Fast and Robust 3-D MRI Brain Structure Segmentation
|
|
|
- Marcia Griffin
- 5 years ago
- Views:
Transcription
1 Fast and Robust 3-D MRI Brain Structure Segmentation Michael Wels 1,4, Yefeng Zheng 2, Gustavo Carneiro 3,, Martin Huber 4, Joachim Hornegger 1, and Dorin Comaniciu 2 1 Chair of Pattern Recognition, Department of Computer Science, University Erlangen-Nuremberg, Germany, michael.wels@informatik.uni-erlangen.de 2 Integrated Data Systems, Siemens Corporate Research, Princeton, NJ, USA 3 Institute for Systems and Robotics, Electrical and Computer Engineering Department, Technical University of Lisbon, Portugal 4 Siemens CT SE5 SCR2, Erlangen, Germany Abstract. We present a novel method for the automatic detection and segmentation of (sub-)cortical gray matter structures in 3-D magnetic resonance images of the human brain. Essentially, the method is a topdown segmentation approach based on the recently introduced concept of Marginal Space Learning (MSL). We show that MSL naturally decomposes the parameter space of anatomy shapes along decreasing levels of geometrical abstraction into subspaces of increasing dimensionality by exploiting parameter invariance. At each level of abstraction, i.e., in each subspace, we build strong discriminative models from annotated training data, and use these models to narrow the range of possible solutions until a final shape can be inferred. Contextual information is introduced into the system by representing candidate shape parameters with high-dimensional vectors of 3-D generalized Haar features and steerable features derived from the observed volume intensities. Our system allows us to detect and segment 8 (sub-)cortical gray matter structures in T1-weighted 3-D MR brain scans from a variety of different scanners in on average 13.9 sec., which is faster than most of the approaches in the literature. In order to ensure comparability of the achieved results and to validate robustness, we evaluate our method on two publicly available gold standard databases consisting of several T1-weighted 3-D brain MR scans from different scanners and sites. The proposed method achieves an accuracy better than most state-of-the-art approaches using standardized distance and overlap metrics. 1 Introduction Currently, many scientific questions in neurology, like the revelation of mechanisms affecting generative or degenerative processes in brain development, require quantitative volumetric analysis of (sub-)cortical gray matter structures G. Carneiro contributed to this work when he was with the Integrated Data Systems Department of Siemens Corporate Research. This work was partially funded by the Health-e-Child project (IST ).
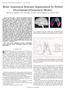
2 2 MSL 3-D Shape Detection and Inference Dynamic Histogram Warping Position Detection Position- Orientation Detection Full Similarity Transformation Detection Shape Inference Fig. 1. The processing pipeline of the proposed 3-D shape detection and inference method. Each image (detection and delineation of the left caudate) schematically represents the input and/or output of individual processing steps. Please view in color. in large populations of patients and healthy controls. For instance, atrophy in the presence of Alzheimer s disease considerably affects morphology of the hippocampus. In addition, 3-D segmentation of various deep gray matter structures facilitates image-based surgical planning, therapy monitoring, and the generation of patient-specific geometrical models from imaging data for further processing. As a result of unclear boundaries, shape complexity, and different anatomical definitions, precise manual delineation is usually time consuming and user dependent. Moreover, typical artifacts present in MR imaging (Rician noise, partial volume effects, and intra-/inter-scan intensity non-uniformities) challenge the consistency of manual delineations. Therefore, a system for the automatic detection and segmentation of (sub-)cortical gray matter structures not only has the potential to increase segmentation consistency, but also has the capability of facilitating large-scale neuromorphological studies. We propose a fully automatic method for the detection and delineation of the following eight (sub-)cortical gray matter structures: the left and right caudate nucleus, hippocampus, globus pallidus, and putamen. Our method consists of two major steps: 1) we standardize the observed MR intensities by non-rigidly aligning their histogram to a template histogram by means of Dynamic Histogram Warping (DHW) [1]; and 2) for each (sub-)cortical structure of interest we detect and infer its position, orientation, scale, and shape in an extended Marginal Space Learning (MSL) framework [2], which explicitly integrates shape inference into the overall MSL formulation. The overall system block diagram is depicted in Fig. 1. In contrast to other methods [3 5] where a partly manually initialized nine parameter registration is part of the approaches we do not require the input volumes to be spatially normalized. In some cases [3, 4], the feature pools used for discriminative model generation are enriched with features explicitly encoding normalized location. In accordance with this observation, the approaches are only evaluated on spatially normalized data sets from one type of MR scanner that are not publicly available [4, 5]. Nevertheless, Morra et al. [3] report state-of-the-art results on data sets that have not been subject to spatial normalization. Apart from that, the mentioned methods make use of machine learning in a similar manner as we do, but follow a bottom up approach ascending from the lowest level of abstraction, i.e., the level of individual voxels, to the level of complete anatomical entities.
3 3 Alignment of a probabilistic atlas by means of an affine registration also plays an important role in further approaches [6, 7]. While sometimes [6] quantitative evaluation is only carried out on simulated data, the method of Akselrod-Ballin et al. [7] is trained and evaluated on only one publicly available dataset that has been subject to a specific preprocessing including intensity standardization. By generating observation or discriminative models based on intensity values without explicitly allowing for inter-scan intensity variations [3 5, 7] the resulting models are at the risk of being over-adapted to specific contrast-characteristics of the data at hand. 2 Methods 2.1 Combined 3-D Shape Detection and Shape Inference For combined 3-D rigid anatomy detection and shape inference we use a method based on the concept of Marginal Space Learning (MSL) [2]. We estimate the structure of interest s center c = (c 1,c 2,c 3 ) R 3, orientation θ = (θ 1,θ 2,θ 3 ) [ π,π] [ π/2,π/2] [ π,π] represented as Euler angles in z x z convention, scale s = (s 1,s 2,s 3 ) {s R 3 s i > 0,i = 1,2,3}, and shape X = (x 1,...,x n ) R 3 n. The latter consists of canonically sampled 3-D points on the surface of an object to be segmented. Note that θ is relative to c, s is relative to c and θ, and X is relative to c, θ, and s. Let V = {1,2,...,N }, N N, be a set of indices to image voxels, Y = (y v ) v V, y v { 1,1}, a binary segmentation of the image voxels into object and non-object voxels, and f be a function with Y = f(i,θ) that provides a binary segmentation of volume I using segmentation parameters Θ = (c,θ,s,x). Let Z = (z Θ ) be a family of high-dimensional feature vectors extracted from a given input volume I = (i v ) v V and associated with different discretized configurations of Θ. In our context Z includes voxel-wise context encoding 3-D generalized Haar features [8] to characterize possible object centers and steerable features [2] that are capable of representing hypothetical orientations and optionally scaling relative to a given object center or shape surface point. These features were chosen for our method because of their fast computation and effective representation [2]. We search for the optimal parameter vector Θ = arg max p(y = Θ 1 Θ,I,M(Θ) ) = arg max p(y = Θ 1 Z,M(Θ) ) (1) maximizing the posterior probability of the presence, i.e., y = 1, of a sought anatomy given the discriminative model M (Θ) and the features Z extracted from the input volume I using a certain set of values for the parameters Θ. Let π (c) (Z), π (c,θ) (Z), π (c,θ,s) (Z), π (c,θ,s,x) (Z) denote the vectors of components of Z associated with individual groups of elements (c), (c,θ), (c,θ,s), and (c,θ,s,x) of the parameter vector Θ. The MSL method avoids exhaustively searching the high-dimensional parameter space spanned by all the possible Θ by exploiting the fact that ideally for any discriminative model for center detection with parameters M (c) working on a restricted amount of possible features c = arg max p(y = 1 π (c) (Z),M (c) ) (2) c
4 4 holds, as the object center c is invariant under relative reorientation, relative rescaling, and relative shape positioning. Similarly, we have θ = arg max p(y = 1 π (c,θ) (Z),M (c,θ) ) (3) θ for combined position-orientation detection with model parameters M (c,θ) where only features π (c,θ) (Z) with c = c are considered. This is due to the fact that position and orientation are invariant under relative rescaling and relative shape positioning. Analogous considerations yield for the object s scaling, and s = arg max p(y = 1 π (c,θ,s) (Z),M (c,θ,s) ) (4) s X = arg max X p(y = 1 π(c,θ,s,x) (Z),M (c,θ,s,x),m (c,θ,s,x) ) (5) for the object s shape where M (c,θ,s,x) are the parameters of a local shape model with respect to individual surface points x and parameters M (c,θ,s,x) represent a global shape model. Equations (2) (5) naturally establish a chain of discriminative models exploiting search space parameter invariance for combined 3-D shape detection and shape inference. It allows us to apply different discriminative models descending along geometrical abstraction as, in our framework, the object center c alone is the most abstract and the complete set of parameters Θ is the least abstract shape representation. Therefore, MSL establishes a hierarchical decomposition of the search space along decreasing levels of geometrical abstraction with increasing dimensionality of the considered parameter subspace D Shape Detection: Similarity Transformation Estimation Let Z be the set of annotated image volumes in their transformed feature representation as mentioned above. We will refer to Z as the training data. In order to find the first parts of the optimal parameter vector Θ describing a nine parameter similarity transformation, i.e., c, θ, and s, we have to learn discriminative models p(y = 1 π (c ) (Z)), p(y = 1 π (c,θ) (Z)), and p(y = 1 π (c,θ,s) (Z)). Following the concept of MSL [2] we generate a set of positive and negative training examples C = {(π (c) (Z),y) Z Z } to train a probabilistic boosting tree (PBT) model [9] for position detection. The feature vectors π (c) (Z) consist of 3-D generalized Haar features [8] encoding voxel context of candidate object centers based on observed intensity values. Decreasing the level of geometric abstraction we analogously train a PBT model for combined position-orientation detection based on an extended set of training examples P = {(π (c,θ) (Z),y) Z Z } where π (c,θ) (Z), associated with (c,θ) and an image volume, is made of steerable features [2]. They allow varying orientation and scaling to be encoded in terms of aligned and scaled intensity sampling patterns. In accordance with this scheme, steerable features are also used to finally train a PBT for full nine parameter similarity transformation detection based on S = {(π (c,θ,s) (Z),y) Z Z } where π (c,θ,s) (Z) is derived from (c,θ,s) and the associated image volume.
5 Table 1. Average segmentation accuracy (left and right structures grouped together) for IBSR 18 of models trained from mutually exclusive training and test data. Except for the Dice coefficient (2 TP/(2 TP + FP + FN) where TP, FP, and FN denote the number of true positive, false positive, and false negative voxels, respectively) see [13] on details on the used accuracy metrics. Structure Overlap Err. Dice Coeff. Volume Diff. Abs. Dist. RMS Dist. Max. Dist. [%] [%] [%] [mm] [mm] [mm] Caudate nucleus ± ± ± ± ± ± 1.82 Hippocampus ± ± ± ± ± ± 1.63 Globus pallidus ± ± ± ± ± ± 1.63 Putamen ± ± ± ± ± ± D Shape Inference under Global Shape Constraints For the final object shape we further decompose ( ) π (c,θ,s,x) (Z) = π (c,θ,s,xi) (Z) i=1,...,n where π (c,θ,s,xi) (Z) are the features associated with an image volume and individual relatively aligned candidate points (c,θ,s,x i ) for the surface of the object of interest. In order to apply discriminative modeling we assume the x i and correspondingly π (c,θ,s,xi) (Z) to be independently and identically distributed (i.i.d.) and approximate X = arg max X arg max X,θ,s,X) p(y = 1 π(c (Z), M (c,θ,s,x), M (c,θ,s,x) ) [ n ] p(y i = 1 π (c,θ,s,x i ) (Z), M (c,θ,s,x) ) p(x c, θ, s, M (c,θ,s,x) ) (6) i=1 in an iterative manner. The term p(y i = 1 π (c,θ,s,xi) (Z)) describes the probability that the relatively aligned point (c,θ,s,x i ) is part of the shape to be inferred, i.e., lies on its surface, and p(x c,θ,s,m (c,θ,s,x) ) is a global shape model [10]. We estimate p(y = 1 π (c,θ,s,xi) (Z)) with a PBT model [9] using steerable features [2] trained on X = {(π c,θ,s,xi (Z),y) i = 1,...,n;Z Z }. An iterative approach for (6) is suitable as, in practice, X R 3 n only varies around the mean shape positioned relatively to the (c,θ,s ) detected before at time t = 0 and the previous most likely anatomy shape in each iteration t = 1,...,T. 3 Material and Experimental Setting For training and quantitative evaluation of our system there were four sets of T1-weighted MRI scans available. The first one is a subset of the Designed Database of MR Brain Images of Healthy Volunteers 5 [11] (DDHV) containing 20 scans. The associated ground-truth annotations were manually recovered 5 The database was collected and made available by the CASILab at the University of North Carolina, Chapel Hill. The images were distributed by the MIDAS Data Server at Kitware, Inc. (insight-journal.org/midas). The authors would like to thank Martin Styner, Clement Vachet, and Paul Pandea for helping to preprocess parts of the data.
6 6 from automatically generated segmentations [12] of the structures of interest. The second collection of 18 MRI scans was provided by the Center of Morphometric Analysis at the Massachusetts General Hospital and is publicly available on the Internet Brain Segmentation Repository 6 (IBSR 18). The scans are accompanied by detailed ground-truth annotations including the (sub-)cortical structures of interest in this paper. 7 A subset 8 of the data provided by the NIH MRI Study of Normal Brain Development 9 consisting of 10 pediatric data sets states another collection (NIH) of annotated MR scans used for model generation. They have been manually annotated by the authors for training purposes. Additionally, we use data provided by the ongoing 3-D Segmentation in the Clinic: A Grand Challenge competition 10 [13] for training and evaluation of the proposed method. The collection consists of several volumetric T1-weighted MR brain scans of varying spatial resolution and size from multiple sources (MICCAI 07 training/testing). The vast majority of data (29 scans) has been provided by the Psychiatry Neuroimaging Laboratory (PNL) at the Brigham and Women s Hospital (BWH), Boston. The other 20 data sets arose from a pediatric study, a Parkinson s Disease study, and a test/re-test study carried out at the University of North Carolina s (UNC) Neuroimaging Laboratory (NIAL), Chapel Hill. A predefined evaluation protocol is carried out fully automatically after uploading the testing fraction of the data to the Cause 07 file server. We refer to Heimann et al. [13] for details on the used evaluation measures and scoring system. All the images were re-oriented to a uniform orientation ( RAI ; right-toleft, anterior-to-posterior, inferior-to-superior) and resampled to isotropic voxel spacing ( mm 3 ) for processing. For increasing the amount of training data we exploited natural brain symmetry and therefore doubled the size of any training data set used for model generation by mirroring all the data sets with respect to the mid-sagittal plane. Throughout all our experiments we ensured that training and testing data are mutually exclusive: We trained on DDHV, We corrected the ground-truth annotations for the left and the right caudate in the IBSR 18 data set to better meet the protocol applied by the 3-D Segmentation in the Clinic: A Grand Challenge competition where the caudate is grouped with the nucleus accumbens in the delineations [13, 14]. 8 The following 10 data sets were used: defaced native 100{2,3,7} V{1,2} t1w r2, defaced native 100{1,4,8} V2 t1w r2, and defaced native 1005 V2 t1w r2. 9 The NIH MRI Study of Normal Brain Development is a multi-site, longitudinal study of typically developing children, from ages newborn through young adulthood, conducted by the Brain Development Cooperative Group and supported by the NICHD, the NIDA, the NIMH, and the NINDS (Contract #s N01-HD , N01-MH9-0002, and N01-NS , -2315, -2316, -2317, and -2320). A listing of the participating sites and a complete listing of the study investigators can be found at centers.html. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH. 10
7 7 Table 2. Average left/right caudate segmentation accuracy for the MICCAI 07 testing data set. The complete results can be found at ( Segmentation Team ). As of 03/10/2009 our method ranks number 2 in the overall ranking list. Cases Overlap Err. Volume Diff. Abs. Dist. RMS Dist. Max. Dist. Total [%] Score [%] Score [mm] Score [mm] Score [mm] Score Score Average UNC Ped Average UNC Eld Average BWH PNL Average All NIH, and IBSR to evaluate on IBSR and on DDHV, NIH, and IBSR to evaluate on IBSR We trained on DDHV, NIH, IBSR 18, and MICCAI 07 training to evaluate on MICCAI 07 testing. As pointed out by Heimann et al. [13] there are differences in the annotation protocols used for annotating the caudate nuclei in data sets originating from the BWH and the UNC. In the former the tail of the caudate is continued much further dorsally. We therefore decided to detect it as a separate structure that can be attached to the caudate if required. We did not try to automatically determine the annotation protocol used from the imaging data itself as this may lead to over-fitted systems. As our real discriminative models are not ideal as assumed for theoretical considerations we keep the top 100 candidates after position detection and the top 25 candidates after position-orientation detection for further processing steps in order to make the full similarity transformation detection more robust. For shape inference we use T = 3 iterations. In an optimized and parallelized C++ implementation of our segmentation method it takes on average 13.9 sec. to detect and segment 8 (sub-)cortical structures in an MRI volume on a Fujitsu Siemens notebook equipped with an Intel Core 2 Duo CPU (2.20 GHz) and 3 GB of memory. Intensity standardization takes 1 2 sec. Our method is therefore faster than other state-of-the-art approaches whose timing is 50 sec. for 8 structures [5], 60 sec. for 1 structure [3], and 8 min. for 8 structures [4]. 4 Experimental Results As can be seen from Table 1 in terms of the Dice coefficient our method achieves better results (80%,73%,75%,82%) for the segmentation of the caudate nuclei, hippocampi, globi pallidi, and putamina on the same IBSR 18 data set than the methods of Akselrod-Ballin et al. [7] (80%, 69%, 74%, 79%) and Gouttard et al. [12] (76%,67%,71%,78%) except for the caudate nuclei in comparison to the method of Akselrod-Ballin et al. [7], where we reach a comparable accuracy. It also reaches a higher score for the caudate nuclei and putamina on IBSR 18 than the method of Bazin and Pham [14] (78%,81%), which does not address segmentation of the hippocampi and globi pallidi. The overall average score in Table 2 shows that for segmenting the caudate nuclei our method performs better than the methods of Morra et al. [3] (73.38), Bazin and Pham [14] (64.73) and Tu et al. [4] (59.71). All the mentioned methods were evaluated on the same MICCAI 07 testing data set.
8 8 5 Conclusions In this paper we integrated shape inference into the overall MSL methodology from the theoretical point of view. We showed that MSL decomposes the parameter space of anatomy shapes along decreasing levels of geometrical abstraction into subspaces of increasing dimensionality and applied MSL to the difficult problem of (sub-)cortical gray matter structure detection and shape inference. In an evaluation on publicly available gold standard databases our method works equally fast, robust, and accurate at a state-of-the-art level. References 1. Cox, I.J., Hingorani, S.L.: Dynamic histogram warping of image pairs for constant image brightness. In: Proc. ICIP, Washington, D.C., USA. (1995) Zheng, Y., Barbu, A., Georgescu, B., Scheuering, M., Comaniciu, D.: Four-chamber heart modeling and automatic segmentation for 3D cardiac CT volumes using marginal space learning and steerable features. IEEE T. Med. Imag. 27(11) (2008) Morra, J.H., Tu, Z., Apostolova, L.G., Green, A.E., Toga, A.W., Thompson, P.M.: Automatic subcortical segmentation using a contextual model. In: Proc. MICCAI, New York, NY, USA. (2008) Tu, Z., Narr, K.L., Dollár, P., Dinov, I., Thompson, P.M., Toga, A.W.: Brain anatomical structure segmentation by hybrid discriminative/generative models. IEEE T. Med. Imag. 27(4) (2008) Corso, J.J., Tu, Z., Yuille, A., Toga, A.: Segmentation of sub-cortical structures by the graph-shifts algorithm. In: Proc. IPMI, Kerkrade, the Netherlands. (2007) Scherrer, B., Forbes, F., Garbay, C., Dojat, M.: Fully Bayesian joint model for MR brain scan and structure segmentation. In: Proc. MICCAI, New York, NY, USA. (2008) Akselrod-Ballin, A., Galun, M., Gomori, J.M., Brandt, A., Basri, R.: Prior knowledge driven multiscale segmentation of brain MRI. In: Proc. MICCAI, Brisbane, Australia. (2007) Tu, Z., Zhou, X.S., Barbu, A., Bogoni, L., Comaniciu, D.: Probabilistic 3D polyp detection in CT images: The role of sample alignment. In: Proc. CVPR, New York, USA. (2006) Tu, Z.: Probabilistic boosting-tree: Learning discriminative models for classification, recognition, and clustering. In: Proc. ICCV, Beijing, China. (2005) Cootes, T.F., Taylor, C.J., Cooper, D.H., Graham, J.: Active shape models their training and application. Comp. Vis. Image Understand. 61(1) (1995) Bullitt, E., Zeng, D., Gerig, G., Aylward, S., Joshi, S., Smith, J.K., Lin, W., Ewend, M.G.: Vessel tortuosity and brain tumor malignancy: A blinded study. Acad. Radiol. 12(10) (2005) Gouttard, S., Styner, M., Joshi, S., Smith, R.G., Cody, H., Gerig, G.: Subcortical structure segmentation using probabilistic atlas priors. In: Proc. SPIE Med. Imag., San Diego, CA, USA. (2007) 65122J Heimann, T., Styner, M., van Ginneken, B.: Workshop on 3D segmentation in the clinic: A grand challenge. In: T. Heimann, M. Styner, B. van Ginneken (Eds.): 3D Segmentation in the Clinic: A Grand Challenge (2007) Bazin, P.L., Pham, D.L.: Homeomorphic brain image segmentation with topological and statistical atlases. Med. Image Anal. 12(5) (2008)
Fast and Robust 3-D MRI Brain Structure Segmentation
 Fast and Robust 3-D MRI Brain Structure Segmentation Michael Wels 1,4, Yefeng Zheng 2, Gustavo Carneiro 3,, Martin Huber 4, Joachim Hornegger 1, and Dorin Comaniciu 2 1 Chair of Pattern Recognition, Department
Fast and Robust 3-D MRI Brain Structure Segmentation Michael Wels 1,4, Yefeng Zheng 2, Gustavo Carneiro 3,, Martin Huber 4, Joachim Hornegger 1, and Dorin Comaniciu 2 1 Chair of Pattern Recognition, Department
Fully Automated Knowledge-Based Segmentation of the Caudate Nuclei in 3-D MRI
 Fully Automated Knowledge-Based Segmentation of the Caudate Nuclei in 3-D MRI Michael Wels 1, Martin Huber 2, and Joachim Hornegger 1 1 Institute of Pattern Recognition, University Erlangen-Nuremberg,
Fully Automated Knowledge-Based Segmentation of the Caudate Nuclei in 3-D MRI Michael Wels 1, Martin Huber 2, and Joachim Hornegger 1 1 Institute of Pattern Recognition, University Erlangen-Nuremberg,
Subcortical Structure Segmentation using Probabilistic Atlas Priors
 Subcortical Structure Segmentation using Probabilistic Atlas Priors Sylvain Gouttard 1, Martin Styner 1,2, Sarang Joshi 3, Brad Davis 2, Rachel G. Smith 1, Heather Cody Hazlett 1, Guido Gerig 1,2 1 Department
Subcortical Structure Segmentation using Probabilistic Atlas Priors Sylvain Gouttard 1, Martin Styner 1,2, Sarang Joshi 3, Brad Davis 2, Rachel G. Smith 1, Heather Cody Hazlett 1, Guido Gerig 1,2 1 Department
Automatic Caudate Segmentation by Hybrid Generative/Discriminative Models
 Automatic Caudate Segmentation by Hybrid Generative/Discriminative Models Zhuowen Tu, Mubeena Mirza, Ivo Dinov, and Arthur W. Toga Lab of Neuro Imaging, School of Medicine University of California, Los
Automatic Caudate Segmentation by Hybrid Generative/Discriminative Models Zhuowen Tu, Mubeena Mirza, Ivo Dinov, and Arthur W. Toga Lab of Neuro Imaging, School of Medicine University of California, Los
Towards Learning-Based Holistic Brain Image Segmentation
 Towards Learning-Based Holistic Brain Image Segmentation Zhuowen Tu Lab of Neuro Imaging University of California, Los Angeles in collaboration with (A. Toga, P. Thompson et al.) Supported by (NIH CCB
Towards Learning-Based Holistic Brain Image Segmentation Zhuowen Tu Lab of Neuro Imaging University of California, Los Angeles in collaboration with (A. Toga, P. Thompson et al.) Supported by (NIH CCB
Marginal Space Learning for Efficient Detection of 2D/3D Anatomical Structures in Medical Images
 Marginal Space Learning for Efficient Detection of 2D/3D Anatomical Structures in Medical Images Yefeng Zheng, Bogdan Georgescu, and Dorin Comaniciu Integrated Data Systems Department, Siemens Corporate
Marginal Space Learning for Efficient Detection of 2D/3D Anatomical Structures in Medical Images Yefeng Zheng, Bogdan Georgescu, and Dorin Comaniciu Integrated Data Systems Department, Siemens Corporate
Automatic Subcortical Segmentation Using a Contextual Model
 Automatic Subcortical Segmentation Using a Contextual Model Jonathan H. Morra 1, Zhuowen Tu 1, Liana G. Apostolova 1,2, Amity E. Green 1,2, Arthur W. Toga 1, and Paul M. Thompson 1 1 Laboratory of Neuro
Automatic Subcortical Segmentation Using a Contextual Model Jonathan H. Morra 1, Zhuowen Tu 1, Liana G. Apostolova 1,2, Amity E. Green 1,2, Arthur W. Toga 1, and Paul M. Thompson 1 1 Laboratory of Neuro
MARS: Multiple Atlases Robust Segmentation
 Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
IOP Publishing Limited Bristol 2011
 This is a preprint of the following published material: Michael Wels, Yefeng Zheng, Martin Huber, Joachim Hornegger and Dorin Comaniciu, "A discriminative modelconstrained EM approach to 3D MRI brain tissue
This is a preprint of the following published material: Michael Wels, Yefeng Zheng, Martin Huber, Joachim Hornegger and Dorin Comaniciu, "A discriminative modelconstrained EM approach to 3D MRI brain tissue
Automatic MS Lesion Segmentation by Outlier Detection and Information Theoretic Region Partitioning Release 0.00
 Automatic MS Lesion Segmentation by Outlier Detection and Information Theoretic Region Partitioning Release 0.00 Marcel Prastawa 1 and Guido Gerig 1 Abstract July 17, 2008 1 Scientific Computing and Imaging
Automatic MS Lesion Segmentation by Outlier Detection and Information Theoretic Region Partitioning Release 0.00 Marcel Prastawa 1 and Guido Gerig 1 Abstract July 17, 2008 1 Scientific Computing and Imaging
GLIRT: Groupwise and Longitudinal Image Registration Toolbox
 Software Release (1.0.1) Last updated: March. 30, 2011. GLIRT: Groupwise and Longitudinal Image Registration Toolbox Guorong Wu 1, Qian Wang 1,2, Hongjun Jia 1, and Dinggang Shen 1 1 Image Display, Enhancement,
Software Release (1.0.1) Last updated: March. 30, 2011. GLIRT: Groupwise and Longitudinal Image Registration Toolbox Guorong Wu 1, Qian Wang 1,2, Hongjun Jia 1, and Dinggang Shen 1 1 Image Display, Enhancement,
Assessment of Reliability of Multi-site Neuroimaging via Traveling Phantom Study
 Assessment of Reliability of Multi-site Neuroimaging via Traveling Phantom Study Sylvain Gouttard 1, Martin Styner 2,3, Marcel Prastawa 1, Joseph Piven 3, and Guido Gerig 1 1 Scientific Computing and Imaging
Assessment of Reliability of Multi-site Neuroimaging via Traveling Phantom Study Sylvain Gouttard 1, Martin Styner 2,3, Marcel Prastawa 1, Joseph Piven 3, and Guido Gerig 1 1 Scientific Computing and Imaging
MARS: Multiple Atlases Robust Segmentation
 Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit
 Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit John Melonakos 1, Ramsey Al-Hakim 1, James Fallon 2 and Allen Tannenbaum 1 1 Georgia Institute of Technology, Atlanta GA 30332,
Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit John Melonakos 1, Ramsey Al-Hakim 1, James Fallon 2 and Allen Tannenbaum 1 1 Georgia Institute of Technology, Atlanta GA 30332,
Auto-contouring the Prostate for Online Adaptive Radiotherapy
 Auto-contouring the Prostate for Online Adaptive Radiotherapy Yan Zhou 1 and Xiao Han 1 Elekta Inc., Maryland Heights, MO, USA yan.zhou@elekta.com, xiao.han@elekta.com, Abstract. Among all the organs under
Auto-contouring the Prostate for Online Adaptive Radiotherapy Yan Zhou 1 and Xiao Han 1 Elekta Inc., Maryland Heights, MO, USA yan.zhou@elekta.com, xiao.han@elekta.com, Abstract. Among all the organs under
Left Ventricle Endocardium Segmentation for Cardiac CT Volumes Using an Optimal Smooth Surface
 Left Ventricle Endocardium Segmentation for Cardiac CT Volumes Using an Optimal Smooth Surface Yefeng Zheng a, Bogdan Georgescu a, Fernando Vega-Higuera b, and Dorin Comaniciu a a Integrated Data Systems
Left Ventricle Endocardium Segmentation for Cardiac CT Volumes Using an Optimal Smooth Surface Yefeng Zheng a, Bogdan Georgescu a, Fernando Vega-Higuera b, and Dorin Comaniciu a a Integrated Data Systems
Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion
 Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion Mattias P. Heinrich Julia A. Schnabel, Mark Jenkinson, Sir Michael Brady 2 Clinical
Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion Mattias P. Heinrich Julia A. Schnabel, Mark Jenkinson, Sir Michael Brady 2 Clinical
NIH Public Access Author Manuscript Proc IEEE Int Symp Biomed Imaging. Author manuscript; available in PMC 2014 November 15.
 NIH Public Access Author Manuscript Published in final edited form as: Proc IEEE Int Symp Biomed Imaging. 2013 April ; 2013: 748 751. doi:10.1109/isbi.2013.6556583. BRAIN TUMOR SEGMENTATION WITH SYMMETRIC
NIH Public Access Author Manuscript Published in final edited form as: Proc IEEE Int Symp Biomed Imaging. 2013 April ; 2013: 748 751. doi:10.1109/isbi.2013.6556583. BRAIN TUMOR SEGMENTATION WITH SYMMETRIC
Comparison Study of Clinical 3D MRI Brain Segmentation Evaluation
 Comparison Study of Clinical 3D MRI Brain Segmentation Evaluation Ting Song 1, Elsa D. Angelini 2, Brett D. Mensh 3, Andrew Laine 1 1 Heffner Biomedical Imaging Laboratory Department of Biomedical Engineering,
Comparison Study of Clinical 3D MRI Brain Segmentation Evaluation Ting Song 1, Elsa D. Angelini 2, Brett D. Mensh 3, Andrew Laine 1 1 Heffner Biomedical Imaging Laboratory Department of Biomedical Engineering,
Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation
 Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Koen Van Leemput 3, Danial Lashkari 4 Marc-André Weber 5, Nicholas Ayache 2, and Polina
Segmenting Glioma in Multi-Modal Images using a Generative Model for Brain Lesion Segmentation Bjoern H. Menze 1,2, Koen Van Leemput 3, Danial Lashkari 4 Marc-André Weber 5, Nicholas Ayache 2, and Polina
A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images
 A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images Jianhua Yao 1, Russell Taylor 2 1. Diagnostic Radiology Department, Clinical Center,
A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images Jianhua Yao 1, Russell Taylor 2 1. Diagnostic Radiology Department, Clinical Center,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy
 The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
Non-rigid Image Registration using Electric Current Flow
 Non-rigid Image Registration using Electric Current Flow Shu Liao, Max W. K. Law and Albert C. S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
Non-rigid Image Registration using Electric Current Flow Shu Liao, Max W. K. Law and Albert C. S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
Assessment of Reliability of Multi-site Neuroimaging Via Traveling Phantom Study
 Assessment of Reliability of Multi-site Neuroimaging Via Traveling Phantom Study Sylvain Gouttard 1, Martin Styner 2,3, Marcel Prastawa 1, Joseph Piven 3, and Guido Gerig 1 1 Scientific Computing and Imaging
Assessment of Reliability of Multi-site Neuroimaging Via Traveling Phantom Study Sylvain Gouttard 1, Martin Styner 2,3, Marcel Prastawa 1, Joseph Piven 3, and Guido Gerig 1 1 Scientific Computing and Imaging
Prototype of Silver Corpus Merging Framework
 www.visceral.eu Prototype of Silver Corpus Merging Framework Deliverable number D3.3 Dissemination level Public Delivery data 30.4.2014 Status Authors Final Markus Krenn, Allan Hanbury, Georg Langs This
www.visceral.eu Prototype of Silver Corpus Merging Framework Deliverable number D3.3 Dissemination level Public Delivery data 30.4.2014 Status Authors Final Markus Krenn, Allan Hanbury, Georg Langs This
Elastically Deforming a Three-Dimensional Atlas to Match Anatomical Brain Images
 University of Pennsylvania ScholarlyCommons Technical Reports (CIS) Department of Computer & Information Science May 1993 Elastically Deforming a Three-Dimensional Atlas to Match Anatomical Brain Images
University of Pennsylvania ScholarlyCommons Technical Reports (CIS) Department of Computer & Information Science May 1993 Elastically Deforming a Three-Dimensional Atlas to Match Anatomical Brain Images
Whole Body MRI Intensity Standardization
 Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Automatic Vascular Tree Formation Using the Mahalanobis Distance
 Automatic Vascular Tree Formation Using the Mahalanobis Distance Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, Department of Radiology The University
Automatic Vascular Tree Formation Using the Mahalanobis Distance Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab, Department of Radiology The University
MR IMAGE SEGMENTATION
 MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
Semi-automated Basal Ganglia Segmentation Using Large Deformation Diffeomorphic Metric Mapping
 Semi-automated Basal Ganglia Segmentation Using Large Deformation Diffeomorphic Metric Mapping Ali Khan 1, Elizabeth Aylward 2, Patrick Barta 3, Michael Miller 4,andM.FaisalBeg 1 1 School of Engineering
Semi-automated Basal Ganglia Segmentation Using Large Deformation Diffeomorphic Metric Mapping Ali Khan 1, Elizabeth Aylward 2, Patrick Barta 3, Michael Miller 4,andM.FaisalBeg 1 1 School of Engineering
3D Brain Segmentation Using Active Appearance Models and Local Regressors
 3D Brain Segmentation Using Active Appearance Models and Local Regressors K.O. Babalola, T.F. Cootes, C.J. Twining, V. Petrovic, and C.J. Taylor Division of Imaging Science and Biomedical Engineering,
3D Brain Segmentation Using Active Appearance Models and Local Regressors K.O. Babalola, T.F. Cootes, C.J. Twining, V. Petrovic, and C.J. Taylor Division of Imaging Science and Biomedical Engineering,
A Generic Probabilistic Active Shape Model for Organ Segmentation
 A Generic Probabilistic Active Shape Model for Organ Segmentation Andreas Wimmer 1,2, Grzegorz Soza 2, and Joachim Hornegger 1 1 Chair of Pattern Recognition, Department of Computer Science, Friedrich-Alexander
A Generic Probabilistic Active Shape Model for Organ Segmentation Andreas Wimmer 1,2, Grzegorz Soza 2, and Joachim Hornegger 1 1 Chair of Pattern Recognition, Department of Computer Science, Friedrich-Alexander
SEGMENTING subcortical structures from 3-D brain images
 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 27, NO. 4, APRIL 2008 495 Brain Anatomical Structure Segmentation by Hybrid Discriminative/Generative Models Zhuowen Tu, Katherine L. Narr, Piotr Dollár, Ivo
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 27, NO. 4, APRIL 2008 495 Brain Anatomical Structure Segmentation by Hybrid Discriminative/Generative Models Zhuowen Tu, Katherine L. Narr, Piotr Dollár, Ivo
Multiple Cortical Surface Correspondence Using Pairwise Shape Similarity
 Multiple Cortical Surface Correspondence Using Pairwise Shape Similarity Pahal Dalal 1, Feng Shi 2, Dinggang Shen 2, and Song Wang 1 1 Department of Computer Science and Engineering, University of South
Multiple Cortical Surface Correspondence Using Pairwise Shape Similarity Pahal Dalal 1, Feng Shi 2, Dinggang Shen 2, and Song Wang 1 1 Department of Computer Science and Engineering, University of South
Subcortical Structure Segmentation using Probabilistic Atlas Priors
 Subcortical Structure Segmentation using Probabilistic Atlas Priors Sylvain Gouttard a, Martin Styner a,b, Sarang Joshi c, Rachel G. Smith a, Heather Cody a, Guido Gerig a,b a Department of Psychiatry,
Subcortical Structure Segmentation using Probabilistic Atlas Priors Sylvain Gouttard a, Martin Styner a,b, Sarang Joshi c, Rachel G. Smith a, Heather Cody a, Guido Gerig a,b a Department of Psychiatry,
Automatic corpus callosum segmentation using a deformable active Fourier contour model
 Automatic corpus callosum segmentation using a deformable active Fourier contour model Clement Vachet a, Benjamin Yvernault a, Kshamta Bhatt a, Rachel G. Smith a,b, Guido Gerig c, Heather Cody Hazlett
Automatic corpus callosum segmentation using a deformable active Fourier contour model Clement Vachet a, Benjamin Yvernault a, Kshamta Bhatt a, Rachel G. Smith a,b, Guido Gerig c, Heather Cody Hazlett
PROSTATE CANCER DETECTION USING LABEL IMAGE CONSTRAINED MULTIATLAS SELECTION
 PROSTATE CANCER DETECTION USING LABEL IMAGE CONSTRAINED MULTIATLAS SELECTION Ms. Vaibhavi Nandkumar Jagtap 1, Mr. Santosh D. Kale 2 1 PG Scholar, 2 Assistant Professor, Department of Electronics and Telecommunication,
PROSTATE CANCER DETECTION USING LABEL IMAGE CONSTRAINED MULTIATLAS SELECTION Ms. Vaibhavi Nandkumar Jagtap 1, Mr. Santosh D. Kale 2 1 PG Scholar, 2 Assistant Professor, Department of Electronics and Telecommunication,
An ITK Filter for Bayesian Segmentation: itkbayesianclassifierimagefilter
 An ITK Filter for Bayesian Segmentation: itkbayesianclassifierimagefilter John Melonakos 1, Karthik Krishnan 2 and Allen Tannenbaum 1 1 Georgia Institute of Technology, Atlanta GA 30332, USA {jmelonak,
An ITK Filter for Bayesian Segmentation: itkbayesianclassifierimagefilter John Melonakos 1, Karthik Krishnan 2 and Allen Tannenbaum 1 1 Georgia Institute of Technology, Atlanta GA 30332, USA {jmelonak,
Automatic Generation of Training Data for Brain Tissue Classification from MRI
 MICCAI-2002 1 Automatic Generation of Training Data for Brain Tissue Classification from MRI Chris A. Cocosco, Alex P. Zijdenbos, and Alan C. Evans McConnell Brain Imaging Centre, Montreal Neurological
MICCAI-2002 1 Automatic Generation of Training Data for Brain Tissue Classification from MRI Chris A. Cocosco, Alex P. Zijdenbos, and Alan C. Evans McConnell Brain Imaging Centre, Montreal Neurological
INTEGRATED DETECTION NETWORK (IDN) FOR POSE AND BOUNDARY ESTIMATION IN MEDICAL IMAGES
 INTEGRATED DETECTION NETWORK (IDN) FOR POSE AND BOUNDARY ESTIMATION IN MEDICAL IMAGES Michal Sofka Kristóf Ralovich Neil Birkbeck Jingdan Zhang S.Kevin Zhou Siemens Corporate Research, 775 College Road
INTEGRATED DETECTION NETWORK (IDN) FOR POSE AND BOUNDARY ESTIMATION IN MEDICAL IMAGES Michal Sofka Kristóf Ralovich Neil Birkbeck Jingdan Zhang S.Kevin Zhou Siemens Corporate Research, 775 College Road
Multi-atlas labeling with population-specific template and non-local patch-based label fusion
 Multi-atlas labeling with population-specific template and non-local patch-based label fusion Vladimir Fonov, Pierrick Coupé, Simon Eskildsen, Jose Manjon, Louis Collins To cite this version: Vladimir
Multi-atlas labeling with population-specific template and non-local patch-based label fusion Vladimir Fonov, Pierrick Coupé, Simon Eskildsen, Jose Manjon, Louis Collins To cite this version: Vladimir
Semantic Context Forests for Learning- Based Knee Cartilage Segmentation in 3D MR Images
 Semantic Context Forests for Learning- Based Knee Cartilage Segmentation in 3D MR Images MICCAI 2013: Workshop on Medical Computer Vision Authors: Quan Wang, Dijia Wu, Le Lu, Meizhu Liu, Kim L. Boyer,
Semantic Context Forests for Learning- Based Knee Cartilage Segmentation in 3D MR Images MICCAI 2013: Workshop on Medical Computer Vision Authors: Quan Wang, Dijia Wu, Le Lu, Meizhu Liu, Kim L. Boyer,
Learning-based Neuroimage Registration
 Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
NIH Public Access Author Manuscript Proc Soc Photo Opt Instrum Eng. Author manuscript; available in PMC 2011 September 7.
 NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2011 March ; 7962: 796225-1 796225-7. doi:10.1117/12.878405. Automatic Skull-stripping of Rat MRI/DTI
NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2011 March ; 7962: 796225-1 796225-7. doi:10.1117/12.878405. Automatic Skull-stripping of Rat MRI/DTI
STIC AmSud Project. Graph cut based segmentation of cardiac ventricles in MRI: a shape-prior based approach
 STIC AmSud Project Graph cut based segmentation of cardiac ventricles in MRI: a shape-prior based approach Caroline Petitjean A joint work with Damien Grosgeorge, Pr Su Ruan, Pr JN Dacher, MD October 22,
STIC AmSud Project Graph cut based segmentation of cardiac ventricles in MRI: a shape-prior based approach Caroline Petitjean A joint work with Damien Grosgeorge, Pr Su Ruan, Pr JN Dacher, MD October 22,
Boundary and Medial Shape Analysis of the Hippocampus in Schizophrenia
 Boundary and Medial Shape Analysis of the Hippocampus in Schizophrenia Martin Styner 1, Jeffrey A. Lieberman 2, and Guido Gerig 2,3 1 M.E. Müller Research Center for Orthopaedic Surgery, Institute for
Boundary and Medial Shape Analysis of the Hippocampus in Schizophrenia Martin Styner 1, Jeffrey A. Lieberman 2, and Guido Gerig 2,3 1 M.E. Müller Research Center for Orthopaedic Surgery, Institute for
Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues
 Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues D. Zikic, B. Glocker, E. Konukoglu, J. Shotton, A. Criminisi, D. H. Ye, C. Demiralp 3, O. M. Thomas 4,5, T. Das 4, R. Jena
Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues D. Zikic, B. Glocker, E. Konukoglu, J. Shotton, A. Criminisi, D. H. Ye, C. Demiralp 3, O. M. Thomas 4,5, T. Das 4, R. Jena
IEEE TRANSACTIONS ON IMAGE PROCESSING, VOL. 26, NO. 10, OCTOBER
 IEEE TRANSACTIONS ON IMAGE PROCESSING, VOL. 26, NO. 10, OCTOBER 2017 4753 Detecting Anatomical Landmarks From Limited Medical Imaging Data Using Two-Stage Task-Oriented Deep Neural Networks Jun Zhang,
IEEE TRANSACTIONS ON IMAGE PROCESSING, VOL. 26, NO. 10, OCTOBER 2017 4753 Detecting Anatomical Landmarks From Limited Medical Imaging Data Using Two-Stage Task-Oriented Deep Neural Networks Jun Zhang,
Manifold Learning: Applications in Neuroimaging
 Your own logo here Manifold Learning: Applications in Neuroimaging Robin Wolz 23/09/2011 Overview Manifold learning for Atlas Propagation Multi-atlas segmentation Challenges LEAP Manifold learning for
Your own logo here Manifold Learning: Applications in Neuroimaging Robin Wolz 23/09/2011 Overview Manifold learning for Atlas Propagation Multi-atlas segmentation Challenges LEAP Manifold learning for
Attribute Similarity and Mutual-Saliency Weighting for Registration and Label Fusion
 Attribute Similarity and Mutual-Saliency Weighting for Registration and Label Fusion Yangming Ou, Jimit Doshi, Guray Erus, and Christos Davatzikos Section of Biomedical Image Analysis (SBIA) Department
Attribute Similarity and Mutual-Saliency Weighting for Registration and Label Fusion Yangming Ou, Jimit Doshi, Guray Erus, and Christos Davatzikos Section of Biomedical Image Analysis (SBIA) Department
Low-Rank to the Rescue Atlas-based Analyses in the Presence of Pathologies
 Low-Rank to the Rescue Atlas-based Analyses in the Presence of Pathologies Xiaoxiao Liu 1, Marc Niethammer 2, Roland Kwitt 3, Matthew McCormick 1, and Stephen Aylward 1 1 Kitware Inc., USA 2 University
Low-Rank to the Rescue Atlas-based Analyses in the Presence of Pathologies Xiaoxiao Liu 1, Marc Niethammer 2, Roland Kwitt 3, Matthew McCormick 1, and Stephen Aylward 1 1 Kitware Inc., USA 2 University
Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging
 1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
Automatic Cerebral Aneurysm Detection in Multimodal Angiographic Images
 Automatic Cerebral Aneurysm Detection in Multimodal Angiographic Images Clemens M. Hentschke, Oliver Beuing, Rosa Nickl and Klaus D. Tönnies Abstract We propose a system to automatically detect cerebral
Automatic Cerebral Aneurysm Detection in Multimodal Angiographic Images Clemens M. Hentschke, Oliver Beuing, Rosa Nickl and Klaus D. Tönnies Abstract We propose a system to automatically detect cerebral
Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit
 Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit John Melonakos 1, Ramsey Al-Hakim 1, James Fallon 2 and Allen Tannenbaum 1 1 Georgia Institute of Technology, Atlanta GA 30332,
Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit John Melonakos 1, Ramsey Al-Hakim 1, James Fallon 2 and Allen Tannenbaum 1 1 Georgia Institute of Technology, Atlanta GA 30332,
Hierarchical Multi structure Segmentation Guided by Anatomical Correlations
 Hierarchical Multi structure Segmentation Guided by Anatomical Correlations Oscar Alfonso Jiménez del Toro oscar.jimenez@hevs.ch Henning Müller henningmueller@hevs.ch University of Applied Sciences Western
Hierarchical Multi structure Segmentation Guided by Anatomical Correlations Oscar Alfonso Jiménez del Toro oscar.jimenez@hevs.ch Henning Müller henningmueller@hevs.ch University of Applied Sciences Western
Automatic Mitral Valve Inflow Measurements from Doppler Echocardiography
 Automatic Mitral Valve Inflow Measurements from Doppler Echocardiography JinHyeong Park 1, S. Kevin Zhou 1, John Jackson 2, and Dorin Comaniciu 1 1 Integrated Data Systems,Siemens Corporate Research, Inc.,
Automatic Mitral Valve Inflow Measurements from Doppler Echocardiography JinHyeong Park 1, S. Kevin Zhou 1, John Jackson 2, and Dorin Comaniciu 1 1 Integrated Data Systems,Siemens Corporate Research, Inc.,
Using Probability Maps for Multi organ Automatic Segmentation
 Using Probability Maps for Multi organ Automatic Segmentation Ranveer Joyseeree 1,2, Óscar Jiménez del Toro1, and Henning Müller 1,3 1 University of Applied Sciences Western Switzerland (HES SO), Sierre,
Using Probability Maps for Multi organ Automatic Segmentation Ranveer Joyseeree 1,2, Óscar Jiménez del Toro1, and Henning Müller 1,3 1 University of Applied Sciences Western Switzerland (HES SO), Sierre,
Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry
 Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry Nivedita Agarwal, MD Nivedita.agarwal@apss.tn.it Nivedita.agarwal@unitn.it Volume and surface morphometry Brain volume White matter
Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry Nivedita Agarwal, MD Nivedita.agarwal@apss.tn.it Nivedita.agarwal@unitn.it Volume and surface morphometry Brain volume White matter
Improvement and Evaluation of a Time-of-Flight-based Patient Positioning System
 Improvement and Evaluation of a Time-of-Flight-based Patient Positioning System Simon Placht, Christian Schaller, Michael Balda, André Adelt, Christian Ulrich, Joachim Hornegger Pattern Recognition Lab,
Improvement and Evaluation of a Time-of-Flight-based Patient Positioning System Simon Placht, Christian Schaller, Michael Balda, André Adelt, Christian Ulrich, Joachim Hornegger Pattern Recognition Lab,
Measuring longitudinal brain changes in humans and small animal models. Christos Davatzikos
 Measuring longitudinal brain changes in humans and small animal models Christos Davatzikos Section of Biomedical Image Analysis University of Pennsylvania (Radiology) http://www.rad.upenn.edu/sbia Computational
Measuring longitudinal brain changes in humans and small animal models Christos Davatzikos Section of Biomedical Image Analysis University of Pennsylvania (Radiology) http://www.rad.upenn.edu/sbia Computational
Medical images, segmentation and analysis
 Medical images, segmentation and analysis ImageLab group http://imagelab.ing.unimo.it Università degli Studi di Modena e Reggio Emilia Medical Images Macroscopic Dermoscopic ELM enhance the features of
Medical images, segmentation and analysis ImageLab group http://imagelab.ing.unimo.it Università degli Studi di Modena e Reggio Emilia Medical Images Macroscopic Dermoscopic ELM enhance the features of
Fiber Selection from Diffusion Tensor Data based on Boolean Operators
 Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge
 Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
NIH Public Access Author Manuscript Conf Proc IEEE Eng Med Biol Soc. Author manuscript; available in PMC 2009 December 11.
 NIH Public Access Author Manuscript Published in final edited form as: Conf Proc IEEE Eng Med Biol Soc. 2004 ; 2: 1521 1524. doi:10.1109/iembs.2004.1403466. An Elliptic PDE Approach for Shape Characterization
NIH Public Access Author Manuscript Published in final edited form as: Conf Proc IEEE Eng Med Biol Soc. 2004 ; 2: 1521 1524. doi:10.1109/iembs.2004.1403466. An Elliptic PDE Approach for Shape Characterization
A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation
 A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation Xiahai Zhuang (PhD) Centre for Medical Image Computing University College London Fields-MITACS Conference on Mathematics
A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation Xiahai Zhuang (PhD) Centre for Medical Image Computing University College London Fields-MITACS Conference on Mathematics
Ischemic Stroke Lesion Segmentation Proceedings 5th October 2015 Munich, Germany
 0111010001110001101000100101010111100111011100100011011101110101101012 Ischemic Stroke Lesion Segmentation www.isles-challenge.org Proceedings 5th October 2015 Munich, Germany Preface Stroke is the second
0111010001110001101000100101010111100111011100100011011101110101101012 Ischemic Stroke Lesion Segmentation www.isles-challenge.org Proceedings 5th October 2015 Munich, Germany Preface Stroke is the second
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION
 CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
CHAPTER 6 MODIFIED FUZZY TECHNIQUES BASED IMAGE SEGMENTATION 6.1 INTRODUCTION Fuzzy logic based computational techniques are becoming increasingly important in the medical image analysis arena. The significant
Automatic Ascending Aorta Detection in CTA Datasets
 Automatic Ascending Aorta Detection in CTA Datasets Stefan C. Saur 1, Caroline Kühnel 2, Tobias Boskamp 2, Gábor Székely 1, Philippe Cattin 1,3 1 Computer Vision Laboratory, ETH Zurich, 8092 Zurich, Switzerland
Automatic Ascending Aorta Detection in CTA Datasets Stefan C. Saur 1, Caroline Kühnel 2, Tobias Boskamp 2, Gábor Székely 1, Philippe Cattin 1,3 1 Computer Vision Laboratory, ETH Zurich, 8092 Zurich, Switzerland
1 Introduction Motivation and Aims Functional Imaging Computational Neuroanatomy... 12
 Contents 1 Introduction 10 1.1 Motivation and Aims....... 10 1.1.1 Functional Imaging.... 10 1.1.2 Computational Neuroanatomy... 12 1.2 Overview of Chapters... 14 2 Rigid Body Registration 18 2.1 Introduction.....
Contents 1 Introduction 10 1.1 Motivation and Aims....... 10 1.1.1 Functional Imaging.... 10 1.1.2 Computational Neuroanatomy... 12 1.2 Overview of Chapters... 14 2 Rigid Body Registration 18 2.1 Introduction.....
Automated segmentation methods for liver analysis in oncology applications
 University of Szeged Department of Image Processing and Computer Graphics Automated segmentation methods for liver analysis in oncology applications Ph. D. Thesis László Ruskó Thesis Advisor Dr. Antal
University of Szeged Department of Image Processing and Computer Graphics Automated segmentation methods for liver analysis in oncology applications Ph. D. Thesis László Ruskó Thesis Advisor Dr. Antal
Where are we now? Structural MRI processing and analysis
 Where are we now? Structural MRI processing and analysis Pierre-Louis Bazin bazin@cbs.mpg.de Leipzig, Germany Structural MRI processing: why bother? Just use the standards? SPM FreeSurfer FSL However:
Where are we now? Structural MRI processing and analysis Pierre-Louis Bazin bazin@cbs.mpg.de Leipzig, Germany Structural MRI processing: why bother? Just use the standards? SPM FreeSurfer FSL However:
Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information
 Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information Andreas Biesdorf 1, Stefan Wörz 1, Hans-Jürgen Kaiser 2, Karl Rohr 1 1 University of Heidelberg, BIOQUANT, IPMB,
Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information Andreas Biesdorf 1, Stefan Wörz 1, Hans-Jürgen Kaiser 2, Karl Rohr 1 1 University of Heidelberg, BIOQUANT, IPMB,
Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search
 Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search Baochun He, Cheng Huang, Fucang Jia Shenzhen Institutes of Advanced Technology, Chinese Academy
Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search Baochun He, Cheng Huang, Fucang Jia Shenzhen Institutes of Advanced Technology, Chinese Academy
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION
 ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
Robust and Accurate Coronary Artery Centerline Extraction in CTA by Combining Model-Driven and Data-Driven Approaches
 Robust and Accurate Coronary Artery Centerline Extraction in CTA by Combining Model-Driven and Data-Driven Approaches Yefeng Zheng, Huseyin Tek, and Gareth Funka-Lea Imaging and Computer Vision, Siemens
Robust and Accurate Coronary Artery Centerline Extraction in CTA by Combining Model-Driven and Data-Driven Approaches Yefeng Zheng, Huseyin Tek, and Gareth Funka-Lea Imaging and Computer Vision, Siemens
Manifold Learning-based Data Sampling for Model Training
 Manifold Learning-based Data Sampling for Model Training Shuqing Chen 1, Sabrina Dorn 2, Michael Lell 3, Marc Kachelrieß 2,Andreas Maier 1 1 Pattern Recognition Lab, FAU Erlangen-Nürnberg 2 German Cancer
Manifold Learning-based Data Sampling for Model Training Shuqing Chen 1, Sabrina Dorn 2, Michael Lell 3, Marc Kachelrieß 2,Andreas Maier 1 1 Pattern Recognition Lab, FAU Erlangen-Nürnberg 2 German Cancer
Segmentation of Brain MR Images via Sparse Patch Representation
 Segmentation of Brain MR Images via Sparse Patch Representation Tong Tong 1, Robin Wolz 1, Joseph V. Hajnal 2, and Daniel Rueckert 1 1 Department of Computing, Imperial College London, London, UK 2 MRC
Segmentation of Brain MR Images via Sparse Patch Representation Tong Tong 1, Robin Wolz 1, Joseph V. Hajnal 2, and Daniel Rueckert 1 1 Department of Computing, Imperial College London, London, UK 2 MRC
2D Vessel Segmentation Using Local Adaptive Contrast Enhancement
 2D Vessel Segmentation Using Local Adaptive Contrast Enhancement Dominik Schuldhaus 1,2, Martin Spiegel 1,2,3,4, Thomas Redel 3, Maria Polyanskaya 1,3, Tobias Struffert 2, Joachim Hornegger 1,4, Arnd Doerfler
2D Vessel Segmentation Using Local Adaptive Contrast Enhancement Dominik Schuldhaus 1,2, Martin Spiegel 1,2,3,4, Thomas Redel 3, Maria Polyanskaya 1,3, Tobias Struffert 2, Joachim Hornegger 1,4, Arnd Doerfler
Multi-Atlas Segmentation of the Cardiac MR Right Ventricle
 Multi-Atlas Segmentation of the Cardiac MR Right Ventricle Yangming Ou, Jimit Doshi, Guray Erus, and Christos Davatzikos Section of Biomedical Image Analysis (SBIA) Department of Radiology, University
Multi-Atlas Segmentation of the Cardiac MR Right Ventricle Yangming Ou, Jimit Doshi, Guray Erus, and Christos Davatzikos Section of Biomedical Image Analysis (SBIA) Department of Radiology, University
BrainParser: Deformable Templates Guided Discriminative Models for Robust 3D Brain MRI Segmentation
 Noname manuscript No. (will be inserted by the editor) BrainParser: Deformable Templates Guided Discriminative Models for Robust 3D Brain MRI Segmentation Cheng-Yi Liu, Juan Eugenio Iglesias, Zhuowen Tu,
Noname manuscript No. (will be inserted by the editor) BrainParser: Deformable Templates Guided Discriminative Models for Robust 3D Brain MRI Segmentation Cheng-Yi Liu, Juan Eugenio Iglesias, Zhuowen Tu,
Robust 3D Organ Localization with Dual Learning Architectures and Fusion
 Robust 3D Organ Localization with Dual Learning Architectures and Fusion Xiaoguang Lu (B), Daguang Xu, and David Liu Medical Imaging Technologies, Siemens Medical Solutions, Inc., Princeton, NJ, USA xiaoguang.lu@siemens.com
Robust 3D Organ Localization with Dual Learning Architectures and Fusion Xiaoguang Lu (B), Daguang Xu, and David Liu Medical Imaging Technologies, Siemens Medical Solutions, Inc., Princeton, NJ, USA xiaoguang.lu@siemens.com
Medical Image Registration by Maximization of Mutual Information
 Medical Image Registration by Maximization of Mutual Information EE 591 Introduction to Information Theory Instructor Dr. Donald Adjeroh Submitted by Senthil.P.Ramamurthy Damodaraswamy, Umamaheswari Introduction
Medical Image Registration by Maximization of Mutual Information EE 591 Introduction to Information Theory Instructor Dr. Donald Adjeroh Submitted by Senthil.P.Ramamurthy Damodaraswamy, Umamaheswari Introduction
Supplementary methods
 Supplementary methods This section provides additional technical details on the sample, the applied imaging and analysis steps and methods. Structural imaging Trained radiographers placed all participants
Supplementary methods This section provides additional technical details on the sample, the applied imaging and analysis steps and methods. Structural imaging Trained radiographers placed all participants
Development in Object Detection. Junyuan Lin May 4th
 Development in Object Detection Junyuan Lin May 4th Line of Research [1] N. Dalal and B. Triggs. Histograms of oriented gradients for human detection, CVPR 2005. HOG Feature template [2] P. Felzenszwalb,
Development in Object Detection Junyuan Lin May 4th Line of Research [1] N. Dalal and B. Triggs. Histograms of oriented gradients for human detection, CVPR 2005. HOG Feature template [2] P. Felzenszwalb,
Atlas Based Segmentation of the prostate in MR images
 Atlas Based Segmentation of the prostate in MR images Albert Gubern-Merida and Robert Marti Universitat de Girona, Computer Vision and Robotics Group, Girona, Spain {agubern,marly}@eia.udg.edu Abstract.
Atlas Based Segmentation of the prostate in MR images Albert Gubern-Merida and Robert Marti Universitat de Girona, Computer Vision and Robotics Group, Girona, Spain {agubern,marly}@eia.udg.edu Abstract.
Accurate Regression-based 4D Mitral Valve Segmentation from 2D MRI Slices
 Accurate Regression-based 4D Mitral Valve Segmentation from 2D MRI Slices Dime Vitanovski 2,3, Alexey Tsymbal 2, Razvan Ioan Ionasec 1, Andreas Greiser 4, Edgar Mueller 4, Xiaoguang Lu 1, Gareth Funka-Lea
Accurate Regression-based 4D Mitral Valve Segmentation from 2D MRI Slices Dime Vitanovski 2,3, Alexey Tsymbal 2, Razvan Ioan Ionasec 1, Andreas Greiser 4, Edgar Mueller 4, Xiaoguang Lu 1, Gareth Funka-Lea
Detecting Anatomical Landmarks from Limited Medical Imaging Data using Two-Stage Task-Oriented Deep Neural Networks
 IEEE TRANSACTIONS ON IMAGE PROCESSING Detecting Anatomical Landmarks from Limited Medical Imaging Data using Two-Stage Task-Oriented Deep Neural Networks Jun Zhang, Member, IEEE, Mingxia Liu, Member, IEEE,
IEEE TRANSACTIONS ON IMAGE PROCESSING Detecting Anatomical Landmarks from Limited Medical Imaging Data using Two-Stage Task-Oriented Deep Neural Networks Jun Zhang, Member, IEEE, Mingxia Liu, Member, IEEE,
Joint Segmentation and Registration for Infant Brain Images
 Joint Segmentation and Registration for Infant Brain Images Guorong Wu 1, Li Wang 1, John Gilmore 2, Weili Lin 1, and Dinggang Shen 1(&) 1 Department of Radiology and BRIC, University of North Carolina,
Joint Segmentation and Registration for Infant Brain Images Guorong Wu 1, Li Wang 1, John Gilmore 2, Weili Lin 1, and Dinggang Shen 1(&) 1 Department of Radiology and BRIC, University of North Carolina,
8/3/2017. Contour Assessment for Quality Assurance and Data Mining. Objective. Outline. Tom Purdie, PhD, MCCPM
 Contour Assessment for Quality Assurance and Data Mining Tom Purdie, PhD, MCCPM Objective Understand the state-of-the-art in contour assessment for quality assurance including data mining-based techniques
Contour Assessment for Quality Assurance and Data Mining Tom Purdie, PhD, MCCPM Objective Understand the state-of-the-art in contour assessment for quality assurance including data mining-based techniques
CHAPTER 2. Morphometry on rodent brains. A.E.H. Scheenstra J. Dijkstra L. van der Weerd
 CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
Rigid and Deformable Vasculature-to-Image Registration : a Hierarchical Approach
 Rigid and Deformable Vasculature-to-Image Registration : a Hierarchical Approach Julien Jomier and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel
Rigid and Deformable Vasculature-to-Image Registration : a Hierarchical Approach Julien Jomier and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel
Automatic Delineation of Left and Right Ventricles in Cardiac MRI Sequences Using a Joint Ventricular Model
 Automatic Delineation of Left and Right Ventricles in Cardiac MRI Sequences Using a Joint Ventricular Model Xiaoguang Lu 1,, Yang Wang 1, Bogdan Georgescu 1, Arne Littman 2, and Dorin Comaniciu 1 1 Siemens
Automatic Delineation of Left and Right Ventricles in Cardiac MRI Sequences Using a Joint Ventricular Model Xiaoguang Lu 1,, Yang Wang 1, Bogdan Georgescu 1, Arne Littman 2, and Dorin Comaniciu 1 1 Siemens
Fully Automatic Model Creation for Object Localization utilizing the Generalized Hough Transform
 Fully Automatic Model Creation for Object Localization utilizing the Generalized Hough Transform Heike Ruppertshofen 1,2,3, Cristian Lorenz 2, Peter Beyerlein 4, Zein Salah 3, Georg Rose 3, Hauke Schramm
Fully Automatic Model Creation for Object Localization utilizing the Generalized Hough Transform Heike Ruppertshofen 1,2,3, Cristian Lorenz 2, Peter Beyerlein 4, Zein Salah 3, Georg Rose 3, Hauke Schramm
Shape-based Diffeomorphic Registration on Hippocampal Surfaces Using Beltrami Holomorphic Flow
 Shape-based Diffeomorphic Registration on Hippocampal Surfaces Using Beltrami Holomorphic Flow Abstract. Finding meaningful 1-1 correspondences between hippocampal (HP) surfaces is an important but difficult
Shape-based Diffeomorphic Registration on Hippocampal Surfaces Using Beltrami Holomorphic Flow Abstract. Finding meaningful 1-1 correspondences between hippocampal (HP) surfaces is an important but difficult
Lilla Zöllei A.A. Martinos Center, MGH; Boston, MA
 Lilla Zöllei lzollei@nmr.mgh.harvard.edu A.A. Martinos Center, MGH; Boston, MA Bruce Fischl Gheorghe Postelnicu Jean Augustinack Anastasia Yendiki Allison Stevens Kristen Huber Sita Kakonoori + the FreeSurfer
Lilla Zöllei lzollei@nmr.mgh.harvard.edu A.A. Martinos Center, MGH; Boston, MA Bruce Fischl Gheorghe Postelnicu Jean Augustinack Anastasia Yendiki Allison Stevens Kristen Huber Sita Kakonoori + the FreeSurfer
NIH Public Access Author Manuscript Proc SPIE. Author manuscript; available in PMC 2013 December 30.
 NIH Public Access Author Manuscript Published in final edited form as: Proc SPIE. 2013 March 12; 8669:. doi:10.1117/12.2006682. Longitudinal Intensity Normalization of Magnetic Resonance Images using Patches
NIH Public Access Author Manuscript Published in final edited form as: Proc SPIE. 2013 March 12; 8669:. doi:10.1117/12.2006682. Longitudinal Intensity Normalization of Magnetic Resonance Images using Patches
Automatic Generation of Training Data for Brain Tissue Classification from MRI
 Automatic Generation of Training Data for Brain Tissue Classification from MRI Chris A. COCOSCO, Alex P. ZIJDENBOS, and Alan C. EVANS http://www.bic.mni.mcgill.ca/users/crisco/ McConnell Brain Imaging
Automatic Generation of Training Data for Brain Tissue Classification from MRI Chris A. COCOSCO, Alex P. ZIJDENBOS, and Alan C. EVANS http://www.bic.mni.mcgill.ca/users/crisco/ McConnell Brain Imaging
Norbert Schuff VA Medical Center and UCSF
 Norbert Schuff Medical Center and UCSF Norbert.schuff@ucsf.edu Medical Imaging Informatics N.Schuff Course # 170.03 Slide 1/67 Objective Learn the principle segmentation techniques Understand the role
Norbert Schuff Medical Center and UCSF Norbert.schuff@ucsf.edu Medical Imaging Informatics N.Schuff Course # 170.03 Slide 1/67 Objective Learn the principle segmentation techniques Understand the role
Detecting Hippocampal Shape Changes in Alzheimer s Disease using Statistical Shape Models
 Detecting Hippocampal Shape Changes in Alzheimer s Disease using Statistical Shape Models Kaikai Shen a,b, Pierrick Bourgeat a, Jurgen Fripp a, Fabrice Meriaudeau b, Olivier Salvado a and the Alzheimer
Detecting Hippocampal Shape Changes in Alzheimer s Disease using Statistical Shape Models Kaikai Shen a,b, Pierrick Bourgeat a, Jurgen Fripp a, Fabrice Meriaudeau b, Olivier Salvado a and the Alzheimer
A Discriminative Model-Constrained Graph Cuts Approach to Fully Automated Pediatric Brain Tumor Segmentation in 3-D MRI
 A Discriminative Model-Constrained Graph Cuts Approach to Fully Automated Pediatric Brain Tumor Segmentation in 3-D MRI Michael Wels 1,4, Gustavo Carneiro 2, Alexander Aplas 3,MartinHuber 4, Joachim Hornegger
A Discriminative Model-Constrained Graph Cuts Approach to Fully Automated Pediatric Brain Tumor Segmentation in 3-D MRI Michael Wels 1,4, Gustavo Carneiro 2, Alexander Aplas 3,MartinHuber 4, Joachim Hornegger
