short Read Alignment Mapping Reads to a Reference Brandi Cantarel, Ph.D. & Daehwan Kim, Ph.D. BICF 05/2018 Introduction to Mapping Short Read Aligners DNA vs RNA Alignment Quality Pitfalls and Improvements
Short Read Alignment Mapping Reads to a Reference Brandi Cantarel, Ph.D. & Daehwan Kim, Ph.D. BICF 05/2018 Introduction to Mapping Short Read Aligners DNA vs RNA Alignment Quality Pitfalls and Improvements
Mar%n Norling. Uppsala, November 15th 2016
 Mar%n Norling Uppsala, November 15th 2016 What can we do with an assembly? Since we can never know the actual sequence, or its varia%ons, valida%ng an assembly is tricky. But once you ve used all the assemblers,
Mar%n Norling Uppsala, November 15th 2016 What can we do with an assembly? Since we can never know the actual sequence, or its varia%ons, valida%ng an assembly is tricky. But once you ve used all the assemblers,
Hashing. Hashing Procedures
 Hashing Hashing Procedures Let us denote the set of all possible key values (i.e., the universe of keys) used in a dictionary application by U. Suppose an application requires a dictionary in which elements
Hashing Hashing Procedures Let us denote the set of all possible key values (i.e., the universe of keys) used in a dictionary application by U. Suppose an application requires a dictionary in which elements
THE RELATIVE EFFICIENCY OF DATA COMPRESSION BY LZW AND LZSS
 THE RELATIVE EFFICIENCY OF DATA COMPRESSION BY LZW AND LZSS Yair Wiseman 1* * 1 Computer Science Department, Bar-Ilan University, Ramat-Gan 52900, Israel Email: wiseman@cs.huji.ac.il, http://www.cs.biu.ac.il/~wiseman
THE RELATIVE EFFICIENCY OF DATA COMPRESSION BY LZW AND LZSS Yair Wiseman 1* * 1 Computer Science Department, Bar-Ilan University, Ramat-Gan 52900, Israel Email: wiseman@cs.huji.ac.il, http://www.cs.biu.ac.il/~wiseman
Category: Informational May DEFLATE Compressed Data Format Specification version 1.3
 Network Working Group P. Deutsch Request for Comments: 1951 Aladdin Enterprises Category: Informational May 1996 DEFLATE Compressed Data Format Specification version 1.3 Status of This Memo This memo provides
Network Working Group P. Deutsch Request for Comments: 1951 Aladdin Enterprises Category: Informational May 1996 DEFLATE Compressed Data Format Specification version 1.3 Status of This Memo This memo provides
Analysis of ChIP-seq data
 Before we start: 1. Log into tak (step 0 on the exercises) 2. Go to your lab space and create a folder for the class (see separate hand out) 3. Connect to your lab space through the wihtdata network and
Before we start: 1. Log into tak (step 0 on the exercises) 2. Go to your lab space and create a folder for the class (see separate hand out) 3. Connect to your lab space through the wihtdata network and
BLAST & Genome assembly
 BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies May 15, 2014 1 BLAST What is BLAST? The algorithm 2 Genome assembly De novo assembly Mapping assembly 3
BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies May 15, 2014 1 BLAST What is BLAST? The algorithm 2 Genome assembly De novo assembly Mapping assembly 3
HARDWARE IMPLEMENTATION OF LOSSLESS LZMA DATA COMPRESSION ALGORITHM
 HARDWARE IMPLEMENTATION OF LOSSLESS LZMA DATA COMPRESSION ALGORITHM Parekar P. M. 1, Thakare S. S. 2 1,2 Department of Electronics and Telecommunication Engineering, Amravati University Government College
HARDWARE IMPLEMENTATION OF LOSSLESS LZMA DATA COMPRESSION ALGORITHM Parekar P. M. 1, Thakare S. S. 2 1,2 Department of Electronics and Telecommunication Engineering, Amravati University Government College
Data Compression. Guest lecture, SGDS Fall 2011
 Data Compression Guest lecture, SGDS Fall 2011 1 Basics Lossy/lossless Alphabet compaction Compression is impossible Compression is possible RLE Variable-length codes Undecidable Pigeon-holes Patterns
Data Compression Guest lecture, SGDS Fall 2011 1 Basics Lossy/lossless Alphabet compaction Compression is impossible Compression is possible RLE Variable-length codes Undecidable Pigeon-holes Patterns
High-throughout sequencing and using short-read aligners. Simon Anders
 High-throughout sequencing and using short-read aligners Simon Anders High-throughput sequencing (HTS) Sequencing millions of short DNA fragments in parallel. a.k.a.: next-generation sequencing (NGS) massively-parallel
High-throughout sequencing and using short-read aligners Simon Anders High-throughput sequencing (HTS) Sequencing millions of short DNA fragments in parallel. a.k.a.: next-generation sequencing (NGS) massively-parallel
Bioinformatics Framework
 Persona: A High-Performance Bioinformatics Framework Stuart Byma 1, Sam Whitlock 1, Laura Flueratoru 2, Ethan Tseng 3, Christos Kozyrakis 4, Edouard Bugnion 1, James Larus 1 EPFL 1, U. Polytehnica of Bucharest
Persona: A High-Performance Bioinformatics Framework Stuart Byma 1, Sam Whitlock 1, Laura Flueratoru 2, Ethan Tseng 3, Christos Kozyrakis 4, Edouard Bugnion 1, James Larus 1 EPFL 1, U. Polytehnica of Bucharest
Sentieon Documentation
 Sentieon Documentation Release 201808.03 Sentieon, Inc Dec 21, 2018 Sentieon Manual 1 Introduction 1 1.1 Description.............................................. 1 1.2 Benefits and Value..........................................
Sentieon Documentation Release 201808.03 Sentieon, Inc Dec 21, 2018 Sentieon Manual 1 Introduction 1 1.1 Description.............................................. 1 1.2 Benefits and Value..........................................
Evolutionary Lossless Compression with GP-ZIP
 Evolutionary Lossless Compression with GP-ZIP Ahmad Kattan and Riccardo Poli Abstract In this paper we propose a new approach for applying Genetic Programming to lossless data compression based on combining
Evolutionary Lossless Compression with GP-ZIP Ahmad Kattan and Riccardo Poli Abstract In this paper we propose a new approach for applying Genetic Programming to lossless data compression based on combining
Ch. 2: Compression Basics Multimedia Systems
 Ch. 2: Compression Basics Multimedia Systems Prof. Ben Lee School of Electrical Engineering and Computer Science Oregon State University Outline Why compression? Classification Entropy and Information
Ch. 2: Compression Basics Multimedia Systems Prof. Ben Lee School of Electrical Engineering and Computer Science Oregon State University Outline Why compression? Classification Entropy and Information
Indexing. CS6200: Information Retrieval. Index Construction. Slides by: Jesse Anderton
 Indexing Index Construction CS6200: Information Retrieval Slides by: Jesse Anderton Motivation: Scale Corpus Terms Docs Entries A term incidence matrix with V terms and D documents has O(V x D) entries.
Indexing Index Construction CS6200: Information Retrieval Slides by: Jesse Anderton Motivation: Scale Corpus Terms Docs Entries A term incidence matrix with V terms and D documents has O(V x D) entries.
discosnp++ Reference-free detection of SNPs and small indels v2.2.2
 discosnp++ Reference-free detection of SNPs and small indels v2.2.2 User's guide November 2015 contact: pierre.peterlongo@inria.fr Table of contents GNU AFFERO GENERAL PUBLIC LICENSE... 1 Publication...
discosnp++ Reference-free detection of SNPs and small indels v2.2.2 User's guide November 2015 contact: pierre.peterlongo@inria.fr Table of contents GNU AFFERO GENERAL PUBLIC LICENSE... 1 Publication...
S 1. Evaluation of Fast-LZ Compressors for Compacting High-Bandwidth but Redundant Streams from FPGA Data Sources
 Evaluation of Fast-LZ Compressors for Compacting High-Bandwidth but Redundant Streams from FPGA Data Sources Author: Supervisor: Luhao Liu Dr. -Ing. Thomas B. Preußer Dr. -Ing. Steffen Köhler 09.10.2014
Evaluation of Fast-LZ Compressors for Compacting High-Bandwidth but Redundant Streams from FPGA Data Sources Author: Supervisor: Luhao Liu Dr. -Ing. Thomas B. Preußer Dr. -Ing. Steffen Köhler 09.10.2014
IMPROVED CONTEXT-ADAPTIVE ARITHMETIC CODING IN H.264/AVC
 17th European Signal Processing Conference (EUSIPCO 2009) Glasgow, Scotland, August 24-28, 2009 IMPROVED CONTEXT-ADAPTIVE ARITHMETIC CODING IN H.264/AVC Damian Karwowski, Marek Domański Poznań University
17th European Signal Processing Conference (EUSIPCO 2009) Glasgow, Scotland, August 24-28, 2009 IMPROVED CONTEXT-ADAPTIVE ARITHMETIC CODING IN H.264/AVC Damian Karwowski, Marek Domański Poznań University
Cycle «Analyse de données de séquençage à haut-débit» Module 1/5 Analyse ADN. Sophie Gallina CNRS Evo-Eco-Paléo (EEP)
 Cycle «Analyse de données de séquençage à haut-débit» Module 1/5 Analyse ADN Sophie Gallina CNRS Evo-Eco-Paléo (EEP) (sophie.gallina@univ-lille1.fr) Module 1/5 Analyse DNA NGS Introduction Galaxy : upload
Cycle «Analyse de données de séquençage à haut-débit» Module 1/5 Analyse ADN Sophie Gallina CNRS Evo-Eco-Paléo (EEP) (sophie.gallina@univ-lille1.fr) Module 1/5 Analyse DNA NGS Introduction Galaxy : upload
Pre-processing and quality control of sequence data. Barbera van Schaik KEBB - Bioinformatics Laboratory
 Pre-processing and quality control of sequence data Barbera van Schaik KEBB - Bioinformatics Laboratory b.d.vanschaik@amc.uva.nl Topic: quality control and prepare data for the interesting stuf Keep Throw
Pre-processing and quality control of sequence data Barbera van Schaik KEBB - Bioinformatics Laboratory b.d.vanschaik@amc.uva.nl Topic: quality control and prepare data for the interesting stuf Keep Throw
Easy visualization of the read coverage using the CoverageView package
 Easy visualization of the read coverage using the CoverageView package Ernesto Lowy European Bioinformatics Institute EMBL June 13, 2018 > options(width=40) > library(coverageview) 1 Introduction This
Easy visualization of the read coverage using the CoverageView package Ernesto Lowy European Bioinformatics Institute EMBL June 13, 2018 > options(width=40) > library(coverageview) 1 Introduction This
The SAM Format Specification (v1.3-r837)
 The SAM Format Specification (v1.3-r837) The SAM Format Specification Working Group November 18, 2010 1 The SAM Format Specification SAM stands for Sequence Alignment/Map format. It is a TAB-delimited
The SAM Format Specification (v1.3-r837) The SAM Format Specification Working Group November 18, 2010 1 The SAM Format Specification SAM stands for Sequence Alignment/Map format. It is a TAB-delimited
JULIA ENABLED COMPUTATION OF MOLECULAR LIBRARY COMPLEXITY IN DNA SEQUENCING
 JULIA ENABLED COMPUTATION OF MOLECULAR LIBRARY COMPLEXITY IN DNA SEQUENCING Larson Hogstrom, Mukarram Tahir, Andres Hasfura Massachusetts Institute of Technology, Cambridge, Massachusetts, USA 18.337/6.338
JULIA ENABLED COMPUTATION OF MOLECULAR LIBRARY COMPLEXITY IN DNA SEQUENCING Larson Hogstrom, Mukarram Tahir, Andres Hasfura Massachusetts Institute of Technology, Cambridge, Massachusetts, USA 18.337/6.338
Performance analysis of parallel de novo genome assembly in shared memory system
 IOP Conference Series: Earth and Environmental Science PAPER OPEN ACCESS Performance analysis of parallel de novo genome assembly in shared memory system To cite this article: Syam Budi Iryanto et al 2018
IOP Conference Series: Earth and Environmental Science PAPER OPEN ACCESS Performance analysis of parallel de novo genome assembly in shared memory system To cite this article: Syam Budi Iryanto et al 2018
SIGNAL COMPRESSION Lecture Lempel-Ziv Coding
 SIGNAL COMPRESSION Lecture 5 11.9.2007 Lempel-Ziv Coding Dictionary methods Ziv-Lempel 77 The gzip variant of Ziv-Lempel 77 Ziv-Lempel 78 The LZW variant of Ziv-Lempel 78 Asymptotic optimality of Ziv-Lempel
SIGNAL COMPRESSION Lecture 5 11.9.2007 Lempel-Ziv Coding Dictionary methods Ziv-Lempel 77 The gzip variant of Ziv-Lempel 77 Ziv-Lempel 78 The LZW variant of Ziv-Lempel 78 Asymptotic optimality of Ziv-Lempel
BIOINFORMATICS. Lossy Compression of Quality Scores in Genomic Data. Rodrigo Cánovas, Alistair Moffat, and Andrew Turpin
 BIOINFORMATICS Vol. 00 no. 00 014 Pages 1 7 Lossy Compression of Quality Scores in Genomic Data Rodrigo Cánovas, Alistair Moffat, and Andrew Turpin NICTA Victoria Research Laboratory, Department of Computing
BIOINFORMATICS Vol. 00 no. 00 014 Pages 1 7 Lossy Compression of Quality Scores in Genomic Data Rodrigo Cánovas, Alistair Moffat, and Andrew Turpin NICTA Victoria Research Laboratory, Department of Computing
Modified SPIHT Image Coder For Wireless Communication
 Modified SPIHT Image Coder For Wireless Communication M. B. I. REAZ, M. AKTER, F. MOHD-YASIN Faculty of Engineering Multimedia University 63100 Cyberjaya, Selangor Malaysia Abstract: - The Set Partitioning
Modified SPIHT Image Coder For Wireless Communication M. B. I. REAZ, M. AKTER, F. MOHD-YASIN Faculty of Engineering Multimedia University 63100 Cyberjaya, Selangor Malaysia Abstract: - The Set Partitioning
Analyzing Variant Call results using EuPathDB Galaxy, Part II
 Analyzing Variant Call results using EuPathDB Galaxy, Part II In this exercise, we will work in groups to examine the results from the SNP analysis workflow that we started yesterday. The first step is
Analyzing Variant Call results using EuPathDB Galaxy, Part II In this exercise, we will work in groups to examine the results from the SNP analysis workflow that we started yesterday. The first step is
Data Preprocessing : Next Generation Sequencing analysis CBS - DTU Next Generation Sequencing Analysis
 Data Preprocessing 27626: Next Generation Sequencing analysis CBS - DTU Generalized NGS analysis Data size Application Assembly: Compare Raw Pre- specific: Question Alignment / samples / Answer? reads
Data Preprocessing 27626: Next Generation Sequencing analysis CBS - DTU Generalized NGS analysis Data size Application Assembly: Compare Raw Pre- specific: Question Alignment / samples / Answer? reads
From the Schnable Lab:
 From the Schnable Lab: Yang Zhang and Daniel Ngu s Pipeline for Processing RNA-seq Data (As of November 17, 2016) yzhang91@unl.edu dngu2@huskers.unl.edu Pre-processing the reads: The alignment software
From the Schnable Lab: Yang Zhang and Daniel Ngu s Pipeline for Processing RNA-seq Data (As of November 17, 2016) yzhang91@unl.edu dngu2@huskers.unl.edu Pre-processing the reads: The alignment software
Exome sequencing. Jong Kyoung Kim
 Exome sequencing Jong Kyoung Kim Genome Analysis Toolkit The GATK is the industry standard for identifying SNPs and indels in germline DNA and RNAseq data. Its scope is now expanding to include somatic
Exome sequencing Jong Kyoung Kim Genome Analysis Toolkit The GATK is the industry standard for identifying SNPs and indels in germline DNA and RNAseq data. Its scope is now expanding to include somatic
Halvade: scalable sequence analysis with MapReduce
 Bioinformatics Advance Access published March 26, 2015 Halvade: scalable sequence analysis with MapReduce Dries Decap 1,5, Joke Reumers 2,5, Charlotte Herzeel 3,5, Pascal Costanza, 4,5 and Jan Fostier
Bioinformatics Advance Access published March 26, 2015 Halvade: scalable sequence analysis with MapReduce Dries Decap 1,5, Joke Reumers 2,5, Charlotte Herzeel 3,5, Pascal Costanza, 4,5 and Jan Fostier
Reads Alignment and Variant Calling
 Reads Alignment and Variant Calling CB2-201 Computational Biology and Bioinformatics February 22, 2016 Emidio Capriotti http://biofold.org/ Institute for Mathematical Modeling of Biological Systems Department
Reads Alignment and Variant Calling CB2-201 Computational Biology and Bioinformatics February 22, 2016 Emidio Capriotti http://biofold.org/ Institute for Mathematical Modeling of Biological Systems Department
Run-Length and Markov Compression Revisited in DNA Sequences
 Run-Length and Markov Compression Revisited in DNA Sequences Saurabh Kadekodi M.S. Computer Science saurabhkadekodi@u.northwestern.edu Efficient and economical storage of DNA sequences has always been
Run-Length and Markov Compression Revisited in DNA Sequences Saurabh Kadekodi M.S. Computer Science saurabhkadekodi@u.northwestern.edu Efficient and economical storage of DNA sequences has always been
Accelerating InDel Detection on Modern Multi-Core SIMD CPU Architecture
 Accelerating InDel Detection on Modern Multi-Core SIMD CPU Architecture Da Zhang Collaborators: Hao Wang, Kaixi Hou, Jing Zhang Advisor: Wu-chun Feng Evolution of Genome Sequencing1 In 20032: 1 human genome
Accelerating InDel Detection on Modern Multi-Core SIMD CPU Architecture Da Zhang Collaborators: Hao Wang, Kaixi Hou, Jing Zhang Advisor: Wu-chun Feng Evolution of Genome Sequencing1 In 20032: 1 human genome
Under the Hood of Alignment Algorithms for NGS Researchers
 Under the Hood of Alignment Algorithms for NGS Researchers April 16, 2014 Gabe Rudy VP of Product Development Golden Helix Questions during the presentation Use the Questions pane in your GoToWebinar window
Under the Hood of Alignment Algorithms for NGS Researchers April 16, 2014 Gabe Rudy VP of Product Development Golden Helix Questions during the presentation Use the Questions pane in your GoToWebinar window
seqcna: A Package for Copy Number Analysis of High-Throughput Sequencing Cancer DNA
 seqcna: A Package for Copy Number Analysis of High-Throughput Sequencing Cancer DNA David Mosen-Ansorena 1 October 30, 2017 Contents 1 Genome Analysis Platform CIC biogune and CIBERehd dmosen.gn@cicbiogune.es
seqcna: A Package for Copy Number Analysis of High-Throughput Sequencing Cancer DNA David Mosen-Ansorena 1 October 30, 2017 Contents 1 Genome Analysis Platform CIC biogune and CIBERehd dmosen.gn@cicbiogune.es
Performing whole genome SNP analysis with mapping performed locally
 BioNumerics Tutorial: Performing whole genome SNP analysis with mapping performed locally 1 Introduction 1.1 An introduction to whole genome SNP analysis A Single Nucleotide Polymorphism (SNP) is a variation
BioNumerics Tutorial: Performing whole genome SNP analysis with mapping performed locally 1 Introduction 1.1 An introduction to whole genome SNP analysis A Single Nucleotide Polymorphism (SNP) is a variation
Read Naming Format Specification
 Read Naming Format Specification Karel Břinda Valentina Boeva Gregory Kucherov Version 0.1.3 (4 August 2015) Abstract This document provides a standard for naming simulated Next-Generation Sequencing (Ngs)
Read Naming Format Specification Karel Břinda Valentina Boeva Gregory Kucherov Version 0.1.3 (4 August 2015) Abstract This document provides a standard for naming simulated Next-Generation Sequencing (Ngs)
Comparison of Phylogenetic Trees of Multiple Protein Sequence Alignment Methods
 Comparison of Phylogenetic Trees of Multiple Protein Sequence Alignment Methods Khaddouja Boujenfa, Nadia Essoussi, and Mohamed Limam International Science Index, Computer and Information Engineering waset.org/publication/482
Comparison of Phylogenetic Trees of Multiple Protein Sequence Alignment Methods Khaddouja Boujenfa, Nadia Essoussi, and Mohamed Limam International Science Index, Computer and Information Engineering waset.org/publication/482
