Fully Automated Knowledge-Based Segmentation of the Caudate Nuclei in 3-D MRI
|
|
|
- Brooke Hensley
- 5 years ago
- Views:
Transcription
1 Fully Automated Knowledge-Based Segmentation of the Caudate Nuclei in 3-D MRI Michael Wels 1, Martin Huber 2, and Joachim Hornegger 1 1 Institute of Pattern Recognition, University Erlangen-Nuremberg, Germany michael.wels@informatik.uni-erlangen.de 2 Siemens AG, CT SE SCR 2, Erlangen, Germany Abstract. In this paper we present a fully automated approach to segmentation of the caudate nuclei in 3-D magnetic resonance images. It is based on the technique of probabilistic boosting trees. As a strategy for supervised learning it is capable to derive a discriminative model for the distinction of object and non-object voxels from expert annotated imaging data and rather rough anatomical prior knowledge provided by a probabilistic anatomical atlas. Training the model involves successively selecting and combining features that best separate the available training samples, i.e., image voxels, and grouping the resulting boosted classifiers in a tree structure. Most of the features used are taken from an intra-axial 2-D context surrounding the voxel of interest and its transformation to a particular set of Haar-like features. The final segmentation is obtained after post-processing the preliminary result by a fast marching approach whose two speed images are seeded at the inner and outer bounds of the object detected. This allows for adaptation to local edges close to the initially detected object s boundary. A detailed quantitative evaluation critically reveals strengths and weaknesses of the proposed method. 1 Introduction Fully automated segmentation tools both for pathologic as wells as non-pathologic structures within the human brain serve as valuable components of clinical decision support systems. Proper and reliable segmentation results are considered critical for the automatic extraction of quantitative or more abstract findings that are diagnostically relevant like, for example, the segmented objects volume or their relative location, respectively. Those semantic features are, in a second step, easily combined with other findings emerging from different examinations a patient undergoes during diagnostics to form the input to integrated decision support systems. In the context of brain tissue classification within magnetic resonance (MR) volume sequences segmentation of particular anatomical entities states a challenging problem for fully automated approaches: the distinction of deep gray matter structures, like the caudate nuclei, and cortical gray matter based on observed intensities only, is virtually impossible. Prior knowledge about the anatomical composition of the human brain has to be integrated to guide the T. Heimann, M. Styner, B. van Ginneken (Eds.): 3D Segmentation in The Clinic: A Grand Challenge, pp , 2007.
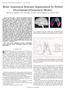
2 Fig. 1. The proposed segmentation framework. segmentation process. Furthermore, segmentation methods need to be robust with regard to the characteristic artifacts of the MR imaging modality: Rician noise, partial volume effects, and intensity inhomogeneities. The contribution of this work is a fully automated knowledge-based method for delineation of both the right as well as the left caudate nuclei in 3-D brain MR images that allows for fast and convenient segmentation of those two anatomical structures. Our method consists of four major steps: First, the whole brain is extracted from its surroundings with the Brain Extraction Tool (BET) [1]. Then, in a pre-processing step, the observed intensities within the area that is detected to be part of the brain itself are normalized to have zero mean and unit variance. Subsequently, two probabilistic boosting trees (PBT) [2] are used in a similar manner to [3] to classify individual voxels of the input data into foreground and background with respect to the right and the left caudate nucleus. This step also incorporates spatial knowledge about the typical location of the caudate nuclei in brain MR scans taken from a probabilistic anatomical atlas. Finally, a twofold fast marching approach that is seeded at the inner and outer bounds of the detected objects is adopted as a post-processing step to better adapt to local object edges apparent in the image. The inner and outer bound of the object necessary for this operation are determined by means of mathematical morphology. Experimental results on several previously unseen real-world data sets demonstrate strengths and weaknesses of the proposed method. The overall system block diagram is depicted in Fig Methods 2.1 Probabilistic Boosting Tree Our method s main component PBT recursively groups boosted ensembles of weak classifiers to a tree structure during learning from expert annotated data. We have chosen Real AdaBoost [4] to be the boosting strategy within each tree node. In this case learning a PBT resembles building a multivariate binary regression tree as the final strong classifier H(x) = T h t (x) (1) t=1 20
3 generated within each inner node for a feature vector x through a combination of real-valued contributions h t (x) of T N weak classifiers asymptotically approaches the additive logistic regression model [4]: H(x) 1 2 ln p(y = +1 x) p(y = 1 x) (2) where y { +1, 1 } denotes the classification outcome. Therefore, at each inner node v of the resulting PBT with boosted strong classifier H v there are current approximations of the posterior probabilities q v (y = +1 x) = e2hv(x) 1 + e 2Hv(x) (3) and q v (y = 1 x) = e 2Hv(x). (4) 1 + e 2Hv(x) While training the classifier, those probabilities are used to successively split the set of training data relative to the prior probability p v (y = +1) associated with the current training (sub-)set into two new subsets. The soft thresholding parameter ɛ > 0 sees to pass on training samples x that are close to the current node s decision boundary, i.e., if q v (y = +1 x) [(1 ɛ)p v (y = +1); (1+ɛ)p v (y = +1)], to both of the resulting subsets and associated subtrees. This differs from the original formulation in [2] that did not explicitly take into account disparities of positive and negative examples within the training (sub-)sets available at each tree node. Apart from that [2] contains a detailed discussion of the overall learning procedure. During classification the values for q v (y = +1 x) are used to guide tree traversing and combined propagation of posteriors in order to get final approximations p v (y x) of the true posterior probabilities p v (y x) at each tree node v: For outgoing arrows rv 1 and rv 1 associated with the possible classifications the approximation p v (y x) can be computed via the recursive formula p β(r 1 v ) (y x) : q v(+1) < (1 ɛ)p v (+1) p v (y x) = p β(r +1 v ) (y x) : q v(+1) (1 ɛ)p v (+1) (5) i p β(rv i ) (y x) q v (i x) : otherwise where β(r) denotes the vertex where arrow r ends. 2.2 Discriminative Features In order to adequately capture discriminative knowledge about image voxels the features used in our method origin from two categories: 21
4 Fig. 2. From left to right, a sagittal, coronal and axial slice from a a subject in the adults BWH group (top), one in the elderly UNC group (middle) and one in the pediatric UNC group (bottom). The outline of the reference standard segmentation is in red, the outline of the segmentation of the method described in this paper is in blue. The by far largest fraction of feature values in the feature vector x i associated with a given image voxel i state 2-D Haar-like features [5] computed on an intra-axial 2-D context surrounding the voxel of interest. They are derived from a subset of the extended set of Haar-like feature prototypes by [6] and are represented only implicitly in memory by so-called (Rotated) Integral Images. This allows for fast re-computation of the features with respect to a given voxel when they are actually assessed. Mainly due to combinatorial complexity we did not consider 3-D Haar-like features and furthermore restricted on 2-D Haar-like features whose centers are aligned with the center of the context considered. 22
5 All Dataset Overlap Err Volume Diff. Abs. Dist. RMS Dist. Max. Dist. Total [%] Score [%] Score [mm] Score [mm] Score [mm] Score Score UNC Ped UNC Ped UNC Ped UNC Ped UNC Ped UNC Eld UNC Eld UNC Eld UNC Eld UNC Eld BWH PNL BWH PNL BWH PNL BWH PNL BWH PNL BWH PNL BWH PNL BWH PNL BWH PNL BWH PNL BWH PNL BWH PNL BWH PNL BWH PNL Average All Average UNC Ped Average UNC Eld Average BWH PNL Table 1. Results of the comparison metrics and corresponding scores for all test cases averaged for the left and right segmentation. The summary rows at the end of the table display the overall average across all test cases, as well as grouped for the three testing groups. The second category of features consists of the three voxel indices in patient space and the voxel s probability to be part of the caudate nuclei. The latter is taken from a probabilistic anatomical atlas [7] which is roughly registered with the current patient data set by aligning orientations and image centers. This approach might seem brute force on the first sight, yet boosting asks for weak classifiers, i.e., individual features in our case. Further, there are certain quality standards for medical image acquisition such that the image region where the anatomical object that is about to be segmented appears in the final image underlies at least rough regularities. 23
6 2.3 Segmentation Refinement As depicted in Fig. 1 the segmentation obtained from PBT needs to be smoothed to get an acceptable final result. In our approach this is done by means of an adversarially seeded fast marching approach: Two speed images are computed assigning detection times to every voxel relative to two sets of seed points. For that the current volume is interpreted as a graph where each voxel becomes a node with connectivity to its six spatially nearest neighbors. The resulting edges between voxels i and j are assigned edge weights w ij = e xi xj 1 (6) where x i and x j denote intensity values of a median filtered version (index radii r x = 1, r y = 1, and r z = 1) of the initial data. Starting with zero detection time for the seed points the voxels detection times are determined by computing shortest paths following the graph edges from the set of seed voxels to the voxels of interest. One set of seed points is obtained by smoothing the current segmentation with a binary median filter with index radii r x = 3, r y = 3, and r z = 3 from the Insight Segmentation and Registration Toolkit (ITK, After morphological erosion this gives a first estimate of voxels that are likely to be inside of the object to segment. The second set of seed points is obtained by morphologically dilating the current segmentation and selecting those voxels that still remain part of the background, which gives an estimation of voxels outside of the object of interest that are not too close to the object boundaries. An adaptation to local image characteristics (edges and homogeneous regions) of the initial segmentation is then achieved through assigning each voxel to that region, i.e., foreground or background, for whoever s seeds it is detected first. Finally the resulting segmentation is again subject to binary median filtering with slightly smaller index radii r x = 2, r y = 2, and r z = Processing Pipeline The whole procedure used for segmentation of either the left or the right caudate nucleus within a given MR volume data set is described in Algorithm Algorithm 1: Left/right caudate nucleus segmentation Input: MR volume Output: Binary voxel classification Brain extraction; Re-sampling of input image to resolution suitable for PBT classifier; Normalization; PBT binary voxel classification; Re-sampling of classification to original image resolution; Adversarial fast marching; Binary median filtering; 24
7 Correl UNC Ped UNC Eld BWH PNL Total Left Right Average Table 2. Pearson correlation for the volume measurements in the three testing groups as well as in total. This coefficient captures how well the volumetric measurements correlate with those of the reference segmentations. 3 Material and Experimental Setting For training and evaluation of the proposed method several volumetric T1- weighted MR brain scans of varying spatial resolution and size from multiple sources were used. The vast majority of data (29 scans) has been provided by the Psychiatry Neuroimaging Laboratory (PNL) at the Brigham and Women s Hospital (BWH), Boston. They all are accompanied by expert annotations. The other 20 data sets arose from several studies carried out mainly at the University of North Carolina s (UNC) Neuroimaging Laboratory (NIAL), Chapel Hill. The studies contain a pediatric study, a Parkinson s Disease study, and a test/re-test study. For the latter no ground-truth was available. The training data for both the classifiers for the right and the left caudate nucleus were taken from the data sets provided by the BWH PNL (serial numbers 1 8). In this paper our method is first of all quantitatively evaluated on the BWH PNL scans 16 29, which are all considered to be routine scans, 5 of the pediatric scans, and 5 scans of patients older than 55 years. (See Tabs. 1-3) Additionally, our algorithm was tested on 10 datasets of the same young healthy person acquired within 60 days on 5 different scanners. (See Tab. 4) The coefficient of variation of the volumetric measurements is an indicator on how stable the method operates in a test/retest situation including scanner variability. We refer to [8] and [9] for details on the used evaluation measures and scoring system. The PBT voxel classifiers were built from approximately one million randomly selected training samples, i.e., voxels inside the head of the patients, uniformly distributed over all the input slices of the training scans. The scans were re-sampled to a voxel spacing of 1.0mm 1.0mm 2.0mm and size , which was mainly due to hardware limitations that our implementation currently faces. Since 2-D contexts of voxels that are very close to each other are likely to be very similar only every second voxel in each of the three spatial dimensions was considered for random selection. The maximum number of features selected by AdaBoost in each tree node were increased level-wise beginning with 2 at the trees root node. The maximum depth of the trees learned was restricted to 10 and soft thresholding was turned off. The 2-D voxel context chosen for computing the 1,908 Haar-like features used per individual voxel sample was of size centered at the voxel of interest. 25
8 Test/Re-Test UNC 03 UNC 04 UNC 09 UNC 11 UNC 17 UNC 18 UNC 21 UNC 22 UNC 24 UNC 25 Mean Stdev COV [mm 3 ] [mm 3 ] [mm 3 ] [mm 3 ] [mm 3 ] [mm 3 ] [mm 3 ] [mm 3 ] [mm 3 ] [mm 3 ] [mm 3 ] [mm 3 ] [%] Left Right Total Table 3. The volumetric measurements of the 10 data sets of the same young adult acquired on 5 different scanners within 60 days. The coefficient of variation (COV = standard deviation / average, last column) indicates the stability of the algorithm in a test/re-test situation including scanner variability. It takes about 2 3 minutes to process one of the MRI volumes in a nonoptimized C++ implementation of our segmentation method on a Fujitsu Siemens Computers notebook equipped with an Intel Pentium M 2.0 GHz processor and 2 GB of memory. Most of the computational cost is due to post-processing the initial segmentation result obtained by the PBT. With the same hardware as above building one classifier takes about 6 hours. 4 Results and Discussion From the performance scores given in Tab. 1 it can be clearly recognized that our algorithm suffers from significant over fitting mainly due to scanner specific contrast characteristics. This may also eclipse potential over fitting to sub-group dependent anatomical characteristics: segmentation results are, with minor exceptions, only acceptable for those data sets that come from the same routine study as the training data. This can also be seen by examining individual segmentations obtained by our method on representative scans from each of the data sub-groups as in Fig. 2. As one of the results of this problem the performance of our method in a test/re-test situation is poor. (See Tab. 3) The reliability of our method when used for volumetric measurements can be estimated from figures in Tab. 2. Due to the very low segmentation performance on the pediatric and the elderly patient datasets associated correlation measures are not very expressive. The observable positive association between volume measurements for the left caudate nuclei in the routine data set is caused by, to some extent, regular under-segmentation provided by our algorithm. However, there is little association for the segmentations of the right caudate nuclei as our method seems to arbitrarily over- and under-segment the object of interest in equal manner. 5 Conclusion In this paper we have presented a fully automatic approach to segmentation of the left and right caudate nuclei in 3-D MR images. The novelty of the proposed method lies both in composition and pipelining of different previously published methods as well as in the specific field of application. Experimental 26
9 results show that acceptable results can be obtained very fast and without any user-interaction if it has been possible to train the classifier on images that are acquired with the same MR scanner and associated settings as those images that are about to be segmented. The problem of over fitting to scanner and acquisition protocol dependent contrast characteristics currently present in the method could possibly be weakened by the integration of more reliable MR inter-scan intensity standardization approaches into the framework. Once the PBT shape detection step has been improved this way, more sophisticated approaches to refine the initial segmentation could be utilized. This may make the presented knowledge-based PBT-approach for fast fully automatic segmentation of the caudate nuclei a valuable tool in clinical practice. References 1. Smith, S.M.: Fast robust automated brain extraction. Hum. Brain Mapp. 17(3) (2002) Tu, Z.: Probabilistic boosting-tree: Learning discriminative models for classification, recognition, and clustering. In: IEEE Intl. Conf. Comput. Vis., Beijing, China. (2005) Tu, Z., Zhou, X., Comaniciu, D., Bogoni, L.: A learning based approach for 3D segmentation and colon detagging. In: Europ. Conf. Comput. Vis., Graz, Austria. (2006) Friedman, J., Hastie, T., Tibshirani, R.: Additive logistic regression: a statistical view of boosting. Stanford University, Department of Statistics, Technical Report (1998) 5. Viola, P., Jones, M.: Robust real-time object detection. In: 2nd International Workshop on Statistical Learning and Computational Theories of Vision, Vancouver, Canada. (2001) Lienhart, R., Kuranov, A., Pisarevsky, V.: Empirical analysis of detection cascades of boosted classifiers for rapid object detection. In Michaelis, B., Krell, G., eds.: DAGM-Symposium. Volume 2781 of Lect. Notes in Comp. Sci., Springer (2003) Rex, D.E., Ma, J.Q., Toga, A.W.: The LONI pipeline processing environment. NeuroImage 19(3) (2003) Gerig, G., Jomier, M., Chakos, M.: Valmet: A new validation tool for assessing and improving 3D object segmentation. In: Int. Conf. Med. Image Comput. and Comp.-Assist. Interv., Utrecht, The Netherlands. (2001) Heimann, T., Styner, M., van Ginneken, B.: Workshop on 3d segmentation in the clinic: A grand challenge. mbi.dkfz-heidelberg.de/grand-challenge2007 (2007) 27
Automatic Caudate Segmentation by Hybrid Generative/Discriminative Models
 Automatic Caudate Segmentation by Hybrid Generative/Discriminative Models Zhuowen Tu, Mubeena Mirza, Ivo Dinov, and Arthur W. Toga Lab of Neuro Imaging, School of Medicine University of California, Los
Automatic Caudate Segmentation by Hybrid Generative/Discriminative Models Zhuowen Tu, Mubeena Mirza, Ivo Dinov, and Arthur W. Toga Lab of Neuro Imaging, School of Medicine University of California, Los
Subcortical Structure Segmentation using Probabilistic Atlas Priors
 Subcortical Structure Segmentation using Probabilistic Atlas Priors Sylvain Gouttard 1, Martin Styner 1,2, Sarang Joshi 3, Brad Davis 2, Rachel G. Smith 1, Heather Cody Hazlett 1, Guido Gerig 1,2 1 Department
Subcortical Structure Segmentation using Probabilistic Atlas Priors Sylvain Gouttard 1, Martin Styner 1,2, Sarang Joshi 3, Brad Davis 2, Rachel G. Smith 1, Heather Cody Hazlett 1, Guido Gerig 1,2 1 Department
Fast and Robust 3-D MRI Brain Structure Segmentation
 Fast and Robust 3-D MRI Brain Structure Segmentation Michael Wels 1,4, Yefeng Zheng 2, Gustavo Carneiro 3,, Martin Huber 4, Joachim Hornegger 1, and Dorin Comaniciu 2 1 Chair of Pattern Recognition, Department
Fast and Robust 3-D MRI Brain Structure Segmentation Michael Wels 1,4, Yefeng Zheng 2, Gustavo Carneiro 3,, Martin Huber 4, Joachim Hornegger 1, and Dorin Comaniciu 2 1 Chair of Pattern Recognition, Department
Machine Learning for Medical Image Analysis. A. Criminisi
 Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Fast and Robust 3-D MRI Brain Structure Segmentation
 Fast and Robust 3-D MRI Brain Structure Segmentation Michael Wels 1,4, Yefeng Zheng 2, Gustavo Carneiro 3,, Martin Huber 4, Joachim Hornegger 1, and Dorin Comaniciu 2 1 Chair of Pattern Recognition, Department
Fast and Robust 3-D MRI Brain Structure Segmentation Michael Wels 1,4, Yefeng Zheng 2, Gustavo Carneiro 3,, Martin Huber 4, Joachim Hornegger 1, and Dorin Comaniciu 2 1 Chair of Pattern Recognition, Department
CHAPTER 2. Morphometry on rodent brains. A.E.H. Scheenstra J. Dijkstra L. van der Weerd
 CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
Postprint.
 http://www.diva-portal.org Postprint This is the accepted version of a paper presented at 14th International Conference of the Biometrics Special Interest Group, BIOSIG, Darmstadt, Germany, 9-11 September,
http://www.diva-portal.org Postprint This is the accepted version of a paper presented at 14th International Conference of the Biometrics Special Interest Group, BIOSIG, Darmstadt, Germany, 9-11 September,
Semantic Context Forests for Learning- Based Knee Cartilage Segmentation in 3D MR Images
 Semantic Context Forests for Learning- Based Knee Cartilage Segmentation in 3D MR Images MICCAI 2013: Workshop on Medical Computer Vision Authors: Quan Wang, Dijia Wu, Le Lu, Meizhu Liu, Kim L. Boyer,
Semantic Context Forests for Learning- Based Knee Cartilage Segmentation in 3D MR Images MICCAI 2013: Workshop on Medical Computer Vision Authors: Quan Wang, Dijia Wu, Le Lu, Meizhu Liu, Kim L. Boyer,
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes
 Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
MR IMAGE SEGMENTATION
 MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
A Discriminative Model-Constrained Graph Cuts Approach to Fully Automated Pediatric Brain Tumor Segmentation in 3-D MRI
 A Discriminative Model-Constrained Graph Cuts Approach to Fully Automated Pediatric Brain Tumor Segmentation in 3-D MRI Michael Wels 1,4, Gustavo Carneiro 2, Alexander Aplas 3,MartinHuber 4, Joachim Hornegger
A Discriminative Model-Constrained Graph Cuts Approach to Fully Automated Pediatric Brain Tumor Segmentation in 3-D MRI Michael Wels 1,4, Gustavo Carneiro 2, Alexander Aplas 3,MartinHuber 4, Joachim Hornegger
Automatic Registration-Based Segmentation for Neonatal Brains Using ANTs and Atropos
 Automatic Registration-Based Segmentation for Neonatal Brains Using ANTs and Atropos Jue Wu and Brian Avants Penn Image Computing and Science Lab, University of Pennsylvania, Philadelphia, USA Abstract.
Automatic Registration-Based Segmentation for Neonatal Brains Using ANTs and Atropos Jue Wu and Brian Avants Penn Image Computing and Science Lab, University of Pennsylvania, Philadelphia, USA Abstract.
Fiber Selection from Diffusion Tensor Data based on Boolean Operators
 Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
A Study of Medical Image Analysis System
 Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
2D Vessel Segmentation Using Local Adaptive Contrast Enhancement
 2D Vessel Segmentation Using Local Adaptive Contrast Enhancement Dominik Schuldhaus 1,2, Martin Spiegel 1,2,3,4, Thomas Redel 3, Maria Polyanskaya 1,3, Tobias Struffert 2, Joachim Hornegger 1,4, Arnd Doerfler
2D Vessel Segmentation Using Local Adaptive Contrast Enhancement Dominik Schuldhaus 1,2, Martin Spiegel 1,2,3,4, Thomas Redel 3, Maria Polyanskaya 1,3, Tobias Struffert 2, Joachim Hornegger 1,4, Arnd Doerfler
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION
 ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
Whole Body MRI Intensity Standardization
 Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge
 Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
doi: /
 Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
Yiting Xie ; Anthony P. Reeves; Single 3D cell segmentation from optical CT microscope images. Proc. SPIE 934, Medical Imaging 214: Image Processing, 9343B (March 21, 214); doi:1.1117/12.243852. (214)
Supervised Learning for Image Segmentation
 Supervised Learning for Image Segmentation Raphael Meier 06.10.2016 Raphael Meier MIA 2016 06.10.2016 1 / 52 References A. Ng, Machine Learning lecture, Stanford University. A. Criminisi, J. Shotton, E.
Supervised Learning for Image Segmentation Raphael Meier 06.10.2016 Raphael Meier MIA 2016 06.10.2016 1 / 52 References A. Ng, Machine Learning lecture, Stanford University. A. Criminisi, J. Shotton, E.
Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging
 1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
IEEE TRANSACTIONS ON IMAGE PROCESSING, VOL. 26, NO. 10, OCTOBER
 IEEE TRANSACTIONS ON IMAGE PROCESSING, VOL. 26, NO. 10, OCTOBER 2017 4753 Detecting Anatomical Landmarks From Limited Medical Imaging Data Using Two-Stage Task-Oriented Deep Neural Networks Jun Zhang,
IEEE TRANSACTIONS ON IMAGE PROCESSING, VOL. 26, NO. 10, OCTOBER 2017 4753 Detecting Anatomical Landmarks From Limited Medical Imaging Data Using Two-Stage Task-Oriented Deep Neural Networks Jun Zhang,
Towards Learning-Based Holistic Brain Image Segmentation
 Towards Learning-Based Holistic Brain Image Segmentation Zhuowen Tu Lab of Neuro Imaging University of California, Los Angeles in collaboration with (A. Toga, P. Thompson et al.) Supported by (NIH CCB
Towards Learning-Based Holistic Brain Image Segmentation Zhuowen Tu Lab of Neuro Imaging University of California, Los Angeles in collaboration with (A. Toga, P. Thompson et al.) Supported by (NIH CCB
Fully Automatic Model Creation for Object Localization utilizing the Generalized Hough Transform
 Fully Automatic Model Creation for Object Localization utilizing the Generalized Hough Transform Heike Ruppertshofen 1,2,3, Cristian Lorenz 2, Peter Beyerlein 4, Zein Salah 3, Georg Rose 3, Hauke Schramm
Fully Automatic Model Creation for Object Localization utilizing the Generalized Hough Transform Heike Ruppertshofen 1,2,3, Cristian Lorenz 2, Peter Beyerlein 4, Zein Salah 3, Georg Rose 3, Hauke Schramm
A Generic Probabilistic Active Shape Model for Organ Segmentation
 A Generic Probabilistic Active Shape Model for Organ Segmentation Andreas Wimmer 1,2, Grzegorz Soza 2, and Joachim Hornegger 1 1 Chair of Pattern Recognition, Department of Computer Science, Friedrich-Alexander
A Generic Probabilistic Active Shape Model for Organ Segmentation Andreas Wimmer 1,2, Grzegorz Soza 2, and Joachim Hornegger 1 1 Chair of Pattern Recognition, Department of Computer Science, Friedrich-Alexander
NIH Public Access Author Manuscript Proc IEEE Int Symp Biomed Imaging. Author manuscript; available in PMC 2014 November 15.
 NIH Public Access Author Manuscript Published in final edited form as: Proc IEEE Int Symp Biomed Imaging. 2013 April ; 2013: 748 751. doi:10.1109/isbi.2013.6556583. BRAIN TUMOR SEGMENTATION WITH SYMMETRIC
NIH Public Access Author Manuscript Published in final edited form as: Proc IEEE Int Symp Biomed Imaging. 2013 April ; 2013: 748 751. doi:10.1109/isbi.2013.6556583. BRAIN TUMOR SEGMENTATION WITH SYMMETRIC
Improvement and Evaluation of a Time-of-Flight-based Patient Positioning System
 Improvement and Evaluation of a Time-of-Flight-based Patient Positioning System Simon Placht, Christian Schaller, Michael Balda, André Adelt, Christian Ulrich, Joachim Hornegger Pattern Recognition Lab,
Improvement and Evaluation of a Time-of-Flight-based Patient Positioning System Simon Placht, Christian Schaller, Michael Balda, André Adelt, Christian Ulrich, Joachim Hornegger Pattern Recognition Lab,
Edge-Preserving Denoising for Segmentation in CT-Images
 Edge-Preserving Denoising for Segmentation in CT-Images Eva Eibenberger, Anja Borsdorf, Andreas Wimmer, Joachim Hornegger Lehrstuhl für Mustererkennung, Friedrich-Alexander-Universität Erlangen-Nürnberg
Edge-Preserving Denoising for Segmentation in CT-Images Eva Eibenberger, Anja Borsdorf, Andreas Wimmer, Joachim Hornegger Lehrstuhl für Mustererkennung, Friedrich-Alexander-Universität Erlangen-Nürnberg
Automatic Generation of Training Data for Brain Tissue Classification from MRI
 MICCAI-2002 1 Automatic Generation of Training Data for Brain Tissue Classification from MRI Chris A. Cocosco, Alex P. Zijdenbos, and Alan C. Evans McConnell Brain Imaging Centre, Montreal Neurological
MICCAI-2002 1 Automatic Generation of Training Data for Brain Tissue Classification from MRI Chris A. Cocosco, Alex P. Zijdenbos, and Alan C. Evans McConnell Brain Imaging Centre, Montreal Neurological
Prototype of Silver Corpus Merging Framework
 www.visceral.eu Prototype of Silver Corpus Merging Framework Deliverable number D3.3 Dissemination level Public Delivery data 30.4.2014 Status Authors Final Markus Krenn, Allan Hanbury, Georg Langs This
www.visceral.eu Prototype of Silver Corpus Merging Framework Deliverable number D3.3 Dissemination level Public Delivery data 30.4.2014 Status Authors Final Markus Krenn, Allan Hanbury, Georg Langs This
Comparison Study of Clinical 3D MRI Brain Segmentation Evaluation
 Comparison Study of Clinical 3D MRI Brain Segmentation Evaluation Ting Song 1, Elsa D. Angelini 2, Brett D. Mensh 3, Andrew Laine 1 1 Heffner Biomedical Imaging Laboratory Department of Biomedical Engineering,
Comparison Study of Clinical 3D MRI Brain Segmentation Evaluation Ting Song 1, Elsa D. Angelini 2, Brett D. Mensh 3, Andrew Laine 1 1 Heffner Biomedical Imaging Laboratory Department of Biomedical Engineering,
Automated segmentation methods for liver analysis in oncology applications
 University of Szeged Department of Image Processing and Computer Graphics Automated segmentation methods for liver analysis in oncology applications Ph. D. Thesis László Ruskó Thesis Advisor Dr. Antal
University of Szeged Department of Image Processing and Computer Graphics Automated segmentation methods for liver analysis in oncology applications Ph. D. Thesis László Ruskó Thesis Advisor Dr. Antal
Semi-automatic Segmentation of Vertebral Bodies in Volumetric MR Images Using a Statistical Shape+Pose Model
 Semi-automatic Segmentation of Vertebral Bodies in Volumetric MR Images Using a Statistical Shape+Pose Model A. Suzani, A. Rasoulian S. Fels, R. N. Rohling P. Abolmaesumi Robotics and Control Laboratory,
Semi-automatic Segmentation of Vertebral Bodies in Volumetric MR Images Using a Statistical Shape+Pose Model A. Suzani, A. Rasoulian S. Fels, R. N. Rohling P. Abolmaesumi Robotics and Control Laboratory,
arxiv: v1 [cs.cv] 6 Jun 2017
![arxiv: v1 [cs.cv] 6 Jun 2017 arxiv: v1 [cs.cv] 6 Jun 2017](/thumbs/96/128307097.jpg) Volume Calculation of CT lung Lesions based on Halton Low-discrepancy Sequences Liansheng Wang a, Shusheng Li a, and Shuo Li b a Department of Computer Science, Xiamen University, Xiamen, China b Dept.
Volume Calculation of CT lung Lesions based on Halton Low-discrepancy Sequences Liansheng Wang a, Shusheng Li a, and Shuo Li b a Department of Computer Science, Xiamen University, Xiamen, China b Dept.
Sampling-Based Ensemble Segmentation against Inter-operator Variability
 Sampling-Based Ensemble Segmentation against Inter-operator Variability Jing Huo 1, Kazunori Okada, Whitney Pope 1, Matthew Brown 1 1 Center for Computer vision and Imaging Biomarkers, Department of Radiological
Sampling-Based Ensemble Segmentation against Inter-operator Variability Jing Huo 1, Kazunori Okada, Whitney Pope 1, Matthew Brown 1 1 Center for Computer vision and Imaging Biomarkers, Department of Radiological
Data mining for neuroimaging data. John Ashburner
 Data mining for neuroimaging data John Ashburner MODELLING The Scientific Process MacKay, David JC. Bayesian interpolation. Neural computation 4, no. 3 (1992): 415-447. Model Selection Search for the best
Data mining for neuroimaging data John Ashburner MODELLING The Scientific Process MacKay, David JC. Bayesian interpolation. Neural computation 4, no. 3 (1992): 415-447. Model Selection Search for the best
Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion
 Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion Mattias P. Heinrich Julia A. Schnabel, Mark Jenkinson, Sir Michael Brady 2 Clinical
Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion Mattias P. Heinrich Julia A. Schnabel, Mark Jenkinson, Sir Michael Brady 2 Clinical
Semi-automated Basal Ganglia Segmentation Using Large Deformation Diffeomorphic Metric Mapping
 Semi-automated Basal Ganglia Segmentation Using Large Deformation Diffeomorphic Metric Mapping Ali Khan 1, Elizabeth Aylward 2, Patrick Barta 3, Michael Miller 4,andM.FaisalBeg 1 1 School of Engineering
Semi-automated Basal Ganglia Segmentation Using Large Deformation Diffeomorphic Metric Mapping Ali Khan 1, Elizabeth Aylward 2, Patrick Barta 3, Michael Miller 4,andM.FaisalBeg 1 1 School of Engineering
City, University of London Institutional Repository
 City Research Online City, University of London Institutional Repository Citation: Doan, H., Slabaugh, G.G., Unal, G.B. & Fang, T. (2006). Semi-Automatic 3-D Segmentation of Anatomical Structures of Brain
City Research Online City, University of London Institutional Repository Citation: Doan, H., Slabaugh, G.G., Unal, G.B. & Fang, T. (2006). Semi-Automatic 3-D Segmentation of Anatomical Structures of Brain
SISCOM (Subtraction Ictal SPECT CO-registered to MRI)
 SISCOM (Subtraction Ictal SPECT CO-registered to MRI) Introduction A method for advanced imaging of epilepsy patients has been developed with Analyze at the Mayo Foundation which uses a combination of
SISCOM (Subtraction Ictal SPECT CO-registered to MRI) Introduction A method for advanced imaging of epilepsy patients has been developed with Analyze at the Mayo Foundation which uses a combination of
2 Michael E. Leventon and Sarah F. F. Gibson a b c d Fig. 1. (a, b) Two MR scans of a person's knee. Both images have high resolution in-plane, but ha
 Model Generation from Multiple Volumes using Constrained Elastic SurfaceNets Michael E. Leventon and Sarah F. F. Gibson 1 MIT Artificial Intelligence Laboratory, Cambridge, MA 02139, USA leventon@ai.mit.edu
Model Generation from Multiple Volumes using Constrained Elastic SurfaceNets Michael E. Leventon and Sarah F. F. Gibson 1 MIT Artificial Intelligence Laboratory, Cambridge, MA 02139, USA leventon@ai.mit.edu
Face Detection Using Look-Up Table Based Gentle AdaBoost
 Face Detection Using Look-Up Table Based Gentle AdaBoost Cem Demirkır and Bülent Sankur Boğaziçi University, Electrical-Electronic Engineering Department, 885 Bebek, İstanbul {cemd,sankur}@boun.edu.tr
Face Detection Using Look-Up Table Based Gentle AdaBoost Cem Demirkır and Bülent Sankur Boğaziçi University, Electrical-Electronic Engineering Department, 885 Bebek, İstanbul {cemd,sankur}@boun.edu.tr
Assessing the Local Credibility of a Medical Image Segmentation
 Assessing the Local Credibility of a Medical Image Segmentation Joshua H. Levy, Robert E. Broadhurst, Surajit Ray, Edward L. Chaney, and Stephen M. Pizer Medical Image Display and Analysis Group (MIDAG),
Assessing the Local Credibility of a Medical Image Segmentation Joshua H. Levy, Robert E. Broadhurst, Surajit Ray, Edward L. Chaney, and Stephen M. Pizer Medical Image Display and Analysis Group (MIDAG),
Estimating Human Pose in Images. Navraj Singh December 11, 2009
 Estimating Human Pose in Images Navraj Singh December 11, 2009 Introduction This project attempts to improve the performance of an existing method of estimating the pose of humans in still images. Tasks
Estimating Human Pose in Images Navraj Singh December 11, 2009 Introduction This project attempts to improve the performance of an existing method of estimating the pose of humans in still images. Tasks
Automatic MS Lesion Segmentation by Outlier Detection and Information Theoretic Region Partitioning Release 0.00
 Automatic MS Lesion Segmentation by Outlier Detection and Information Theoretic Region Partitioning Release 0.00 Marcel Prastawa 1 and Guido Gerig 1 Abstract July 17, 2008 1 Scientific Computing and Imaging
Automatic MS Lesion Segmentation by Outlier Detection and Information Theoretic Region Partitioning Release 0.00 Marcel Prastawa 1 and Guido Gerig 1 Abstract July 17, 2008 1 Scientific Computing and Imaging
Skull Segmentation of MR images based on texture features for attenuation correction in PET/MR
 Skull Segmentation of MR images based on texture features for attenuation correction in PET/MR CHAIBI HASSEN, NOURINE RACHID ITIO Laboratory, Oran University Algeriachaibih@yahoo.fr, nourine@yahoo.com
Skull Segmentation of MR images based on texture features for attenuation correction in PET/MR CHAIBI HASSEN, NOURINE RACHID ITIO Laboratory, Oran University Algeriachaibih@yahoo.fr, nourine@yahoo.com
The organization of the human cerebral cortex estimated by intrinsic functional connectivity
 1 The organization of the human cerebral cortex estimated by intrinsic functional connectivity Journal: Journal of Neurophysiology Author: B. T. Thomas Yeo, et al Link: https://www.ncbi.nlm.nih.gov/pubmed/21653723
1 The organization of the human cerebral cortex estimated by intrinsic functional connectivity Journal: Journal of Neurophysiology Author: B. T. Thomas Yeo, et al Link: https://www.ncbi.nlm.nih.gov/pubmed/21653723
Assessment of Reliability of Multi-site Neuroimaging via Traveling Phantom Study
 Assessment of Reliability of Multi-site Neuroimaging via Traveling Phantom Study Sylvain Gouttard 1, Martin Styner 2,3, Marcel Prastawa 1, Joseph Piven 3, and Guido Gerig 1 1 Scientific Computing and Imaging
Assessment of Reliability of Multi-site Neuroimaging via Traveling Phantom Study Sylvain Gouttard 1, Martin Styner 2,3, Marcel Prastawa 1, Joseph Piven 3, and Guido Gerig 1 1 Scientific Computing and Imaging
Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues
 Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues D. Zikic, B. Glocker, E. Konukoglu, J. Shotton, A. Criminisi, D. H. Ye, C. Demiralp 3, O. M. Thomas 4,5, T. Das 4, R. Jena
Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues D. Zikic, B. Glocker, E. Konukoglu, J. Shotton, A. Criminisi, D. H. Ye, C. Demiralp 3, O. M. Thomas 4,5, T. Das 4, R. Jena
Analysis of CMR images within an integrated healthcare framework for remote monitoring
 Analysis of CMR images within an integrated healthcare framework for remote monitoring Abstract. We present a software for analyzing Cardiac Magnetic Resonance (CMR) images. This tool has been developed
Analysis of CMR images within an integrated healthcare framework for remote monitoring Abstract. We present a software for analyzing Cardiac Magnetic Resonance (CMR) images. This tool has been developed
Automatic Optimization of Segmentation Algorithms Through Simultaneous Truth and Performance Level Estimation (STAPLE)
 Automatic Optimization of Segmentation Algorithms Through Simultaneous Truth and Performance Level Estimation (STAPLE) Mahnaz Maddah, Kelly H. Zou, William M. Wells, Ron Kikinis, and Simon K. Warfield
Automatic Optimization of Segmentation Algorithms Through Simultaneous Truth and Performance Level Estimation (STAPLE) Mahnaz Maddah, Kelly H. Zou, William M. Wells, Ron Kikinis, and Simon K. Warfield
A MORPHOLOGY-BASED FILTER STRUCTURE FOR EDGE-ENHANCING SMOOTHING
 Proceedings of the 1994 IEEE International Conference on Image Processing (ICIP-94), pp. 530-534. (Austin, Texas, 13-16 November 1994.) A MORPHOLOGY-BASED FILTER STRUCTURE FOR EDGE-ENHANCING SMOOTHING
Proceedings of the 1994 IEEE International Conference on Image Processing (ICIP-94), pp. 530-534. (Austin, Texas, 13-16 November 1994.) A MORPHOLOGY-BASED FILTER STRUCTURE FOR EDGE-ENHANCING SMOOTHING
Scene-Based Segmentation of Multiple Muscles from MRI in MITK
 Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
Detecting Anatomical Landmarks from Limited Medical Imaging Data using Two-Stage Task-Oriented Deep Neural Networks
 IEEE TRANSACTIONS ON IMAGE PROCESSING Detecting Anatomical Landmarks from Limited Medical Imaging Data using Two-Stage Task-Oriented Deep Neural Networks Jun Zhang, Member, IEEE, Mingxia Liu, Member, IEEE,
IEEE TRANSACTIONS ON IMAGE PROCESSING Detecting Anatomical Landmarks from Limited Medical Imaging Data using Two-Stage Task-Oriented Deep Neural Networks Jun Zhang, Member, IEEE, Mingxia Liu, Member, IEEE,
Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model
 Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model Pascal A. Dufour 12,HannanAbdillahi 3, Lala Ceklic 3,Ute Wolf-Schnurrbusch 23,JensKowal 12 1 ARTORG Center
Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model Pascal A. Dufour 12,HannanAbdillahi 3, Lala Ceklic 3,Ute Wolf-Schnurrbusch 23,JensKowal 12 1 ARTORG Center
UNC 4D Infant Cortical Surface Atlases, from Neonate to 6 Years of Age
 UNC 4D Infant Cortical Surface Atlases, from Neonate to 6 Years of Age Version 1.0 UNC 4D infant cortical surface atlases from neonate to 6 years of age contain 11 time points, including 1, 3, 6, 9, 12,
UNC 4D Infant Cortical Surface Atlases, from Neonate to 6 Years of Age Version 1.0 UNC 4D infant cortical surface atlases from neonate to 6 years of age contain 11 time points, including 1, 3, 6, 9, 12,
Normalization for clinical data
 Normalization for clinical data Christopher Rorden, Leonardo Bonilha, Julius Fridriksson, Benjamin Bender, Hans-Otto Karnath (2012) Agespecific CT and MRI templates for spatial normalization. NeuroImage
Normalization for clinical data Christopher Rorden, Leonardo Bonilha, Julius Fridriksson, Benjamin Bender, Hans-Otto Karnath (2012) Agespecific CT and MRI templates for spatial normalization. NeuroImage
GLIRT: Groupwise and Longitudinal Image Registration Toolbox
 Software Release (1.0.1) Last updated: March. 30, 2011. GLIRT: Groupwise and Longitudinal Image Registration Toolbox Guorong Wu 1, Qian Wang 1,2, Hongjun Jia 1, and Dinggang Shen 1 1 Image Display, Enhancement,
Software Release (1.0.1) Last updated: March. 30, 2011. GLIRT: Groupwise and Longitudinal Image Registration Toolbox Guorong Wu 1, Qian Wang 1,2, Hongjun Jia 1, and Dinggang Shen 1 1 Image Display, Enhancement,
Face Tracking in Video
 Face Tracking in Video Hamidreza Khazaei and Pegah Tootoonchi Afshar Stanford University 350 Serra Mall Stanford, CA 94305, USA I. INTRODUCTION Object tracking is a hot area of research, and has many practical
Face Tracking in Video Hamidreza Khazaei and Pegah Tootoonchi Afshar Stanford University 350 Serra Mall Stanford, CA 94305, USA I. INTRODUCTION Object tracking is a hot area of research, and has many practical
SEGMENTING subcortical structures from 3-D brain images
 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 27, NO. 4, APRIL 2008 495 Brain Anatomical Structure Segmentation by Hybrid Discriminative/Generative Models Zhuowen Tu, Katherine L. Narr, Piotr Dollár, Ivo
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 27, NO. 4, APRIL 2008 495 Brain Anatomical Structure Segmentation by Hybrid Discriminative/Generative Models Zhuowen Tu, Katherine L. Narr, Piotr Dollár, Ivo
Manifold Learning-based Data Sampling for Model Training
 Manifold Learning-based Data Sampling for Model Training Shuqing Chen 1, Sabrina Dorn 2, Michael Lell 3, Marc Kachelrieß 2,Andreas Maier 1 1 Pattern Recognition Lab, FAU Erlangen-Nürnberg 2 German Cancer
Manifold Learning-based Data Sampling for Model Training Shuqing Chen 1, Sabrina Dorn 2, Michael Lell 3, Marc Kachelrieß 2,Andreas Maier 1 1 Pattern Recognition Lab, FAU Erlangen-Nürnberg 2 German Cancer
Using Probability Maps for Multi organ Automatic Segmentation
 Using Probability Maps for Multi organ Automatic Segmentation Ranveer Joyseeree 1,2, Óscar Jiménez del Toro1, and Henning Müller 1,3 1 University of Applied Sciences Western Switzerland (HES SO), Sierre,
Using Probability Maps for Multi organ Automatic Segmentation Ranveer Joyseeree 1,2, Óscar Jiménez del Toro1, and Henning Müller 1,3 1 University of Applied Sciences Western Switzerland (HES SO), Sierre,
Bioimage Informatics
 Bioimage Informatics Lecture 13, Spring 2012 Bioimage Data Analysis (IV) Image Segmentation (part 2) Lecture 13 February 29, 2012 1 Outline Review: Steger s line/curve detection algorithm Intensity thresholding
Bioimage Informatics Lecture 13, Spring 2012 Bioimage Data Analysis (IV) Image Segmentation (part 2) Lecture 13 February 29, 2012 1 Outline Review: Steger s line/curve detection algorithm Intensity thresholding
Segmentation of Bony Structures with Ligament Attachment Sites
 Segmentation of Bony Structures with Ligament Attachment Sites Heiko Seim 1, Hans Lamecker 1, Markus Heller 2, Stefan Zachow 1 1 Visualisierung und Datenanalyse, Zuse-Institut Berlin (ZIB), 14195 Berlin
Segmentation of Bony Structures with Ligament Attachment Sites Heiko Seim 1, Hans Lamecker 1, Markus Heller 2, Stefan Zachow 1 1 Visualisierung und Datenanalyse, Zuse-Institut Berlin (ZIB), 14195 Berlin
Utilizing Salient Region Features for 3D Multi-Modality Medical Image Registration
 Utilizing Salient Region Features for 3D Multi-Modality Medical Image Registration Dieter Hahn 1, Gabriele Wolz 2, Yiyong Sun 3, Frank Sauer 3, Joachim Hornegger 1, Torsten Kuwert 2 and Chenyang Xu 3 1
Utilizing Salient Region Features for 3D Multi-Modality Medical Image Registration Dieter Hahn 1, Gabriele Wolz 2, Yiyong Sun 3, Frank Sauer 3, Joachim Hornegger 1, Torsten Kuwert 2 and Chenyang Xu 3 1
Adaptive Fuzzy Connectedness-Based Medical Image Segmentation
 Adaptive Fuzzy Connectedness-Based Medical Image Segmentation Amol Pednekar Ioannis A. Kakadiaris Uday Kurkure Visual Computing Lab, Dept. of Computer Science, Univ. of Houston, Houston, TX, USA apedneka@bayou.uh.edu
Adaptive Fuzzy Connectedness-Based Medical Image Segmentation Amol Pednekar Ioannis A. Kakadiaris Uday Kurkure Visual Computing Lab, Dept. of Computer Science, Univ. of Houston, Houston, TX, USA apedneka@bayou.uh.edu
The Insight Toolkit. Image Registration Algorithms & Frameworks
 The Insight Toolkit Image Registration Algorithms & Frameworks Registration in ITK Image Registration Framework Multi Resolution Registration Framework Components PDE Based Registration FEM Based Registration
The Insight Toolkit Image Registration Algorithms & Frameworks Registration in ITK Image Registration Framework Multi Resolution Registration Framework Components PDE Based Registration FEM Based Registration
Mouse Pointer Tracking with Eyes
 Mouse Pointer Tracking with Eyes H. Mhamdi, N. Hamrouni, A. Temimi, and M. Bouhlel Abstract In this article, we expose our research work in Human-machine Interaction. The research consists in manipulating
Mouse Pointer Tracking with Eyes H. Mhamdi, N. Hamrouni, A. Temimi, and M. Bouhlel Abstract In this article, we expose our research work in Human-machine Interaction. The research consists in manipulating
GeoInterp: Contour Interpolation with Geodesic Snakes Release 1.00
 GeoInterp: Contour Interpolation with Geodesic Snakes Release 1.00 Rohit R. Saboo, Julian G. Rosenman and Stephen M. Pizer July 1, 2006 University of North Carolina at Chapel Hill Abstract The process
GeoInterp: Contour Interpolation with Geodesic Snakes Release 1.00 Rohit R. Saboo, Julian G. Rosenman and Stephen M. Pizer July 1, 2006 University of North Carolina at Chapel Hill Abstract The process
Where are we now? Structural MRI processing and analysis
 Where are we now? Structural MRI processing and analysis Pierre-Louis Bazin bazin@cbs.mpg.de Leipzig, Germany Structural MRI processing: why bother? Just use the standards? SPM FreeSurfer FSL However:
Where are we now? Structural MRI processing and analysis Pierre-Louis Bazin bazin@cbs.mpg.de Leipzig, Germany Structural MRI processing: why bother? Just use the standards? SPM FreeSurfer FSL However:
A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation
 A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation Xiahai Zhuang (PhD) Centre for Medical Image Computing University College London Fields-MITACS Conference on Mathematics
A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation Xiahai Zhuang (PhD) Centre for Medical Image Computing University College London Fields-MITACS Conference on Mathematics
Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search
 Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search Baochun He, Cheng Huang, Fucang Jia Shenzhen Institutes of Advanced Technology, Chinese Academy
Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search Baochun He, Cheng Huang, Fucang Jia Shenzhen Institutes of Advanced Technology, Chinese Academy
Depth-Layer-Based Patient Motion Compensation for the Overlay of 3D Volumes onto X-Ray Sequences
 Depth-Layer-Based Patient Motion Compensation for the Overlay of 3D Volumes onto X-Ray Sequences Jian Wang 1,2, Anja Borsdorf 2, Joachim Hornegger 1,3 1 Pattern Recognition Lab, Friedrich-Alexander-Universität
Depth-Layer-Based Patient Motion Compensation for the Overlay of 3D Volumes onto X-Ray Sequences Jian Wang 1,2, Anja Borsdorf 2, Joachim Hornegger 1,3 1 Pattern Recognition Lab, Friedrich-Alexander-Universität
Towards Whole Brain Segmentation by a Hybrid Model
 Towards Whole Brain Segmentation by a Hybrid Model Zhuowen Tu and Arthur W. Toga Lab of Neuro Imaging, School of Medicine University of California, Los Angeles, USA Abstract. Segmenting cortical and sub-cortical
Towards Whole Brain Segmentation by a Hybrid Model Zhuowen Tu and Arthur W. Toga Lab of Neuro Imaging, School of Medicine University of California, Los Angeles, USA Abstract. Segmenting cortical and sub-cortical
Manifold Learning: Applications in Neuroimaging
 Your own logo here Manifold Learning: Applications in Neuroimaging Robin Wolz 23/09/2011 Overview Manifold learning for Atlas Propagation Multi-atlas segmentation Challenges LEAP Manifold learning for
Your own logo here Manifold Learning: Applications in Neuroimaging Robin Wolz 23/09/2011 Overview Manifold learning for Atlas Propagation Multi-atlas segmentation Challenges LEAP Manifold learning for
Image Segmentation and Registration
 Image Segmentation and Registration Dr. Christine Tanner (tanner@vision.ee.ethz.ch) Computer Vision Laboratory, ETH Zürich Dr. Verena Kaynig, Machine Learning Laboratory, ETH Zürich Outline Segmentation
Image Segmentation and Registration Dr. Christine Tanner (tanner@vision.ee.ethz.ch) Computer Vision Laboratory, ETH Zürich Dr. Verena Kaynig, Machine Learning Laboratory, ETH Zürich Outline Segmentation
Elastic registration of medical images using finite element meshes
 Elastic registration of medical images using finite element meshes Hartwig Grabowski Institute of Real-Time Computer Systems & Robotics, University of Karlsruhe, D-76128 Karlsruhe, Germany. Email: grabow@ira.uka.de
Elastic registration of medical images using finite element meshes Hartwig Grabowski Institute of Real-Time Computer Systems & Robotics, University of Karlsruhe, D-76128 Karlsruhe, Germany. Email: grabow@ira.uka.de
A Design Toolbox to Generate Complex Phantoms for the Evaluation of Medical Image Processing Algorithms
 A Design Toolbox to Generate Complex Phantoms for the Evaluation of Medical Image Processing Algorithms Omar Hamo, Georg Nelles, Gudrun Wagenknecht Central Institute for Electronics, Research Center Juelich,
A Design Toolbox to Generate Complex Phantoms for the Evaluation of Medical Image Processing Algorithms Omar Hamo, Georg Nelles, Gudrun Wagenknecht Central Institute for Electronics, Research Center Juelich,
Ensemble registration: Combining groupwise registration and segmentation
 PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
Automatic Subthalamic Nucleus Targeting for Deep Brain Stimulation. A Validation Study
 Automatic Subthalamic Nucleus Targeting for Deep Brain Stimulation. A Validation Study F. Javier Sánchez Castro a, Claudio Pollo a,b, Jean-Guy Villemure b, Jean-Philippe Thiran a a École Polytechnique
Automatic Subthalamic Nucleus Targeting for Deep Brain Stimulation. A Validation Study F. Javier Sánchez Castro a, Claudio Pollo a,b, Jean-Guy Villemure b, Jean-Philippe Thiran a a École Polytechnique
Fmri Spatial Processing
 Educational Course: Fmri Spatial Processing Ray Razlighi Jun. 8, 2014 Spatial Processing Spatial Re-alignment Geometric distortion correction Spatial Normalization Smoothing Why, When, How, Which Why is
Educational Course: Fmri Spatial Processing Ray Razlighi Jun. 8, 2014 Spatial Processing Spatial Re-alignment Geometric distortion correction Spatial Normalization Smoothing Why, When, How, Which Why is
HepaTux A Semiautomatic Liver Segmentation System
 HepaTux A Semiautomatic Liver Segmentation System Andreas Beck and Volker Aurich Institut für Informatik, Heinrich-Heine-Universität Düsseldorf, D-40225 Düsseldorf becka-miccai@acs.uni-duesseldorf.de Abstract.
HepaTux A Semiautomatic Liver Segmentation System Andreas Beck and Volker Aurich Institut für Informatik, Heinrich-Heine-Universität Düsseldorf, D-40225 Düsseldorf becka-miccai@acs.uni-duesseldorf.de Abstract.
Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model
 Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model Pascal A. Dufour 1,2, Hannan Abdillahi 3, Lala Ceklic 3, Ute Wolf-Schnurrbusch 2,3, and Jens Kowal 1,2 1 ARTORG
Pathology Hinting as the Combination of Automatic Segmentation with a Statistical Shape Model Pascal A. Dufour 1,2, Hannan Abdillahi 3, Lala Ceklic 3, Ute Wolf-Schnurrbusch 2,3, and Jens Kowal 1,2 1 ARTORG
Effect of age and dementia on topology of brain functional networks. Paul McCarthy, Luba Benuskova, Liz Franz University of Otago, New Zealand
 Effect of age and dementia on topology of brain functional networks Paul McCarthy, Luba Benuskova, Liz Franz University of Otago, New Zealand 1 Structural changes in aging brain Age-related changes in
Effect of age and dementia on topology of brain functional networks Paul McCarthy, Luba Benuskova, Liz Franz University of Otago, New Zealand 1 Structural changes in aging brain Age-related changes in
Assessing the Skeletal Age From a Hand Radiograph: Automating the Tanner-Whitehouse Method
 Assessing the Skeletal Age From a Hand Radiograph: Automating the Tanner-Whitehouse Method M. Niemeijer a, B. van Ginneken a, C.A. Maas a, F.J.A. Beek b and M.A. Viergever a a Image Sciences Institute,
Assessing the Skeletal Age From a Hand Radiograph: Automating the Tanner-Whitehouse Method M. Niemeijer a, B. van Ginneken a, C.A. Maas a, F.J.A. Beek b and M.A. Viergever a a Image Sciences Institute,
Level Set Evolution with Region Competition: Automatic 3-D Segmentation of Brain Tumors
 1 Level Set Evolution with Region Competition: Automatic 3-D Segmentation of Brain Tumors 1 Sean Ho, 2 Elizabeth Bullitt, and 1,3 Guido Gerig 1 Department of Computer Science, 2 Department of Surgery,
1 Level Set Evolution with Region Competition: Automatic 3-D Segmentation of Brain Tumors 1 Sean Ho, 2 Elizabeth Bullitt, and 1,3 Guido Gerig 1 Department of Computer Science, 2 Department of Surgery,
Assessment of Reliability of Multi-site Neuroimaging Via Traveling Phantom Study
 Assessment of Reliability of Multi-site Neuroimaging Via Traveling Phantom Study Sylvain Gouttard 1, Martin Styner 2,3, Marcel Prastawa 1, Joseph Piven 3, and Guido Gerig 1 1 Scientific Computing and Imaging
Assessment of Reliability of Multi-site Neuroimaging Via Traveling Phantom Study Sylvain Gouttard 1, Martin Styner 2,3, Marcel Prastawa 1, Joseph Piven 3, and Guido Gerig 1 1 Scientific Computing and Imaging
Marginal Space Learning for Efficient Detection of 2D/3D Anatomical Structures in Medical Images
 Marginal Space Learning for Efficient Detection of 2D/3D Anatomical Structures in Medical Images Yefeng Zheng, Bogdan Georgescu, and Dorin Comaniciu Integrated Data Systems Department, Siemens Corporate
Marginal Space Learning for Efficient Detection of 2D/3D Anatomical Structures in Medical Images Yefeng Zheng, Bogdan Georgescu, and Dorin Comaniciu Integrated Data Systems Department, Siemens Corporate
Global Journal of Engineering Science and Research Management
 ADVANCED K-MEANS ALGORITHM FOR BRAIN TUMOR DETECTION USING NAIVE BAYES CLASSIFIER Veena Bai K*, Dr. Niharika Kumar * MTech CSE, Department of Computer Science and Engineering, B.N.M. Institute of Technology,
ADVANCED K-MEANS ALGORITHM FOR BRAIN TUMOR DETECTION USING NAIVE BAYES CLASSIFIER Veena Bai K*, Dr. Niharika Kumar * MTech CSE, Department of Computer Science and Engineering, B.N.M. Institute of Technology,
PROSTATE CANCER DETECTION USING LABEL IMAGE CONSTRAINED MULTIATLAS SELECTION
 PROSTATE CANCER DETECTION USING LABEL IMAGE CONSTRAINED MULTIATLAS SELECTION Ms. Vaibhavi Nandkumar Jagtap 1, Mr. Santosh D. Kale 2 1 PG Scholar, 2 Assistant Professor, Department of Electronics and Telecommunication,
PROSTATE CANCER DETECTION USING LABEL IMAGE CONSTRAINED MULTIATLAS SELECTION Ms. Vaibhavi Nandkumar Jagtap 1, Mr. Santosh D. Kale 2 1 PG Scholar, 2 Assistant Professor, Department of Electronics and Telecommunication,
Elastically Deforming a Three-Dimensional Atlas to Match Anatomical Brain Images
 University of Pennsylvania ScholarlyCommons Technical Reports (CIS) Department of Computer & Information Science May 1993 Elastically Deforming a Three-Dimensional Atlas to Match Anatomical Brain Images
University of Pennsylvania ScholarlyCommons Technical Reports (CIS) Department of Computer & Information Science May 1993 Elastically Deforming a Three-Dimensional Atlas to Match Anatomical Brain Images
Joint Tumor Segmentation and Dense Deformable Registration of Brain MR Images
 Joint Tumor Segmentation and Dense Deformable Registration of Brain MR Images Sarah Parisot 1,2,3, Hugues Duffau 4, Stéphane Chemouny 3, Nikos Paragios 1,2 1. Center for Visual Computing, Ecole Centrale
Joint Tumor Segmentation and Dense Deformable Registration of Brain MR Images Sarah Parisot 1,2,3, Hugues Duffau 4, Stéphane Chemouny 3, Nikos Paragios 1,2 1. Center for Visual Computing, Ecole Centrale
NIH Public Access Author Manuscript Conf Proc IEEE Eng Med Biol Soc. Author manuscript; available in PMC 2009 December 11.
 NIH Public Access Author Manuscript Published in final edited form as: Conf Proc IEEE Eng Med Biol Soc. 2004 ; 2: 1521 1524. doi:10.1109/iembs.2004.1403466. An Elliptic PDE Approach for Shape Characterization
NIH Public Access Author Manuscript Published in final edited form as: Conf Proc IEEE Eng Med Biol Soc. 2004 ; 2: 1521 1524. doi:10.1109/iembs.2004.1403466. An Elliptic PDE Approach for Shape Characterization
Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit
 Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit John Melonakos 1, Ramsey Al-Hakim 1, James Fallon 2 and Allen Tannenbaum 1 1 Georgia Institute of Technology, Atlanta GA 30332,
Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit John Melonakos 1, Ramsey Al-Hakim 1, James Fallon 2 and Allen Tannenbaum 1 1 Georgia Institute of Technology, Atlanta GA 30332,
Quantitative Measurement of Kidney and Cyst Sizes in Patients with Autosomal Dominant Polycystic Kidney Disease (ADPKD)
 Quantitative Measurement of Kidney and Cyst Sizes in Patients with Autosomal Dominant Polycystic Kidney Disease (ADPKD) V Daum 1, H Helbig 1, R Janka 3,K-U Eckardt 2 and R Zeltner 2 1 Friedrich Alexander
Quantitative Measurement of Kidney and Cyst Sizes in Patients with Autosomal Dominant Polycystic Kidney Disease (ADPKD) V Daum 1, H Helbig 1, R Janka 3,K-U Eckardt 2 and R Zeltner 2 1 Friedrich Alexander
Fast 3D Mean Shift Filter for CT Images
 Fast 3D Mean Shift Filter for CT Images Gustavo Fernández Domínguez, Horst Bischof, and Reinhard Beichel Institute for Computer Graphics and Vision, Graz University of Technology Inffeldgasse 16/2, A-8010,
Fast 3D Mean Shift Filter for CT Images Gustavo Fernández Domínguez, Horst Bischof, and Reinhard Beichel Institute for Computer Graphics and Vision, Graz University of Technology Inffeldgasse 16/2, A-8010,
Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs
 Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs Yonghong Shi 1 and Dinggang Shen 2,*1 1 Digital Medical Research Center, Fudan University, Shanghai, 232, China
Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs Yonghong Shi 1 and Dinggang Shen 2,*1 1 Digital Medical Research Center, Fudan University, Shanghai, 232, China
Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry
 Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry Nivedita Agarwal, MD Nivedita.agarwal@apss.tn.it Nivedita.agarwal@unitn.it Volume and surface morphometry Brain volume White matter
Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry Nivedita Agarwal, MD Nivedita.agarwal@apss.tn.it Nivedita.agarwal@unitn.it Volume and surface morphometry Brain volume White matter
Big Data Methods. Chapter 5: Machine learning. Big Data Methods, Chapter 5, Slide 1
 Big Data Methods Chapter 5: Machine learning Big Data Methods, Chapter 5, Slide 1 5.1 Introduction to machine learning What is machine learning? Concerned with the study and development of algorithms that
Big Data Methods Chapter 5: Machine learning Big Data Methods, Chapter 5, Slide 1 5.1 Introduction to machine learning What is machine learning? Concerned with the study and development of algorithms that
Available Online through
 Available Online through www.ijptonline.com ISSN: 0975-766X CODEN: IJPTFI Research Article ANALYSIS OF CT LIVER IMAGES FOR TUMOUR DIAGNOSIS BASED ON CLUSTERING TECHNIQUE AND TEXTURE FEATURES M.Krithika
Available Online through www.ijptonline.com ISSN: 0975-766X CODEN: IJPTFI Research Article ANALYSIS OF CT LIVER IMAGES FOR TUMOUR DIAGNOSIS BASED ON CLUSTERING TECHNIQUE AND TEXTURE FEATURES M.Krithika
