NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2014 May 21.
|
|
|
- Darren Walker
- 5 years ago
- Views:
Transcription
1 NIH Public Access Author Manuscript Published in final edited form as: Med Image Comput Comput Assist Interv ; 16(0 1): On Describing Human White Matter Anatomy: The White Matter Query Language Demian Wassermann 1,*, Nikos Makris 2, Yogesh Rathi 1, Martha Shenton 1, Ron Kikinis 1, Marek Kubicki 1, and Carl-Fredrik Westin 1 Demian Wassermann: demian@bwh.harvard.edu 1 Brigham and Women s Hospital and Harvard Medical School, Boston, MA, USA 2 Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA Abstract The main contribution of this work is the careful syntactical definition of major white matter tracts in the human brain based on a neuroanatomist s expert knowledge. We present a technique to formally describe white matter tracts and to automatically extract them from diffusion MRI data. The framework is based on a novel query language with a near-to-english textual syntax. This query language allows us to construct a dictionary of anatomical definitions describing white matter tracts. The definitions include adjacent gray and white matter regions, and rules for spatial relations. This enables automated coherent labeling of white matter anatomy across subjects. We use our method to encode anatomical knowledge in human white matter describing 10 association and 8 projection tracts per hemisphere and 7 commissural tracts. The technique is shown to be comparable in accuracy to manual labeling. We present results applying this framework to create a white matter atlas from 77 healthy subjects, and we use this atlas in a proof-of-concept study to detect tract changes specific to schizophrenia. 1 Introduction Diffusion magnetic resonance imaging (dmri) is a technique that allows to probe the structure of white matter the human brain in vivo. dmri streamline tractography has provided the opportunity for non-invasive investigation of white matter anatomy. Most common methods for isolating fiber bundles based on streamline tractography require the manual placement of multiple regions of interest (ROIs). These methods include an approach that starts from seed points within a predefined region of interest, and then calculates and preserves only tracts that touch other predefined ROIs [1]. A different approach creates seed points throughout the entire brain (whole brain tractography) keeping tracts that pass through conjunctions, disjunctions or exclusions of ROIs either off-line [2] or interactively [3]. An alternative method to manual placement of ROIs is to use a clustering approach [4 6]. Clustering methods are in general fully automatic, unguided, and take advantage of the similarity of fiber paths. However, incorporating precise information about human anatomy into a clustering method is difficult. * Camera ready version for the Medical Image Computing and Computed Assisted Intervention 2013 conference
2 Wassermann et al. Page 2 2 Methods The ability to target specific tracts for analyses compared to whole brain studies increases the statistical power and sensitivity of the study, and simplifies the interpretation of results, but requires precise consistent delineation of the tracts across subjects. Although several fascicules, such as the cingulum bundle, are widely recognized and well defined in field of neuroanatomy, there are others which existence or subdivisions is still a matter of discussion. Examples are the different systems to define and subdivide the superior longitudinal fasciculus (SLF) proposed by Catani [7] and Makris [8], and the discussion regarding existence of the inferior occipito-frontal fasciculus (IOFF) in humans [9]. Three major challenges make it difficult to extend and reproduce tractography-based dissections and tract-specific analyses: 1) the anatomical dissection of the white matter into specific fascicles is currently in discussion; 2) comparing fascicle definitions across atlases is a difficult task due to the lack of a system to unify the definitions; 3) semi-automated approaches are usually based on a fixed set of fascicles difficult to extend due to amount of technical knowledge needed for the task. The main contribution of this paper is a system to express anatomical descriptions of white matter tracts in a near-to-english textual language, and we believe this work can help address the above challenges. We designed this textual language to be human-readable to make the tract descriptions easy to change and extend without the need of an engineering background and to be used for automated white matter virtual dissections. The white matter query language (WMQL) proposed in this paper has several applications. For example Wakana s and Catani s definition of tracts using regions of interest (ROIs) [10, 2], can readily be represented using WMQL definitions. This will yield an human readable definition that also can be extended by an anatomist for finer division. Another interesting application is to post-process clustering results and automatically label clusters as anatomically known tracts [4 6]. In this paper we present definitions of different tracts from current literature and formulated WMQL descriptions. Our anatomy dictionary currently contains descriptions of 10 association and 8 projection tracts per hemisphere, and 7 commissural tracts. An implementation of WMQL as well as the definitions specified in this publication can be downloaded at We designed the queries in white matter query language (WMQL) to formalize current descriptions in anatomy literature. Such descriptions are constructed in terms of different relationships between gyri, sulci, subcortical structures or white matter areas.the operations of WMQL, can be divided into 3 groups: 1) anatomical terms stating if a tract traverses or ends in a certain brain structure, 2) relative position terms indicating whether the tracts are, for instance, medial or frontal to a structure like the amygdala, and 3) logical operations like conjunction, disjunction or exclusion of the previous two types of clauses. We illustrate these three types of operations in fig. 1. To apply WMQL queries to dissect dmri-based tractographies, like in fig. 1, we need to situate gyri, sulci and other structures used for the queries relative to the tractography. For this, we overlay an atlas of the brain structures on top the tractography, in this work we used the cortical and white matter parcellation based on the Desikan atlas as described by [11]
3 Wassermann et al. Page 3 and the neuroanatomic structure segmentation by [12] which are readily available in FreeSurfer ( We provide the details of this process in the section MRI Data Acquisition and Processing. It is worth noting that WMQL does not depend on a particular atlas. To implement the white matter query language, we group the tracts in sets representing whether they have an endpoint in each anatomical label (fig. 1b); traverse anatomical label (fig. 1c); or their position relative to each anatomical label (fig. 1d f). Then, for each anatomical label, like the amygdala, we have six sets: 1. endpoints_in(amygdala): all tracts with at least an endpoint in the amygdala. 2. amygdala: all the tracts traversing the amygdala. 3. anterior_of(amygdala), posterior_of(amygdala), medial_of(amygdala), lateral_of(amygdala), superior_of(amygdala), inferior_of(amygdala) : containing the tracts traversing brain areas delimited by their relative position to the amygdala We calculate the endpoint sets (1) by computing which label is the tract touching at each endpoint and adding the tract to the corresponding set. The sets representing label traversals (2) are computed by following each tract and adding it to the set corresponding to a particular label, like the amygdala, if tract traverses it. Finally, we compute the 6 relative positioning sets (3) by checking the points the tract traverses with respect to a label. For instance, every tract that traverses the area anterior to the amygdala, shown in orange in fig. 1e, will be added to the set anterior_of(amygdala). We implement this efficiently using a tree of axis-aligned bounding boxes of the structures as a spatial index of the labels and then computing the relative positions of the tracts with respect to the bounding boxes composing each label [13]. For two sets of tracts a and b and the set of all tracts L, we formalize the WQML logical operations as follows: We have illustrated these operations in fig. 1 a), c); and g)-i). Tracts in WMQL are defined by using an assignment operation, for instance, we defined the left UF in fig. 1i as assigning a meaning to UF.left: UF.left = insula.left and (lateral_frontal.left or medial_frontal.left or orbito_frontal.left) and endpoints_in(temporal.left and anterior_of(amygdala.left)) not in hemisphere.right
4 Wassermann et al. Page 4 3 Results To simplify the definitions of tracts in both hemispheres with a single assignment, we included the suffixes.side and.opposite to WMQL. Creating a bihemispheric definition becomes: UF.side = insula.side and (lateral_frontal.side or medial_frontal.side or orbito_frontal.side) and endpoints_in(temporal.side and anterior_of(amygdala.side)) not in hemisphere.opposite In this way WMQL allows a single definition for tracts found in both hemispheres simultaneously. The formalization of WMQL as the basic set operations allows us to define white matter tracts using all the flexibility and expressiveness power of set theory and propositional logic. Finally, to implement the WMQL language, we defined the WMQL in Backus normal form; we used an LL(1) parser for this grammar which transforms WMQL expressions into an abstract syntax tree; and we implemented an algorithm to traverse the tree evaluating the WMQL operations. 3.1 Development of WMQL We are using WMQL to formalize tract descriptions from classic neuroanatomy textbooks and current literature on the anatomy of the white matter [14, 10, 8, 15]. In table 1, we show the queries for 10 association tracts. In the following, for each tract we describe in WMQL, we provide a description derived from anatomy literature and the derived WMQL query. The population results of these queries are shown in fig. 2 along with the commisural and projection tracts whose definitions we did not include for space reasons. Using the WMQL, we have constructed a comprehensive atlas based on high angular MRI (HARDI) tractography [16]. The combination of WMQL with current tractography technologies has enabled us to generalize previous atlases [10, 2] and extend them with white matter tracts not included in these like the middle longitudinal fasciculus (MdLF) or the three different superior longitudinal fasciculi. In total, our atlas includes 10 long association tracts; 8 projection tracts per hemisphere (7 cortico-striatal and the Cortico-spinal tract) and the 7 sections of the corpus callosum according to [15]. 3.2 Application of WMQL to Tract Extraction and Statistical Analyses Diffusion-weighted images (DWI) from 77 healthy subjects (HS) (32.9 ± 12.4 years old of age; 64 males; right handed) and 20 male schizophrenic subjects (SZ) paired with the HS were acquired. DWI data were acquired on a GE Signa HDxt 3.0T (51 directions with b=900
5 Wassermann et al. Page 5 s/mm2, 8 b=0 s/mm2 images, 1.7 mm 3 isotropic voxels). A T1 MRI acquisition was also performed (25.6cm2 field of view, 1mm 3 isotropic voxels). For each DWI image, we obtained a 2-tensor full-brain tractography placing ten seeds per voxel and obtaining an average of one million tracts per subject. We overlaid a parcellation of the cortical and subcortical structures and the white matter on DWI images by processing the T1 images of each subject using FreeSurfer and registering the results to the DWI images using ANTS [17]. For each subject, this resulted in the subcortical structures labeled and the cortex the white matter parcellated [11]. Validation To validate our extraction protocol, 2 different experts segmented 5 tracts (IOFF, UF, ILF, CST and AF) in each hemisphere of 10 healthy subjects following the protocol in [10]. Overlap between those and the ones extracted was calculated with the kappa measure [2]. For all tracts k >.7 which is considered good agreement, the worse being the left AF (k =.71) and the best the left CST (k =.89). White Matter Atlas Generation with WMQL For each subject we extracted 37 white matter tracts using WMQL queries derived from classical and current literature of the human brain white matter. Some of these are detailed in table 1. Processing each full brain tractography took 5 ±.25 min. to initialize and 7 ±.02 sec. per query. To generate a tract atlas, we normalized all the FA maps to MNI space, created a population FA template using ANTS and then generated group effect maps for each tract [7]. To obtain the group effect maps, we started by calculating a binary visitation map for each tract of each subject. This map is a mask in MNI space where a voxel has a value of one if the tract traverses that voxel and 0 if it doesn t. We create the group effect map for each tract through voxel-wise statistics. The group effect map assesses the probability that a tract traverses each voxel. For this, we rejected the null hypothesis that that voxel is not traversed by such tract. We first smoothed the visitation maps with a 2mm (FWHM) isotropic Gaussian to approximate the distribution of the data to a Gaussian one [18]. Then, we rejected the hypothesis that the voxel does not belong to the tract, i.e. the mean traversal value over all subjects on that voxel is different to 0, by using a voxel wise t-test for a one-sample mean. We calculated the corrected significance using permutation testing (10,000 iterations) to avoid a high dependence on Gaussian assumptions [19]. We set the significance threshold for considering that a voxel belongs to the tract to p-value < corrected for multiple comparisons. Group Differences in Schizophrenia As a proof of concept, we analyzed the tracts that are not included in other atlases, but found in our WM atlas only: MdLF, SLF I, II and III in 20 SZ subjects and controls extracted from the HS paired by age and gender. We used the group maps to obtain a weighted average of FA resulting in one value per subject and performed a t-test for each tract correcting for age. We show these results in fig. 3. Results show agreement with recent discoveries in SZ in the MdLF tract [20]
6 Wassermann et al. Page 6 4 Conclusion Acknowledgments References In this work we have introduced the White Matter Query Language: a tool to represent anatomical knowledge of white matter tracts and extract them from full-brain tractographies. This tool is a complement to current semi-automated approaches in tract extraction based in clustering and can be used to implement ROI-based approaches. The utility of the tool was illustrated by constructing an atlas of white matter tracts from 77 healthy subjects and then performing a statistical analysis in schizophrenia. The textual descriptions in WMQL add transparency between the conceptualization of white matter fiber tracts and their extraction from a dmri. This work has been supported by NIH grants: R01MH074794, R01MH092862, P41RR013218, R01MH097979, P41EB and Swedish Research Council (VR) grant Mori S, van Zijl PCM. Fiber tracking: principles and strategies-a technical review. NMR in Biomedicine Wakana S, Caprihan A, Panzenboeck MM, Fallon JH, Perry M, Gollub RL, Hua K, Zhang J, Jiang H, Dubey P, Blitz A, van Zijl PC, Mori S. Reproducibility of quantitative tractography methods applied to cerebral white matter. NImg Akers D, Sherbondy A, Mackenzie R, Dougherty R, Wandell B. Exploration of the brain s white matter pathways with dynamic queries. IEEE Viz Wassermann D, Bloy L, Kanterakis E, Verma R, Deriche R. Unsupervised white matter fiber clustering and tract probability map generation: Applications of a Gaussian process framework for white matter fibers. NImg O Donnell LJ, Westin CF. Automatic Tractography Segmentation Using a High-Dimensional White Matter Atlas. TMI Wang X, Grimson WEL, Westin CF. Tractography segmentation using a hierarchical Dirichlet processes mixture model. NImg Thiebaut de Schotten M, Ffytche DH, Bizzi A, Dell acqua F, Allin M, Walshe M, Murray R, Williams SC, Murphy DG, Catani M. Atlasing location, asymmetry and inter-subject variability of white matter tracts in the human brain with MR diffusion tractography. NImg Makris N, Kennedy DN, McInerney S, Sorensen AG, Wang R, Caviness VS, Pandya D. Segmentation of Subcomponents within the Superior Longitudinal Fascicle in Humans: A Quantitative, In Vivo, DT-MRI Study. Cerebral Cortex Schmahmann JD, Pandya D. The Complex History of the Fronto-Occipital Fasciculus. Journal of the History of the Neurosciences Catani M, Thiebaut de Schotten M. A diffusion tensor imaging tractography atlas for virtual in vivo dissections. Cortex Salat DH, Greve DN, Pacheco JL, Quinn BT, Helmer KG, Buckner RL, Fischl B. Regional white matter volume differences in nondemented aging and Alzheimer s disease. NImg Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole Brain Segmentation: Automated Labeling of Neuroanatomical Structures in the Human Brain. Neuron Bergen, Gvd. Efficient Collision Detection of Complex Deformable Models using AABB Trees. Journal of Graphic Tools Parent, A. Carpenter s Human Neuroanatomy. Williams & Wilkins; 1996.
7 Wassermann et al. Page Witelson SF, Kigar DL. Anatomical development of the corpus callosum in humans: A review with reference to sex and cognition. Brain Lateralization in children: Developmental implications Malcolm JG, Michailovich O, Bouix S, Westin CF, Shenton ME, Rathi Y. A filtered approach to neural tractography using the Watson directional function. TMI Avants B, Epstein C, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. MIA Ashburner J, Friston KJ. Voxel-Based Morphometry The Methods. NImg Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. HBM Asami T, Saito Y, Whitford TJ, Makris N, Niznikiewicz M, McCarley RW, Shenton ME, Kubicki M. Abnormalities of middle longitudinal fascicle and disorganization in patients with schizophrenia. Schizophrenia Research. 2013
8 Wassermann et al. Page 8 Fig. 1. WMQL Terms (a f) along with an example construction of a WMQL query (g i). Regions in (g i): insula (cyan); the orbito-frontal (purple), middle-frontal (brown) and inferiorfrontal (dark green) gyri. h) shows the anterior temporal lobe (light green) defined as the section of the temporal lobe anterior to the amygdala (yellow).
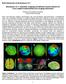
9 Wassermann et al. Page 9 Fig. 2. Iso-surfaces in blue (p-value = 0.01, corrected for multiple comparisons) shows 11 association, 8 projection and 7 commissural tracts.
10 Wassermann et al. Page 10 Fig. 3. Compared mean FA over different tracts between healthy subjects (HS) and schizophrenic subjects (SZ). The p-value was computed using a t-test correcting for age. We observe significant differences on the left and right MdLF, and the left and right SLF II and III tracts. Notation: the * means p-value <.05 and ** p-value <.01
11 Wassermann et al. Page 11 Table 1 Association Tract Definitions in WMQL: Cingulum bundle (CG); Extreme Capsule (EmC); and the fascicules: Superior Longitudinal (SLF) sections I to III; Arcuate (AF); Inferior occipito frontal (IOFF); Middle Longitudinal (MdLF); Uncinate (UF) CB.side = only((cingular.side or cingular_cortex.side) and (middle_frontal.side or cuneus.side or entorhinal.side orsuperior_frontal.side or inferior_parietal.side or fusiform.side or medial_orbitofrontal.side or lateral_orbitofrontal.side or parahippocampal.side or precuneus.side or lingual.side or centrum_semiovale.side)) EmC.side = (endpoints_in(inferior_frontal.side or middle_frontal.side) andendpoints_in(inferior_parietal_lobule.side) andtemporal.side and insula.side)not in hemisphere.opposite SLF_I.side = (superior_parietal.side and precuneus.side and superior_frontal.side) or(superior_parietal.side and precuneus.side and superior_frontal.side and lateral_occipital.side) not in cingular.side not in temporal.side not in subcortical.side not in hemisphere.opposite SLF_II.side =(inferior_parietal.side or supramarginal.side or lateral_occipital.side) and endpoints_in(middle_frontal.side)) not in temporal.side not in subcortical.side not in hemisphere.opposite SLF_III.side = ((inferior_parietal.side or supramarginal.side or lateral_occipital.side) and endpoints_in(inferior_frontal.side)) not in temporal.side not in subcortical.side not in hemisphere.opposite AF.side = (inferior_frontal.side or middle_frontal.side or precentral.side) and (superior_temporal.side or middle_temporal.side) not in medial_of(supramarginal.side)not in subcortical.side not in hemisphere.opposite IOFF.side = (lateralorbitofrontal.side and occipital.side) and temporal.side not in subcortical.side not in cingular.side not in superior_parietal_lobule.side not in hemisphere.opposite ILF.side = only(temporal.side and occipital.side) and anterior_of(hippocampus.side) not in parahippocampal.side MdLF.side = only((temporal_pole.side or superior_temporal.side) and (inferior_parietal.side or superior_parietal.side or supramarginal.side or precuneus.side or (centrum_semiovale.side and superior_parietal.side) or (centrum_semiovale.side and inferior_parietal.side)) UF.side = insula.side and (inferior_frontal.side or middle_frontal.side or orbito_frontal.side) and endpoints_in(temporal.side and anterior_of(amygdala.side))
H. Mirzaalian, L. Ning, P. Savadjiev, O. Pasternak, S. Bouix, O. Michailovich, M. Kubicki, C-F Westin, M.E.Shenton, Y. Rathi
 Harmonizing diffusion MRI data from multiple scanners H. Mirzaalian, L. Ning, P. Savadjiev, O. Pasternak, S. Bouix, O. Michailovich, M. Kubicki, C-F Westin, M.E.Shenton, Y. Rathi Synopsis Diffusion MRI
Harmonizing diffusion MRI data from multiple scanners H. Mirzaalian, L. Ning, P. Savadjiev, O. Pasternak, S. Bouix, O. Michailovich, M. Kubicki, C-F Westin, M.E.Shenton, Y. Rathi Synopsis Diffusion MRI
Introduc)on to FreeSurfer h0p://surfer.nmr.mgh.harvard.edu. Jenni Pacheco.
 Introduc)on to FreeSurfer h0p://surfer.nmr.mgh.harvard.edu Jenni Pacheco jpacheco@mail.utexas.edu Outline FreeSurfer ROI Terminology ROI Sta)s)cs Files ROI Studies Volumetric/Area Intensity FreeSurfer
Introduc)on to FreeSurfer h0p://surfer.nmr.mgh.harvard.edu Jenni Pacheco jpacheco@mail.utexas.edu Outline FreeSurfer ROI Terminology ROI Sta)s)cs Files ROI Studies Volumetric/Area Intensity FreeSurfer
A Tract-Specific Framework for White Matter Morphometry Combining Macroscopic and Microscopic Tract Features
 A Tract-Specific Framework for White Matter Morphometry Combining Macroscopic and Microscopic Tract Features Hui Zhang, Suyash P. Awate, Sandhitsu R. Das, John H. Woo, Elias R. Melhem, James C. Gee, and
A Tract-Specific Framework for White Matter Morphometry Combining Macroscopic and Microscopic Tract Features Hui Zhang, Suyash P. Awate, Sandhitsu R. Das, John H. Woo, Elias R. Melhem, James C. Gee, and
Spatial Distribution of Temporal Lobe Subregions in First Episode Schizophrenia: Statistical Probability Atlas Approach
 Spatial Distribution of Temporal Lobe Subregions in First Episode Schizophrenia: Statistical Probability Atlas Approach Hae-Jeong Park, Martha E. Shenton, James Levitt, Dean Salisbury, Marek Kubicki, Ferenc
Spatial Distribution of Temporal Lobe Subregions in First Episode Schizophrenia: Statistical Probability Atlas Approach Hae-Jeong Park, Martha E. Shenton, James Levitt, Dean Salisbury, Marek Kubicki, Ferenc
Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry
 Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry Nivedita Agarwal, MD Nivedita.agarwal@apss.tn.it Nivedita.agarwal@unitn.it Volume and surface morphometry Brain volume White matter
Neuroimaging and mathematical modelling Lesson 2: Voxel Based Morphometry Nivedita Agarwal, MD Nivedita.agarwal@apss.tn.it Nivedita.agarwal@unitn.it Volume and surface morphometry Brain volume White matter
Fiber Selection from Diffusion Tensor Data based on Boolean Operators
 Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Lilla Zöllei A.A. Martinos Center, MGH; Boston, MA
 Lilla Zöllei lzollei@nmr.mgh.harvard.edu A.A. Martinos Center, MGH; Boston, MA Bruce Fischl Gheorghe Postelnicu Jean Augustinack Anastasia Yendiki Allison Stevens Kristen Huber Sita Kakonoori + the FreeSurfer
Lilla Zöllei lzollei@nmr.mgh.harvard.edu A.A. Martinos Center, MGH; Boston, MA Bruce Fischl Gheorghe Postelnicu Jean Augustinack Anastasia Yendiki Allison Stevens Kristen Huber Sita Kakonoori + the FreeSurfer
SURFACE RECONSTRUCTION OF EX-VIVO HUMAN V1 THROUGH IDENTIFICATION OF THE STRIA OF GENNARI USING MRI AT 7T
 SURFACE RECONSTRUCTION OF EX-VIVO HUMAN V1 THROUGH IDENTIFICATION OF THE STRIA OF GENNARI USING MRI AT 7T Oliver P. Hinds 1, Jonathan R. Polimeni 2, Megan L. Blackwell 3, Christopher J. Wiggins 3, Graham
SURFACE RECONSTRUCTION OF EX-VIVO HUMAN V1 THROUGH IDENTIFICATION OF THE STRIA OF GENNARI USING MRI AT 7T Oliver P. Hinds 1, Jonathan R. Polimeni 2, Megan L. Blackwell 3, Christopher J. Wiggins 3, Graham
Evaluation of multiple voxel-based morphometry approaches and applications in the analysis of white matter changes in temporal lobe epilepsy
 Evaluation of multiple voxel-based morphometry approaches and applications in the analysis of white matter changes in temporal lobe epilepsy Wenjing Li a, Huiguang He a, Jingjing Lu b, Bin Lv a, Meng Li
Evaluation of multiple voxel-based morphometry approaches and applications in the analysis of white matter changes in temporal lobe epilepsy Wenjing Li a, Huiguang He a, Jingjing Lu b, Bin Lv a, Meng Li
An Introduction To Automatic Tissue Classification Of Brain MRI. Colm Elliott Mar 2014
 An Introduction To Automatic Tissue Classification Of Brain MRI Colm Elliott Mar 2014 Tissue Classification Tissue classification is part of many processing pipelines. We often want to classify each voxel
An Introduction To Automatic Tissue Classification Of Brain MRI Colm Elliott Mar 2014 Tissue Classification Tissue classification is part of many processing pipelines. We often want to classify each voxel
Building an Average Population HARDI Atlas
 Building an Average Population HARDI Atlas Sylvain Bouix 1, Yogesh Rathi 1, and Mert Sabuncu 2 1 Psychiatry Neuroimaging Laboratory, Brigham and Women s Hospital, Harvard Medical School, Boston, MA, USA.
Building an Average Population HARDI Atlas Sylvain Bouix 1, Yogesh Rathi 1, and Mert Sabuncu 2 1 Psychiatry Neuroimaging Laboratory, Brigham and Women s Hospital, Harvard Medical School, Boston, MA, USA.
Quantitative MRI of the Brain: Investigation of Cerebral Gray and White Matter Diseases
 Quantities Measured by MR - Quantitative MRI of the Brain: Investigation of Cerebral Gray and White Matter Diseases Static parameters (influenced by molecular environment): T, T* (transverse relaxation)
Quantities Measured by MR - Quantitative MRI of the Brain: Investigation of Cerebral Gray and White Matter Diseases Static parameters (influenced by molecular environment): T, T* (transverse relaxation)
Methods for data preprocessing
 Methods for data preprocessing John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. Overview Voxel-Based Morphometry Morphometry in general Volumetrics VBM preprocessing
Methods for data preprocessing John Ashburner Wellcome Trust Centre for Neuroimaging, 12 Queen Square, London, UK. Overview Voxel-Based Morphometry Morphometry in general Volumetrics VBM preprocessing
syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets
 syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets Julien Gervais; Lisa Chuah Siemens Healthcare, Magnetic
syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets Julien Gervais; Lisa Chuah Siemens Healthcare, Magnetic
White matter multi-resolution segmentation using fuzzy set theory
 White matter multi-resolution segmentation using fuzzy set theory Alessandro Delmonte, Corentin Mercier, Johan Pallud, Isabelle Bloch, Pietro Gori To cite this version: Alessandro Delmonte, Corentin Mercier,
White matter multi-resolution segmentation using fuzzy set theory Alessandro Delmonte, Corentin Mercier, Johan Pallud, Isabelle Bloch, Pietro Gori To cite this version: Alessandro Delmonte, Corentin Mercier,
On Classifying Disease-Induced Patterns in the Brain Using Diffusion Tensor Images
 On Classifying Disease-Induced Patterns in the Brain Using Diffusion Tensor Images Peng Wang 1,2 and Ragini Verma 1 1 Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania,
On Classifying Disease-Induced Patterns in the Brain Using Diffusion Tensor Images Peng Wang 1,2 and Ragini Verma 1 1 Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania,
Surface-based Analysis: Inter-subject Registration and Smoothing
 Surface-based Analysis: Inter-subject Registration and Smoothing Outline Exploratory Spatial Analysis Coordinate Systems 3D (Volumetric) 2D (Surface-based) Inter-subject registration Volume-based Surface-based
Surface-based Analysis: Inter-subject Registration and Smoothing Outline Exploratory Spatial Analysis Coordinate Systems 3D (Volumetric) 2D (Surface-based) Inter-subject registration Volume-based Surface-based
Appendix E1. Supplementary Methods. MR Image Acquisition. MR Image Analysis
 RSNA, 2015 10.1148/radiol.2015150532 Appendix E1 Supplementary Methods MR Image Acquisition By using a 1.5-T system (Avanto, Siemens Medical, Erlangen, Germany) under a program of regular maintenance (no
RSNA, 2015 10.1148/radiol.2015150532 Appendix E1 Supplementary Methods MR Image Acquisition By using a 1.5-T system (Avanto, Siemens Medical, Erlangen, Germany) under a program of regular maintenance (no
Evaluation of Local Filter Approaches for Diffusion Tensor based Fiber Tracking
 Evaluation of Local Filter Approaches for Diffusion Tensor based Fiber Tracking D. Merhof 1, M. Buchfelder 2, C. Nimsky 3 1 Visual Computing, University of Konstanz, Konstanz 2 Department of Neurosurgery,
Evaluation of Local Filter Approaches for Diffusion Tensor based Fiber Tracking D. Merhof 1, M. Buchfelder 2, C. Nimsky 3 1 Visual Computing, University of Konstanz, Konstanz 2 Department of Neurosurgery,
NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2012 February 27.
 NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2012 February 27. Published in final edited form as: Med Image Comput Comput Assist Interv.
NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2012 February 27. Published in final edited form as: Med Image Comput Comput Assist Interv.
Measuring longitudinal brain changes in humans and small animal models. Christos Davatzikos
 Measuring longitudinal brain changes in humans and small animal models Christos Davatzikos Section of Biomedical Image Analysis University of Pennsylvania (Radiology) http://www.rad.upenn.edu/sbia Computational
Measuring longitudinal brain changes in humans and small animal models Christos Davatzikos Section of Biomedical Image Analysis University of Pennsylvania (Radiology) http://www.rad.upenn.edu/sbia Computational
ACCEPTED MANUSCRIPT. Inter-site and inter-scanner diffusion MRI data harmonization
 Inter-site and inter-scanner diffusion MRI data harmonization H. Mirzaalian 1, L. Ning 1, P. Savadjiev 1, O. Pasternak 1, S. Bouix 1, O. Michailovich 2, M. Kubicki 1, C-F Westin 1, M.E.Shenton 1, Y. Rathi
Inter-site and inter-scanner diffusion MRI data harmonization H. Mirzaalian 1, L. Ning 1, P. Savadjiev 1, O. Pasternak 1, S. Bouix 1, O. Michailovich 2, M. Kubicki 1, C-F Westin 1, M.E.Shenton 1, Y. Rathi
The organization of the human cerebral cortex estimated by intrinsic functional connectivity
 1 The organization of the human cerebral cortex estimated by intrinsic functional connectivity Journal: Journal of Neurophysiology Author: B. T. Thomas Yeo, et al Link: https://www.ncbi.nlm.nih.gov/pubmed/21653723
1 The organization of the human cerebral cortex estimated by intrinsic functional connectivity Journal: Journal of Neurophysiology Author: B. T. Thomas Yeo, et al Link: https://www.ncbi.nlm.nih.gov/pubmed/21653723
Corpus Callosum Subdivision based on a Probabilistic Model of Inter-Hemispheric Connectivity
 Corpus Callosum Subdivision based on a Probabilistic Model of Inter-Hemispheric Connectivity Martin A. Styner 1,2, Ipek Oguz 1, Rachel Gimpel Smith 2, Carissa Cascio 2, and Matthieu Jomier 1 1 Dept. of
Corpus Callosum Subdivision based on a Probabilistic Model of Inter-Hemispheric Connectivity Martin A. Styner 1,2, Ipek Oguz 1, Rachel Gimpel Smith 2, Carissa Cascio 2, and Matthieu Jomier 1 1 Dept. of
HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING. Francesca Pizzorni Ferrarese
 FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING Francesca Pizzorni Ferrarese Pipeline overview WM and GM Segmentation Registration Data reconstruction Tractography
FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING Francesca Pizzorni Ferrarese Pipeline overview WM and GM Segmentation Registration Data reconstruction Tractography
Structural connectivity of the brain measured by diffusion tensor imaging [DTI]
![Structural connectivity of the brain measured by diffusion tensor imaging [DTI] Structural connectivity of the brain measured by diffusion tensor imaging [DTI]](/thumbs/74/71065400.jpg) Structural connectivity of the brain measured by diffusion tensor imaging [DTI] Lars T. Westlye CSHC / Centre for Advanced Study PSY4320 06.05.2012 l.t.westlye@psykologi.uio.no ~ 40 % of the brain is white
Structural connectivity of the brain measured by diffusion tensor imaging [DTI] Lars T. Westlye CSHC / Centre for Advanced Study PSY4320 06.05.2012 l.t.westlye@psykologi.uio.no ~ 40 % of the brain is white
NEURO M203 & BIOMED M263 WINTER 2014
 NEURO M203 & BIOMED M263 WINTER 2014 MRI Lab 2: Neuroimaging Connectivity Lab In today s lab we will work with sample diffusion imaging data and the group averaged fmri data collected during your scanning
NEURO M203 & BIOMED M263 WINTER 2014 MRI Lab 2: Neuroimaging Connectivity Lab In today s lab we will work with sample diffusion imaging data and the group averaged fmri data collected during your scanning
Incorporating Non-rigid Registration into Expectation Maximization Algorithm to Segment MR Images
 Incorporating Non-rigid Registration into Expectation Maximization Algorithm to Segment MR Images Kilian M Pohl 1, William M. Wells 2, Alexandre Guimond 3, Kiyoto Kasai 4, Martha E. Shenton 2,4, Ron Kikinis
Incorporating Non-rigid Registration into Expectation Maximization Algorithm to Segment MR Images Kilian M Pohl 1, William M. Wells 2, Alexandre Guimond 3, Kiyoto Kasai 4, Martha E. Shenton 2,4, Ron Kikinis
Diffusion Tensor Processing and Visualization
 NA-MIC National Alliance for Medical Image Computing http://na-mic.org Diffusion Tensor Processing and Visualization Guido Gerig University of Utah Martin Styner, UNC NAMIC: National Alliance for Medical
NA-MIC National Alliance for Medical Image Computing http://na-mic.org Diffusion Tensor Processing and Visualization Guido Gerig University of Utah Martin Styner, UNC NAMIC: National Alliance for Medical
A combined Surface And VOlumetric Registration (SAVOR) framework to study cortical biomarkers and volumetric imaging data
 A combined Surface And VOlumetric Registration (SAVOR) framework to study cortical biomarkers and volumetric imaging data Eli Gibson 1, Ali R. Khan 1, and Mirza Faisal Beg 1 School of Engineering Science,
A combined Surface And VOlumetric Registration (SAVOR) framework to study cortical biomarkers and volumetric imaging data Eli Gibson 1, Ali R. Khan 1, and Mirza Faisal Beg 1 School of Engineering Science,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy
 The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
Reconstruction of Fiber Trajectories via Population-Based Estimation of Local Orientations
 IDEA Reconstruction of Fiber Trajectories via Population-Based Estimation of Local Orientations Pew-Thian Yap, John H. Gilmore, Weili Lin, Dinggang Shen Email: ptyap@med.unc.edu 2011-09-21 Poster: P2-46-
IDEA Reconstruction of Fiber Trajectories via Population-Based Estimation of Local Orientations Pew-Thian Yap, John H. Gilmore, Weili Lin, Dinggang Shen Email: ptyap@med.unc.edu 2011-09-21 Poster: P2-46-
Fiber Selection from Diffusion Tensor Data based on Boolean Operators
 Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhofl, G. Greiner 2, M. Buchfelder 3, C. Nimsky4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhofl, G. Greiner 2, M. Buchfelder 3, C. Nimsky4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
THE identification and delineation of brain structures from
 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 26, NO. 4, APRIL 2007 479 Atlas Renormalization for Improved Brain MR Image Segmentation Across Scanner Platforms Xiao Han and Bruce Fischl* Abstract Atlas-based
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 26, NO. 4, APRIL 2007 479 Atlas Renormalization for Improved Brain MR Image Segmentation Across Scanner Platforms Xiao Han and Bruce Fischl* Abstract Atlas-based
An Intensity Consistent Approach to the Cross Sectional Analysis of Deformation Tensor Derived Maps of Brain Shape
 An Intensity Consistent Approach to the Cross Sectional Analysis of Deformation Tensor Derived Maps of Brain Shape C. Studholme, V. Cardenas, A. Maudsley, and M. Weiner U.C.S.F., Dept of Radiology, VAMC
An Intensity Consistent Approach to the Cross Sectional Analysis of Deformation Tensor Derived Maps of Brain Shape C. Studholme, V. Cardenas, A. Maudsley, and M. Weiner U.C.S.F., Dept of Radiology, VAMC
UNC 4D Infant Cortical Surface Atlases, from Neonate to 6 Years of Age
 UNC 4D Infant Cortical Surface Atlases, from Neonate to 6 Years of Age Version 1.0 UNC 4D infant cortical surface atlases from neonate to 6 years of age contain 11 time points, including 1, 3, 6, 9, 12,
UNC 4D Infant Cortical Surface Atlases, from Neonate to 6 Years of Age Version 1.0 UNC 4D infant cortical surface atlases from neonate to 6 years of age contain 11 time points, including 1, 3, 6, 9, 12,
Diffusion Tensor Imaging and Reading Development
 Diffusion Tensor Imaging and Reading Development Bob Dougherty Stanford Institute for Reading and Learning Reading and Anatomy Every brain is different... Not all brains optimized for highly proficient
Diffusion Tensor Imaging and Reading Development Bob Dougherty Stanford Institute for Reading and Learning Reading and Anatomy Every brain is different... Not all brains optimized for highly proficient
Identifying white-matter fiber bundles in DTI data using an automated proximity-based fiber-clustering method
 1 Identifying white-matter fiber bundles in DTI data using an automated proximity-based fiber-clustering method Song Zhang 1, Stephen Correia 2, and David H. Laidlaw 3 1 Department of Computer Science
1 Identifying white-matter fiber bundles in DTI data using an automated proximity-based fiber-clustering method Song Zhang 1, Stephen Correia 2, and David H. Laidlaw 3 1 Department of Computer Science
CHAPTER 2. Morphometry on rodent brains. A.E.H. Scheenstra J. Dijkstra L. van der Weerd
 CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
CHAPTER 2 Morphometry on rodent brains A.E.H. Scheenstra J. Dijkstra L. van der Weerd This chapter was adapted from: Volumetry and other quantitative measurements to assess the rodent brain, In vivo NMR
A Method for Registering Diffusion Weighted Magnetic Resonance Images
 A Method for Registering Diffusion Weighted Magnetic Resonance Images Xiaodong Tao and James V. Miller GE Research, Niskayuna, New York, USA Abstract. Diffusion weighted magnetic resonance (DWMR or DW)
A Method for Registering Diffusion Weighted Magnetic Resonance Images Xiaodong Tao and James V. Miller GE Research, Niskayuna, New York, USA Abstract. Diffusion weighted magnetic resonance (DWMR or DW)
NeuroImage. Tract-based morphometry for white matter group analysis. Lauren J. O'Donnell a,b,, Carl-Fredrik Westin b, Alexandra J.
 NeuroImage 45 (2009) 832 844 Contents lists available at ScienceDirect NeuroImage journal homepage: www.elsevier.com/locate/ynimg Tract-based morphometry for white matter group analysis Lauren J. O'Donnell
NeuroImage 45 (2009) 832 844 Contents lists available at ScienceDirect NeuroImage journal homepage: www.elsevier.com/locate/ynimg Tract-based morphometry for white matter group analysis Lauren J. O'Donnell
Automatic Registration-Based Segmentation for Neonatal Brains Using ANTs and Atropos
 Automatic Registration-Based Segmentation for Neonatal Brains Using ANTs and Atropos Jue Wu and Brian Avants Penn Image Computing and Science Lab, University of Pennsylvania, Philadelphia, USA Abstract.
Automatic Registration-Based Segmentation for Neonatal Brains Using ANTs and Atropos Jue Wu and Brian Avants Penn Image Computing and Science Lab, University of Pennsylvania, Philadelphia, USA Abstract.
NIH Public Access Author Manuscript Conf Proc IEEE Eng Med Biol Soc. Author manuscript; available in PMC 2009 December 11.
 NIH Public Access Author Manuscript Published in final edited form as: Conf Proc IEEE Eng Med Biol Soc. 2004 ; 2: 1521 1524. doi:10.1109/iembs.2004.1403466. An Elliptic PDE Approach for Shape Characterization
NIH Public Access Author Manuscript Published in final edited form as: Conf Proc IEEE Eng Med Biol Soc. 2004 ; 2: 1521 1524. doi:10.1109/iembs.2004.1403466. An Elliptic PDE Approach for Shape Characterization
Morphological Analysis of Brain Structures Using Spatial Normalization
 Morphological Analysis of Brain Structures Using Spatial Normalization C. Davatzikos 1, M. Vaillant 1, S. Resnick 2, J.L. Prince 3;1, S. Letovsky 1, and R.N. Bryan 1 1 Department of Radiology, Johns Hopkins
Morphological Analysis of Brain Structures Using Spatial Normalization C. Davatzikos 1, M. Vaillant 1, S. Resnick 2, J.L. Prince 3;1, S. Letovsky 1, and R.N. Bryan 1 1 Department of Radiology, Johns Hopkins
Spline-Based Probabilistic Model for Anatomical Landmark Detection
 Spline-Based Probabilistic Model for Anatomical Landmark Detection Camille Izard 1,2, Bruno Jedynak 1,2, and Craig E.L. Stark 3 1 Laboratoire Paul Painlevé, Université des Sciences et Technologies de Lille,
Spline-Based Probabilistic Model for Anatomical Landmark Detection Camille Izard 1,2, Bruno Jedynak 1,2, and Craig E.L. Stark 3 1 Laboratoire Paul Painlevé, Université des Sciences et Technologies de Lille,
Diffusion Imaging Models 1: from DTI to HARDI models
 Diffusion Imaging Models 1: from DTI to HARDI models Flavio Dell Acqua, PhD. www.natbrainlab.com flavio.dellacqua@kcl.ac.uk @flaviodellacqua Diffusion Tensor Imaging (DTI) z λ 1 λ 2 The profile of the
Diffusion Imaging Models 1: from DTI to HARDI models Flavio Dell Acqua, PhD. www.natbrainlab.com flavio.dellacqua@kcl.ac.uk @flaviodellacqua Diffusion Tensor Imaging (DTI) z λ 1 λ 2 The profile of the
Atelier 2 : Calcul Haute Performance et Sciences du Vivant Forum er juillet, Paris, France
 From Diffusion MR Image Analysis to Whole Brain Connectivity Simulation Jean-Philippe Thiran EPFL Lausanne, Switzerland EPFL - Lausanne HPC in life sciences at EPFL The Blue Brain project: create a biologically
From Diffusion MR Image Analysis to Whole Brain Connectivity Simulation Jean-Philippe Thiran EPFL Lausanne, Switzerland EPFL - Lausanne HPC in life sciences at EPFL The Blue Brain project: create a biologically
Cocozza S., et al. : ALTERATIONS OF FUNCTIONAL CONNECTIVITY OF THE MOTOR CORTEX IN FABRY'S DISEASE: AN RS-FMRI STUDY
 ALTERATIONS OF FUNCTIONAL CONNECTIVITY OF THE MOTOR CORTEX IN FABRY'S DISEASE: AN RS-FMRI STUDY SUPPLEMENTARY MATERIALS Sirio Cocozza, MD 1*, Antonio Pisani, MD, PhD 2, Gaia Olivo, MD 1, Francesco Saccà,
ALTERATIONS OF FUNCTIONAL CONNECTIVITY OF THE MOTOR CORTEX IN FABRY'S DISEASE: AN RS-FMRI STUDY SUPPLEMENTARY MATERIALS Sirio Cocozza, MD 1*, Antonio Pisani, MD, PhD 2, Gaia Olivo, MD 1, Francesco Saccà,
Diffusion Tractography of the Corticospinal Tract with Multi-fiber Orientation Filtering
 Diffusion Tractography of the Corticospinal Tract with Multi-fiber Orientation Filtering Ryan P. Cabeen, David H. Laidlaw Computer Science Department Brown University Providence, RI, USA {cabeen,dhl}@cs.brown.edu
Diffusion Tractography of the Corticospinal Tract with Multi-fiber Orientation Filtering Ryan P. Cabeen, David H. Laidlaw Computer Science Department Brown University Providence, RI, USA {cabeen,dhl}@cs.brown.edu
BrainSpace V2.1: Geometric mapping and diffusion-based software for cross-subject multimodality brain imaging informatics
 Brief Introduction to BrainSpace V2.1 BrainSpace V2.1: Geometric mapping and diffusion-based software for cross-subject multimodality brain imaging informatics Graphics and Imaging Laboratory Wayne State
Brief Introduction to BrainSpace V2.1 BrainSpace V2.1: Geometric mapping and diffusion-based software for cross-subject multimodality brain imaging informatics Graphics and Imaging Laboratory Wayne State
NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2014 August 15.
 NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2014 August 15. Published in final edited form as: Med Image Comput Comput Assist Interv.
NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2014 August 15. Published in final edited form as: Med Image Comput Comput Assist Interv.
Stereoscopic Atlas of Intrinsic Brain Networks (SAIBN)
 Stereoscopic Atlas of Intrinsic Brain Networks (SAIBN) Gonzalo M. Rojas, Marcelo Gálvez, Daniel Margulies, Cameron Craddock, Xavier Castellanos, Michael Milham 1.- Introduction Stereoscopic Atlas of Intrinsic
Stereoscopic Atlas of Intrinsic Brain Networks (SAIBN) Gonzalo M. Rojas, Marcelo Gálvez, Daniel Margulies, Cameron Craddock, Xavier Castellanos, Michael Milham 1.- Introduction Stereoscopic Atlas of Intrinsic
Supplementary methods
 Supplementary methods This section provides additional technical details on the sample, the applied imaging and analysis steps and methods. Structural imaging Trained radiographers placed all participants
Supplementary methods This section provides additional technical details on the sample, the applied imaging and analysis steps and methods. Structural imaging Trained radiographers placed all participants
Feasibility and Advantages of Diffusion Weighted Imaging Atlas Construction in Q-Space
 Feasibility and Advantages of Diffusion Weighted Imaging Atlas Construction in Q-Space Thijs Dhollander 1,2, Jelle Veraart 3,WimVanHecke 1,4,5, Frederik Maes 1,2, Stefan Sunaert 1,4,JanSijbers 3, and Paul
Feasibility and Advantages of Diffusion Weighted Imaging Atlas Construction in Q-Space Thijs Dhollander 1,2, Jelle Veraart 3,WimVanHecke 1,4,5, Frederik Maes 1,2, Stefan Sunaert 1,4,JanSijbers 3, and Paul
Diffusion-MRI processing for group analysis
 Diffusion-MRI processing for group analysis Felix Renard IRMaGe: Inserm US 17 / CNRS UMS 3552 University Hospital of Grenoble - France 25/09/2015 felixrenard@gmail.com 1 Diffusion-MRI processing for group
Diffusion-MRI processing for group analysis Felix Renard IRMaGe: Inserm US 17 / CNRS UMS 3552 University Hospital of Grenoble - France 25/09/2015 felixrenard@gmail.com 1 Diffusion-MRI processing for group
Hierarchical Fiber Clustering Based on Multi-scale Neuroanatomical Features
 Hierarchical Fiber Clustering Based on Multi-scale Neuroanatomical Features Qian Wang 1, 2, Pew-Thian Yap 2, Hongjun Jia 2, Guorong Wu 2, and Dinggang Shen 2 1 Department of Computer Science, University
Hierarchical Fiber Clustering Based on Multi-scale Neuroanatomical Features Qian Wang 1, 2, Pew-Thian Yap 2, Hongjun Jia 2, Guorong Wu 2, and Dinggang Shen 2 1 Department of Computer Science, University
Diffusion MRI. Introduction and Modern Methods. John Plass. Department Of Psychology
 Diffusion MRI Introduction and Modern Methods John Plass Department Of Psychology Diffusion MRI Introduction and Modern Methods John Plass Department Of Psychology Overview I. Why use diffusion MRI? II.
Diffusion MRI Introduction and Modern Methods John Plass Department Of Psychology Diffusion MRI Introduction and Modern Methods John Plass Department Of Psychology Overview I. Why use diffusion MRI? II.
Kernel Regression Estimation of Fiber Orientation Mixtures in Di usion MRI
 Ryan P. Cabeen, Mark E. Bastin, David H. Laidlaw, Kernel regression estimation of fiber orientation mixtures in diffusion MRI, NeuroImage, Volume 127, 15 February 2016, Pages 158-172, ISSN 1053-8119, http://dx.doi.org/10.1016/j.neuroimage.2015.11.061
Ryan P. Cabeen, Mark E. Bastin, David H. Laidlaw, Kernel regression estimation of fiber orientation mixtures in diffusion MRI, NeuroImage, Volume 127, 15 February 2016, Pages 158-172, ISSN 1053-8119, http://dx.doi.org/10.1016/j.neuroimage.2015.11.061
Structural MRI analysis
 Structural MRI analysis volumetry and voxel-based morphometry cortical thickness measurements structural covariance network mapping Boris Bernhardt, PhD Department of Social Neuroscience, MPI-CBS bernhardt@cbs.mpg.de
Structural MRI analysis volumetry and voxel-based morphometry cortical thickness measurements structural covariance network mapping Boris Bernhardt, PhD Department of Social Neuroscience, MPI-CBS bernhardt@cbs.mpg.de
NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2009 December 4.
 NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2009 December 4. Published in final edited form as: Med Image Comput Comput Assist Interv.
NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2009 December 4. Published in final edited form as: Med Image Comput Comput Assist Interv.
Estimation of Extracellular Volume from Regularized Multi-shell Diffusion MRI
 Estimation of Extracellular Volume from Regularized Multi-shell Diffusion MRI The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters
Estimation of Extracellular Volume from Regularized Multi-shell Diffusion MRI The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters
Automatic Quantification of DTI Parameters along Fiber Bundles
 Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein 1, Simon Hermann 1, Olaf Konrad 1, Horst K. Hahn 1, and Heinz-Otto Peitgen 1 1 MeVis Research, 28359 Bremen Email: klein@mevis.de
Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein 1, Simon Hermann 1, Olaf Konrad 1, Horst K. Hahn 1, and Heinz-Otto Peitgen 1 1 MeVis Research, 28359 Bremen Email: klein@mevis.de
Supplementary Online Content
 Supplementary Online Content Sun H, Lui S, Yao L, et al. Two patterns of white matter abnormalities in medication-naive patients with first-episode schizophrenia revealed by diffusion tensor imaging and
Supplementary Online Content Sun H, Lui S, Yao L, et al. Two patterns of white matter abnormalities in medication-naive patients with first-episode schizophrenia revealed by diffusion tensor imaging and
IS MRI Based Structure a Mediator for Lead s Effect on Cognitive Function?
 IS MRI Based Structure a Mediator for Lead s Effect on Cognitive Function? Brian Caffo, Sining Chen, Brian Schwartz Department of Biostatistics and Environmental Health Sciences Johns Hopkins University
IS MRI Based Structure a Mediator for Lead s Effect on Cognitive Function? Brian Caffo, Sining Chen, Brian Schwartz Department of Biostatistics and Environmental Health Sciences Johns Hopkins University
Direct Registration of White Matter Tractographies with Application to Atlas Construction
 Direct Registration of White Matter Tractographies with Application to Atlas Construction Arnaldo Mayer, Hayit Greenspan Medical image processing lab, Tel-Aviv University, Ramat-Aviv, Israel arnakdom@post.tau.ac.il,
Direct Registration of White Matter Tractographies with Application to Atlas Construction Arnaldo Mayer, Hayit Greenspan Medical image processing lab, Tel-Aviv University, Ramat-Aviv, Israel arnakdom@post.tau.ac.il,
Group Statistics of DTI Fiber Bundles Using Spatial Functions of Tensor Measures
 Group Statistics of DTI Fiber Bundles Using Spatial Functions of Tensor Measures Casey B. Goodlett 1,2, P. Thomas Fletcher 1,2, John H. Gilmore 3, and Guido Gerig 1,2 1 School of Computing, University
Group Statistics of DTI Fiber Bundles Using Spatial Functions of Tensor Measures Casey B. Goodlett 1,2, P. Thomas Fletcher 1,2, John H. Gilmore 3, and Guido Gerig 1,2 1 School of Computing, University
Reproducibility of Whole-brain Structural Connectivity Networks
 Reproducibility of Whole-brain Structural Connectivity Networks Christopher Parker Thesis for Masters of Research in Medical and Biomedical Imaging Supervised by Prof. Sebastien Ourselin and Dr Jonathan
Reproducibility of Whole-brain Structural Connectivity Networks Christopher Parker Thesis for Masters of Research in Medical and Biomedical Imaging Supervised by Prof. Sebastien Ourselin and Dr Jonathan
Development of High Angular Resolution Diffusion Imaging Analysis Paradigms for the Investigation of Neuropathology
 University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 1-1-2012 Development of High Angular Resolution Diffusion Imaging Analysis Paradigms for the Investigation of Neuropathology
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 1-1-2012 Development of High Angular Resolution Diffusion Imaging Analysis Paradigms for the Investigation of Neuropathology
Automatic Generation of Training Data for Brain Tissue Classification from MRI
 MICCAI-2002 1 Automatic Generation of Training Data for Brain Tissue Classification from MRI Chris A. Cocosco, Alex P. Zijdenbos, and Alan C. Evans McConnell Brain Imaging Centre, Montreal Neurological
MICCAI-2002 1 Automatic Generation of Training Data for Brain Tissue Classification from MRI Chris A. Cocosco, Alex P. Zijdenbos, and Alan C. Evans McConnell Brain Imaging Centre, Montreal Neurological
Dual Tensor Atlas Generation Based on a Cohort of Coregistered non-hardi Datasets
 Dual Tensor Atlas Generation Based on a Cohort of Coregistered non-hardi Datasets Matthan Caan 1,2, Caroline Sage 3, Maaike van der Graaf 1, Cornelis Grimbergen 1, Stefan Sunaert 3, Lucas van Vliet 2,
Dual Tensor Atlas Generation Based on a Cohort of Coregistered non-hardi Datasets Matthan Caan 1,2, Caroline Sage 3, Maaike van der Graaf 1, Cornelis Grimbergen 1, Stefan Sunaert 3, Lucas van Vliet 2,
Multiple Cortical Surface Correspondence Using Pairwise Shape Similarity
 Multiple Cortical Surface Correspondence Using Pairwise Shape Similarity Pahal Dalal 1, Feng Shi 2, Dinggang Shen 2, and Song Wang 1 1 Department of Computer Science and Engineering, University of South
Multiple Cortical Surface Correspondence Using Pairwise Shape Similarity Pahal Dalal 1, Feng Shi 2, Dinggang Shen 2, and Song Wang 1 1 Department of Computer Science and Engineering, University of South
Fmri Spatial Processing
 Educational Course: Fmri Spatial Processing Ray Razlighi Jun. 8, 2014 Spatial Processing Spatial Re-alignment Geometric distortion correction Spatial Normalization Smoothing Why, When, How, Which Why is
Educational Course: Fmri Spatial Processing Ray Razlighi Jun. 8, 2014 Spatial Processing Spatial Re-alignment Geometric distortion correction Spatial Normalization Smoothing Why, When, How, Which Why is
Automatic Quantification of DTI Parameters along Fiber Bundles
 Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein, Simon Hermann, Olaf Konrad, Horst K. Hahn, Heinz-Otto Peitgen MeVis Research, 28359 Bremen Email: klein@mevis.de Abstract. We introduce
Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein, Simon Hermann, Olaf Konrad, Horst K. Hahn, Heinz-Otto Peitgen MeVis Research, 28359 Bremen Email: klein@mevis.de Abstract. We introduce
Automatic Optimization of Segmentation Algorithms Through Simultaneous Truth and Performance Level Estimation (STAPLE)
 Automatic Optimization of Segmentation Algorithms Through Simultaneous Truth and Performance Level Estimation (STAPLE) Mahnaz Maddah, Kelly H. Zou, William M. Wells, Ron Kikinis, and Simon K. Warfield
Automatic Optimization of Segmentation Algorithms Through Simultaneous Truth and Performance Level Estimation (STAPLE) Mahnaz Maddah, Kelly H. Zou, William M. Wells, Ron Kikinis, and Simon K. Warfield
Detecting Changes In Non-Isotropic Images
 Detecting Changes In Non-Isotropic Images K.J. Worsley 1, M. Andermann 1, T. Koulis 1, D. MacDonald, 2 and A.C. Evans 2 August 4, 1999 1 Department of Mathematics and Statistics, 2 Montreal Neurological
Detecting Changes In Non-Isotropic Images K.J. Worsley 1, M. Andermann 1, T. Koulis 1, D. MacDonald, 2 and A.C. Evans 2 August 4, 1999 1 Department of Mathematics and Statistics, 2 Montreal Neurological
Performance Issues in Shape Classification
 Performance Issues in Shape Classification Samson J. Timoner 1, Pollina Golland 1, Ron Kikinis 2, Martha E. Shenton 3, W. Eric L. Grimson 1, and William M. Wells III 1,2 1 MIT AI Laboratory, Cambridge
Performance Issues in Shape Classification Samson J. Timoner 1, Pollina Golland 1, Ron Kikinis 2, Martha E. Shenton 3, W. Eric L. Grimson 1, and William M. Wells III 1,2 1 MIT AI Laboratory, Cambridge
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge
 Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
Automatic segmentation of the cortical grey and white matter in MRI using a Region Growing approach based on anatomical knowledge Christian Wasserthal 1, Karin Engel 1, Karsten Rink 1 und André Brechmann
The Allen Human Brain Atlas offers three types of searches to allow a user to: (1) obtain gene expression data for specific genes (or probes) of
 Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
DIFFUSION TENSOR IMAGING ANALYSIS. Using Analyze
 DIFFUSION TENSOR IMAGING ANALYSIS Using Analyze 2 Table of Contents 1. Introduction page 3 2. Loading DTI Data page 4 3. Computing DTI Maps page 5 4. Defining ROIs for Fiber Tracking page 6 5. Visualizing
DIFFUSION TENSOR IMAGING ANALYSIS Using Analyze 2 Table of Contents 1. Introduction page 3 2. Loading DTI Data page 4 3. Computing DTI Maps page 5 4. Defining ROIs for Fiber Tracking page 6 5. Visualizing
Title: Wired for function: Anatomical connectivity patterns predict face-selectivity in the
 Title: Wired for function: Anatomical connectivity patterns predict face-selectivity in the fusiform gyrus Authors: Zeynep M. Saygin *, David E. Osher *, Kami Koldewyn, Gretchen Reynolds, John D.E. Gabrieli,
Title: Wired for function: Anatomical connectivity patterns predict face-selectivity in the fusiform gyrus Authors: Zeynep M. Saygin *, David E. Osher *, Kami Koldewyn, Gretchen Reynolds, John D.E. Gabrieli,
Diffusion model fitting and tractography: A primer
 Diffusion model fitting and tractography: A primer Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 03/18/10 Why n how Diffusion model fitting and tractography 0/18 Why
Diffusion model fitting and tractography: A primer Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 03/18/10 Why n how Diffusion model fitting and tractography 0/18 Why
MITK-DI. A new Diffusion Imaging Component for MITK. Klaus Fritzsche, Hans-Peter Meinzer
 MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
MITK Global Tractography
 MITK Global Tractography Peter F. Neher a, Bram Stieltjes b, Marco Reisert c, Ignaz Reicht a, Hans-Peter Meinzer a, Klaus H. Fritzsche a,b a German Cancer Research Center, Medical and Biological Informatics,
MITK Global Tractography Peter F. Neher a, Bram Stieltjes b, Marco Reisert c, Ignaz Reicht a, Hans-Peter Meinzer a, Klaus H. Fritzsche a,b a German Cancer Research Center, Medical and Biological Informatics,
THE brain is a massively interconnected organ. Individual
 IEEE TRANSACTIONS ON VISUALIZATION AND COMPUTER GRAPHICS 1 Exploring Connectivity of the Brain s White Matter with Dynamic Queries Anthony Sherbondy, David Akers, Rachel Mackenzie, Robert Dougherty, and
IEEE TRANSACTIONS ON VISUALIZATION AND COMPUTER GRAPHICS 1 Exploring Connectivity of the Brain s White Matter with Dynamic Queries Anthony Sherbondy, David Akers, Rachel Mackenzie, Robert Dougherty, and
Image-Driven Population Analysis Through Mixture Modeling Mert R. Sabuncu*, Member, IEEE, Serdar K. Balci, Martha E. Shenton, and Polina Golland
 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 28, NO. 9, SEPTEMBER 2009 1473 Image-Driven Population Analysis Through Mixture Modeling Mert R. Sabuncu*, Member, IEEE, Serdar K. Balci, Martha E. Shenton, and
IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 28, NO. 9, SEPTEMBER 2009 1473 Image-Driven Population Analysis Through Mixture Modeling Mert R. Sabuncu*, Member, IEEE, Serdar K. Balci, Martha E. Shenton, and
Tech Report Computer Science Department, UNC Chapel Hill
 Tech Report Computer Science Department, UNC Chapel Hill Brain morphometry by distance measurement in a non-euclidean, curvilinear space Martin Styner 1, Thomas Coradi 2 and Guido Gerig 1 1 Dept. of Computer
Tech Report Computer Science Department, UNC Chapel Hill Brain morphometry by distance measurement in a non-euclidean, curvilinear space Martin Styner 1, Thomas Coradi 2 and Guido Gerig 1 1 Dept. of Computer
HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
Description of files included in the IIT Human Brain Atlas (v.4.1)
 Description of files included in the IIT Human Brain Atlas (v.4.1) Summary: The IIT Human Brain Atlas contains anatomical, DTI, and HARDI templates, probabilistic gray matter labels, probabilistic connectivity-based
Description of files included in the IIT Human Brain Atlas (v.4.1) Summary: The IIT Human Brain Atlas contains anatomical, DTI, and HARDI templates, probabilistic gray matter labels, probabilistic connectivity-based
What is a network? Network Analysis
 What is a network? Network Analysis Valerie Cardenas Nicolson Associate Adjunct Professor Department of Radiology and Biomedical Imaging Complex weblike structures Cell is network of chemicals connected
What is a network? Network Analysis Valerie Cardenas Nicolson Associate Adjunct Professor Department of Radiology and Biomedical Imaging Complex weblike structures Cell is network of chemicals connected
A Novel Contrast for DTI Visualization for Thalamus Delineation
 A Novel Contrast for DTI Visualization for Thalamus Delineation Xian Fan a, Meredith Thompson a,b, John A. Bogovic a, Pierre-Louis Bazin c, Jerry L. Prince a,c a Johns Hopkins University, Baltimore, MD,
A Novel Contrast for DTI Visualization for Thalamus Delineation Xian Fan a, Meredith Thompson a,b, John A. Bogovic a, Pierre-Louis Bazin c, Jerry L. Prince a,c a Johns Hopkins University, Baltimore, MD,
BrainPrint: Identifying Subjects by Their Brain
 BrainPrint: Identifying Subjects by Their Brain The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Wachinger,
BrainPrint: Identifying Subjects by Their Brain The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Wachinger,
Automatic corpus callosum segmentation using a deformable active Fourier contour model
 Automatic corpus callosum segmentation using a deformable active Fourier contour model Clement Vachet a, Benjamin Yvernault a, Kshamta Bhatt a, Rachel G. Smith a,b, Guido Gerig c, Heather Cody Hazlett
Automatic corpus callosum segmentation using a deformable active Fourier contour model Clement Vachet a, Benjamin Yvernault a, Kshamta Bhatt a, Rachel G. Smith a,b, Guido Gerig c, Heather Cody Hazlett
TRACULA: Troubleshooting, visualization, and group analysis
 TRACULA: Troubleshooting, visualization, and group analysis Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 18/11/13 TRACULA: troubleshooting, visualization, group analysis
TRACULA: Troubleshooting, visualization, and group analysis Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 18/11/13 TRACULA: troubleshooting, visualization, group analysis
Preprocessing II: Between Subjects John Ashburner
 Preprocessing II: Between Subjects John Ashburner Pre-processing Overview Statistics or whatever fmri time-series Anatomical MRI Template Smoothed Estimate Spatial Norm Motion Correct Smooth Coregister
Preprocessing II: Between Subjects John Ashburner Pre-processing Overview Statistics or whatever fmri time-series Anatomical MRI Template Smoothed Estimate Spatial Norm Motion Correct Smooth Coregister
Introduction to Neuroimaging Janaina Mourao-Miranda
 Introduction to Neuroimaging Janaina Mourao-Miranda Neuroimaging techniques have changed the way neuroscientists address questions about functional anatomy, especially in relation to behavior and clinical
Introduction to Neuroimaging Janaina Mourao-Miranda Neuroimaging techniques have changed the way neuroscientists address questions about functional anatomy, especially in relation to behavior and clinical
White Matter Tractography Using Diffusion Tensor Deflection
 Human Brain Mapping 18:306 321(2003) White Matter Tractography Using Diffusion Tensor Deflection Mariana Lazar, 1 David M. Weinstein, 2 Jay S. Tsuruda, 3 Khader M. Hasan, 4,8 Konstantinos Arfanakis, 4
Human Brain Mapping 18:306 321(2003) White Matter Tractography Using Diffusion Tensor Deflection Mariana Lazar, 1 David M. Weinstein, 2 Jay S. Tsuruda, 3 Khader M. Hasan, 4,8 Konstantinos Arfanakis, 4
Bias in Resampling-Based Thresholding of Statistical Maps in fmri
 Bias in Resampling-Based Thresholding of Statistical Maps in fmri Ola Friman and Carl-Fredrik Westin Laboratory of Mathematics in Imaging, Department of Radiology Brigham and Women s Hospital, Harvard
Bias in Resampling-Based Thresholding of Statistical Maps in fmri Ola Friman and Carl-Fredrik Westin Laboratory of Mathematics in Imaging, Department of Radiology Brigham and Women s Hospital, Harvard
MRI Tissue Classification with Neighborhood Statistics: A Nonparametric, Entropy-Minimizing Approach
 MRI Tissue Classification with Neighborhood Statistics: A Nonparametric, Entropy-Minimizing Approach Tolga Tasdizen 1, Suyash P. Awate 1, Ross T. Whitaker 1, and Norman L. Foster 2 1 School of Computing,
MRI Tissue Classification with Neighborhood Statistics: A Nonparametric, Entropy-Minimizing Approach Tolga Tasdizen 1, Suyash P. Awate 1, Ross T. Whitaker 1, and Norman L. Foster 2 1 School of Computing,
MR Diffusion-Based Inference of a Fiber Bundle Model from a Population of Subjects
 MR Diffusion-Based Inference of a Fiber Bundle Model from a Population of Subjects V. El Kouby 1,3, Y. Cointepas 1,3, C. Poupon 1,3, D. Rivière 1,3, N. Golestani 1,2, J.-B. Poline 4, D. Le Bihan 1,3, and
MR Diffusion-Based Inference of a Fiber Bundle Model from a Population of Subjects V. El Kouby 1,3, Y. Cointepas 1,3, C. Poupon 1,3, D. Rivière 1,3, N. Golestani 1,2, J.-B. Poline 4, D. Le Bihan 1,3, and
