Guide to Reviewing and Approving Custom Designs
|
|
|
- Corey Hoover
- 6 years ago
- Views:
Transcription
1 Guide to Reviewing and Approving Custom Designs SeqCap EZ Designs, v4.1 Overview This document describes how to review and approve the proposed custom SeqCap EZ and SeqCap EZ Prime designs based on the customer-provided genomic regions. The purpose of reviewing designs is to verify that all desired regions are covered adequately and that the design meets your needs. This is part of our process, and must be completed by the customer prior to manufacturing of the design. If the regions vital to your experiment are not adequately covered, you may need to modify the parameters of your design. Before You Begin Roche provides design file(s) in three or four formats: BED (.bed) files: Viewable in the Roche SignalMap software or other genome browsers, such as in the Internet-based UCSC Genome browser, the Broad Institute's Integrative Genomics Viewer or ENSEMBL. For information about SignalMap and to download software, go to Coverage Summary Files (.txt) files: Viewable in any text editor. (Conditional) bedgraph (.bedgraph) file: Viewable in several genome browsers, such as the Internetbased UCSC Genome browser, the Broad Institute s Integrative Genomics Viewer, or ENSEMBL. BedGraph files are only included if targeted replication has been requested. This file type is not properly rendered in SignalMap. Design Summary (.pdf) file: This file contains relevant information on your design and can be opened in any PDF viewer. For Research Use Only. Not for use in diagnostic procedures.
2 Keep the following in mind while reviewing your design: For the purposes of visualizing coverage, use the three BED files provided with the design: primary_targets.bed, capture_targets.bed, and predicted_no_coverage_regions.bed. These files are described in detail in the appendix. If targeted replication was requested for the design, an additional file, a bedgraph, was included to visualize relative probe abundance. When reviewing the design files, focus on gaps in the capture_targets track that do not provide coverage for the primary_targets track. These gaps represent portions of your target region not covered by the design. Review the region-by-region coverage file for detailed coverage for each region. If two or more of your target regions overlap, Roche merges them into one region. In addition, for a SeqCap EZ or SeqCap EZ Prime design, regions that are less than 100 bp are padded to a minimum size of 100 bp. Regions not covered by the design are usually repetitive regions which, if included, cause capture of other homologous regions in the genome and decrease capture efficiency. Therefore, most Roche SeqCap experiments benefit from excluding these regions in the design. Check the region-by-region coverage file for information on regions not covered due to repetitive regions. The stringency filter Roche uses by default will not include low copy repeats in the design. If these regions are necessary to your research and you are working with the custom design group, indicate this requirement in the to Roche (refer to Step 3. Approve or Request Changes to the Design on page 9). Roche will use less stringent criteria in generating the design. Be aware that the less stringent design may provide more genomic coverage but at the cost of a decrease in capture efficiency and you may see more off-target reads when the captured DNA is sequenced. Please note that this parameter selection is not available in NimbleDesign. If you have questions, contact Roche Technical Support (refer to page 13 for contact details.) 2 Guide to Reviewing and Approving Custom Designs: SeqCap EZ Designs, v4.1
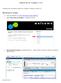
3 Review and Approve Step 1. Review the Design Summary File and Coverage Files The design summary files and coverage files describe the properties of your SeqCap custom design. 1. Open the Design Summary File (PDF), and check that all information is correct for the design specification requested. For example: Figure 1: Example Design Summary File. 2. Open the coverage_summary.txt file using a text editor, such as WordPad, Notepad, or Microsoft Excel. 3. Review each field to ensure the design meets the specifications for your Sequence Capture project. 4. Refer to the appendix to become familiar with the definitions of each field. Guide to Reviewing and Approving Custom Designs: SeqCap EZ Designs v4.1 3
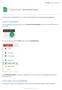
4 Figure 2: Example Coverage Summary File Displayed in Excel. 5. Open the coverage.txt file. It is recommended that you view this file with a spreadsheet program, such as Microsoft Excel. Figure 3: Example Region-By-Region Coverage File Displayed in Excel. 6. This file displays region-by-region coverage information, with some details on why regions might not have full coverage. Review this file thoroughly to ensure the design meets the specifications for your Sequence Capture project. This file may be sorted by percent coverage to rapidly identify regions with little or no coverage in the design. See the Appendix for a detailed description of each column in the file. If two or more of your target regions overlap, Roche merges them into one region. In addition, for a SeqCap EZ or SeqCap EZ Prime design, regions that are less than 100 bp are padded to a minimum size of 100 bp. 4 Guide to Reviewing and Approving Custom Designs: SeqCap EZ Designs, v4.1
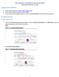
5 Step 2. Visual Review of the Design Refer to the appropriate instructions for the software you are using to review the design files. Using SignalMap Software to Review the Design 1. Save the bed files provided via (if working with a designer) or the Review tab in NimbleDesign to your computer or a network drive. 2. Open SignalMap software. 3. Select File -> New to open the primary_targets.bed file. 4. Select File -> Import to import the capture_targets.bed, and, if desired, the predicted_uncovered_targets.bed files into the same panel. Figure 4 shows example primary and capture target files displayed in the SignalMap window. Figure 4: Example Design Files Displayed in the SignalMap Window. Review the design using the SignalMap software s functions, such as the following: Zoom: Click the magnifier button on the toolbar, and then click and drag to draw a bounding box around the region to magnify. Alternatively, hold down Ctrl and press + to zoom in or - to zoom out. Arrange chromosomes: View data tracks in many different ways by chromosome. Use the pane selector field below the toolbar to change the view to either all chromosomes (All Tracks) or a selected chromosome (e.g. chr2). Go to specific genomic location: Select View -> Go to position, select the chromosome, and enter the start and end positions. Refer to the SignalMap User s Guide available at for additional details about the software s capabilities. Guide to Reviewing and Approving Custom Designs: SeqCap EZ Designs v4.1 5
6 Using UCSC Genome Browser to Review the Design The images and results used in this document reflect the UCSC Genome Browser interface as of December Save the primary_targets.bed, capture_targets.bed, and predicted_uncovered_targets.bed files provided via to your computer or a network drive. If a bedgraph was provided, save that as well. 2. Open the UCSC Genome browser home page: 3. In the left pane of the UCSC Genome browser home page, click Genomes. 4. In the Genome Browser Gateway page, choose the appropriate options in the genome and assembly list boxes for your design. This example uses Human for the build and Feb (GRCh37/hg19) as the assembly. 5. Click the add custom tracks button. If the Gateway page displays the manage custom tracks button, you may need to delete some of the existing tracks before viewing new data. 6. In the Add Custom Tracks page: a. Click the Choose file button to select and upload the BED file. b. Click the Submit button. 7. In the Manage Custom Tracks page: The Manage Custom Tracks page provides a link to name and describe your tracks. If you are loading multiple tracks, this will be necessary to help visualize the data. a. Change the configuration of existing tracks. Figure 5 shows an example of changing a track configuration. Figure 5: Example of Changing the Track Configuration in the UCSC Genome Browser. 6 Guide to Reviewing and Approving Custom Designs: SeqCap EZ Designs, v4.1
7 b. Add additional BED files as in Step 6, above. c. Click the go button to view the custom track(s). Figure 6 shows an example design displayed in the UCSC Genome browser. Figure 6: Example Design File Displayed in the UCSC Genome Browser. 8. Review the design using the UCSC Genome browser s functions, such as the following: Zoom: Click the zoom in and zoom out buttons to zoom in or out on the center of the annotation tracks window by 1.5-, 3-, or 10-fold. Scroll: Click the move buttons to scroll to the left or right. Display a different position in the genome: Enter the coordinates in the position/search text box and click the jump button. View base composition: Click the base button to view the base composition of the sequence underlying the current annotation track display. Useful UCSC-provided tracks to load include mapability and RepeatMasker tracks. These can be used to diagnose regions uncovered due to repeats. 9. Click the Help link for additional details about the browser s capabilities. 10. If your design included per-region probe copy number adjustment (aka targeted replication ), you will have a bedgraph file in your deliverables. You may visualize this data in UCSC s genome browser, as follows: Please note, SignalMap treats bedgraph files as a BED file and does not currently plot the 4 th column. a. Load the bedgraph file as you would any other custom track (see above steps 4-6). Guide to Reviewing and Approving Custom Designs: SeqCap EZ Designs v4.1 7
8 b. Rename the track to something unique, or it may be replaced by any subsequent track additions (see above step 7). c. Click the go button to view custom track(s). Figure 7 shows an example bedgraph track displayed in the UCSC Genome browser. Note the bar height corresponds to the actual relative replicate value for the region. Figure 7: Example bedgraph File Displayed in the UCSC Genome Browser. 8 Guide to Reviewing and Approving Custom Designs: SeqCap EZ Designs, v4.1
9 Step 3. Approve or Request Changes to the Design For approving designs generated with NimbleDesign, please see the NimbleDesign User's Guide, available at For tracking purposes, reply to the that included the deliverables with the proposed design. In the reply, indicate you approve the design or specify any changes needed for approval. Roche Designers cannot finalize the design and send to production to manufacture the SeqCap probes until we receive your written approval. Guide to Reviewing and Approving Custom Designs: SeqCap EZ Designs v4.1 9
10 Appendix: Included files capture_targets.bed Overlapping probe coverage regions where each base is covered by at least one probe. This is a three column, tab-delimited coordinate file, with no header, in BED format ( suitable for loading into various genome browsers. primary_targets.bed Requested regions, padded to a minimal length and with overlapping ranges merged. This is a three column, tab-delimited coordinate file, with no header, in BED format ( suitable for loading into various genome browsers. Only regions with at least one selected capture probe appear in this file. predicted_no_coverage_regions.bed.bedgraph All positions from the primary_targets.bed that are not within 100 base pairs of a capture probe. This is a three column, tab-delimited coordinate file, with no header, in BED format ( suitable for loading into various genome browsers. All positions from the capture_targets.bed have associated values indicating the relative number of probes. This is a four column, tab-delimited coordinate file, with a header indicating track type, in BEDGRAPH format ( suitable for loading into various genome browsers where it will be rendered as a bar graph. coverage_summary.txt The global coverage properties of your SeqCap EZ Choice design. This file is tab-delimited text and can be opened in Excel or a text editor. A description of the fields included in this file can be found below. Genome build: Genome and build targeted by the design (e.g. GRCh37/hg19). Number of regions: Number of regions after padding and consolidation. Length of regions: Sum total of all region sizes (in basepairs) after padding and consolidation. Probe_Coverage column: Direct coverage; for the padded and consolidated regions, these values represent bases, covered or uncovered, directly by a capture probe. 10 Guide to Reviewing and Approving Custom Designs: SeqCap EZ Designs, v4.1
11 Estimated_Coverage column: Indirect or adjacent coverage; for the padded and consolidated regions, these values represent bases, covered or uncovered, that are within 100 bp of a capture probe s sequence position. This is an estimate of the actual amount of sequence that may be captured by a capture probe, determined in empirical tests, reflecting that capture probes may hybridize to the end of library insert and extend coverage away from the probe. The 100 bp capture padding was validated with Illumina dual-end sequencing, using a typical library size of ~200 bp. This number may not be accurate for libraries with much larger or smaller insert sizes, or single end reads. Target bases covered: Sum of all bases covered (in basepairs) by at least one capture probe or by predicted captured sequence. Calculations for Probe_Coverage and Estimated_Coverage are provided. Percent target bases covered: Percentage of all bases from the padded and consolidated regions which are covered by one or more capture probe or by predicted captured sequence. Calculations for Probe_Coverage and Estimated_Coverage are provided. Targets with no coverage: Number of padded and consolidated regions with no probes or no predicted captured sequence. Calculations for Probe_Coverage and Estimated_Coverage are provided. Target Bases Not Covered: The number of target bases from the padded and consolidated regions that are not covered by a capture probe or captured sequence. Calculations for Probe_Coverage and Estimated_Coverage are provided. Target Bases Not Covered (due to N's): The number of target bases from the padded and consolidated regions that are not covered by a capture probe or captured sequence due to the source genome having N s or ambiguous bases within the target range. Calculations for Probe_Coverage and Estimated_Coverage are provided. Target Bases Not Covered (due to repeats): The number of target bases from the padded and consolidated regions that are not covered by a capture probe or captured sequence due to the source genome having low complexity or highly repetitive DNA within the target range. Roche avoids selecting Sequence Capture probes in regions of low complexity or high repeat content to reduce the chance of capturing off-target sequence.calculations for Probe_Coverage and Estimated_Coverage are provided. Percent Target Bases Not Covered: The percentage of target bases from the padded and consolidated regions that are not covered by a capture probe or captured sequence. Calculations for Probe_Coverage and Estimated_Coverage are provided. Percent Target Bases Not Covered (due to N's): The percentage of target bases from the padded and consolidated regions that are not covered by a capture probe or captured sequence due to the source genome having N s or ambiguous bases within the target range. Calculations for Probe_Coverage and Estimated_Coverage are provided. Percent Target Bases Not Covered (due to repeats): The percentage of target bases from the padded and consolidated regions that are not covered by a capture probe or captured sequence due to the source genome having low complexity or highly repetitive DNA within the target range. Roche avoids selecting Sequence Capture probes in regions of low complexity or high repeat content to reduce the chance of capturing off-target sequence. Calculations for Probe_Coverage and Estimated_Coverage are provided. Total capture targets: The total number of regions in the capture target files. Total capture space (bp): The total number of bases covered by the capture targets. This can be very different from the primary target space, and provides an idea of the total amount of sequencing that will be needed for each sample. Guide to Reviewing and Approving Custom Designs: SeqCap EZ Designs v4.1 11
12 coverage.txt A tab-delimited text file show region-by-region coverage information of the padded an consolidated customer input regions. The headers are as follows: REGION_NAME: The customer-provided name from the 4th column of their input BED file(s). If no 4th column was provided, then a default name of the selection region will be used instead. This name takes the format of CHROMOSOME:START-STOP. CHROMOSOME: The target chromosome for the region. START: The region start coordinate. STOP: The region stop coordinate. LENGTH: The length of the region. BASES_PROBE_COVERAGE: The number of bases in the region which are directly covered by a capture probe. FRAC_PROBE_COVERAGE: The percent coverage of the region, as a fraction of 1, using direct coverage. For example, a value means that every base of the target is covered by one or more capture probes. A value of means that 46% of the region is covered by one or more capture probes. BASES_ESTIMATE_COVERAGE: The number of bases in the region directly covered by a probe or by indirect/adjacent coverage. This is an estimate of the actual amount of sequence that may be captured by a capture probe, determined in empirical tests, reflecting that capture probes may hybridize to the end of library insert and extend coverage away from the probe. The 100 bp capture padding was validated with Illumina dual-end sequencing, using a typical library size of ~200 bp. This number may not be accurate for libraries with much larger or smaller insert sizes, or single end reads. FRAC_ESTIMATED_COVERAGE: The percent coverage of the region, as a fraction of 1, using indirect/adjacent coverage. For example, a value means that 98.2% of the target is covered indirectly by one or more capture probes. PREDICTED_NO_COVERAGE_BASES: Number of bases in the region that are not covered indirectly and are likely to be missed during capture. BASES_W_NO_PROBE_COV: The number of bases in the region which are not directly covered by a capture probe. BASES_W_NO_PROBE_COV_DUE_TO_N: Number of bases in the region that are not covered directly by probes due to the region containing N s or ambiguous bases in the source. Roche cannot design probes against sequence containing Ns or non-acgt characters. BASES_W_NO_PROBE_COV_DUE_TO_REPEATS: Number of bases in the region that are not covered directly by probes due to the region containing low complexity or highly repetitive sequence. Roche avoids selecting Sequence Capture probes in regions of low complexity or high repeat content for the purposes of reducing off-target sequencing results. BASES_W_NO_EST_COV: The number of bases in the region not directly covered by a probe or by indirect/adjacent coverage. BASES_W_NO_EST_COV_DUE_TO_N: Number of bases in the region that are not covered indirectly due to the region containing N s or ambiguous bases in the source. BASES_W_NO_EST_COV_DUE_TO_REPEATS: Number of bases in the region that are not covered indirectly due to the region containing repetitive sequence. 12 Guide to Reviewing and Approving Custom Designs: SeqCap EZ Designs, v4.1
13 Contact Information Customer Service If you have questions, contact Roche Customer Service: Technical Support If you have technical questions, contact your local Roche Technical Support. Please go to for contact information. Patent License Limitations For patent license limitations for individual products, please refer to /17 For Research Use Only. Not for use in diagnostic procedures. NIMBLEDESIGN and SEQCAP are trademarks of Roche. All other product names and trademarks are the property of their respective owners. Roche Sequencing Solutions, Inc Hacienda Drive Pleasanton, CA USA sequencing.roche.com/support.html Roche Sequencing Solutions, Inc. All rights reserved. Guide to Reviewing and Approving Custom Designs: SeqCap Designs v4.1 13
Design and Annotation Files
 Design and Annotation Files Release Notes SeqCap EZ Exome Target Enrichment System The design and annotation files provide information about genomic regions covered by the capture probes and the genes
Design and Annotation Files Release Notes SeqCap EZ Exome Target Enrichment System The design and annotation files provide information about genomic regions covered by the capture probes and the genes
NimbleDesign Software User s Guide Version 4.3
 NimbleDesign Software User s Guide Version 4.3 For Research Use Only. Not for use in diagnostic procedures. Copyright 2015-2017 Roche Sequencing Solutions, Inc. All rights reserved. Roche Sequencing Solutions,
NimbleDesign Software User s Guide Version 4.3 For Research Use Only. Not for use in diagnostic procedures. Copyright 2015-2017 Roche Sequencing Solutions, Inc. All rights reserved. Roche Sequencing Solutions,
ChIP-Seq Tutorial on Galaxy
 1 Introduction ChIP-Seq Tutorial on Galaxy 2 December 2010 (modified April 6, 2017) Rory Stark The aim of this practical is to give you some experience handling ChIP-Seq data. We will be working with data
1 Introduction ChIP-Seq Tutorial on Galaxy 2 December 2010 (modified April 6, 2017) Rory Stark The aim of this practical is to give you some experience handling ChIP-Seq data. We will be working with data
A short Introduction to UCSC Genome Browser
 A short Introduction to UCSC Genome Browser Elodie Girard, Nicolas Servant Institut Curie/INSERM U900 Bioinformatics, Biostatistics, Epidemiology and computational Systems Biology of Cancer 1 Why using
A short Introduction to UCSC Genome Browser Elodie Girard, Nicolas Servant Institut Curie/INSERM U900 Bioinformatics, Biostatistics, Epidemiology and computational Systems Biology of Cancer 1 Why using
How to use earray to create custom content for the SureSelect Target Enrichment platform. Page 1
 How to use earray to create custom content for the SureSelect Target Enrichment platform Page 1 Getting Started Access earray Access earray at: https://earray.chem.agilent.com/earray/ Log in to earray,
How to use earray to create custom content for the SureSelect Target Enrichment platform Page 1 Getting Started Access earray Access earray at: https://earray.chem.agilent.com/earray/ Log in to earray,
Agilent Genomic Workbench Lite Edition 6.5
 Agilent Genomic Workbench Lite Edition 6.5 SureSelect Quality Analyzer User Guide For Research Use Only. Not for use in diagnostic procedures. Agilent Technologies Notices Agilent Technologies, Inc. 2010
Agilent Genomic Workbench Lite Edition 6.5 SureSelect Quality Analyzer User Guide For Research Use Only. Not for use in diagnostic procedures. Agilent Technologies Notices Agilent Technologies, Inc. 2010
Click on "+" button Select your VCF data files (see #Input Formats->1 above) Remove file from files list:
 CircosVCF: CircosVCF is a web based visualization tool of genome-wide variant data described in VCF files using circos plots. The provided visualization capabilities, gives a broad overview of the genomic
CircosVCF: CircosVCF is a web based visualization tool of genome-wide variant data described in VCF files using circos plots. The provided visualization capabilities, gives a broad overview of the genomic
ValuePRO Tutorial Custom ExcelLINK Template
 ValuePRO Tutorial Custom ExcelLINK Template Table of Contents Contents 1. Setting up the template... 1 1. In Microsoft Excel... 1 2. Creating the report... 2 1. In ValuePRO... 2 1. Home Screen... 2 2.
ValuePRO Tutorial Custom ExcelLINK Template Table of Contents Contents 1. Setting up the template... 1 1. In Microsoft Excel... 1 2. Creating the report... 2 1. In ValuePRO... 2 1. Home Screen... 2 2.
Creating and Using Genome Assemblies Tutorial
 Creating and Using Genome Assemblies Tutorial Release 8.1 Golden Helix, Inc. March 18, 2014 Contents 1. Create a Genome Assembly for Danio rerio 2 2. Building Annotation Sources 5 A. Creating a Reference
Creating and Using Genome Assemblies Tutorial Release 8.1 Golden Helix, Inc. March 18, 2014 Contents 1. Create a Genome Assembly for Danio rerio 2 2. Building Annotation Sources 5 A. Creating a Reference
Tutorial 1: Exploring the UCSC Genome Browser
 Last updated: May 12, 2011 Tutorial 1: Exploring the UCSC Genome Browser Open the homepage of the UCSC Genome Browser at: http://genome.ucsc.edu/ In the blue bar at the top, click on the Genomes link.
Last updated: May 12, 2011 Tutorial 1: Exploring the UCSC Genome Browser Open the homepage of the UCSC Genome Browser at: http://genome.ucsc.edu/ In the blue bar at the top, click on the Genomes link.
Advanced UCSC Browser Functions
 Advanced UCSC Browser Functions Dr. Thomas Randall tarandal@email.unc.edu bioinformatics.unc.edu UCSC Browser: genome.ucsc.edu Overview Custom Tracks adding your own datasets Utilities custom tools for
Advanced UCSC Browser Functions Dr. Thomas Randall tarandal@email.unc.edu bioinformatics.unc.edu UCSC Browser: genome.ucsc.edu Overview Custom Tracks adding your own datasets Utilities custom tools for
GenomeStudio Software Release Notes
 GenomeStudio Software 2009.2 Release Notes 1. GenomeStudio Software 2009.2 Framework... 1 2. Illumina Genome Viewer v1.5...2 3. Genotyping Module v1.5... 4 4. Gene Expression Module v1.5... 6 5. Methylation
GenomeStudio Software 2009.2 Release Notes 1. GenomeStudio Software 2009.2 Framework... 1 2. Illumina Genome Viewer v1.5...2 3. Genotyping Module v1.5... 4 4. Gene Expression Module v1.5... 6 5. Methylation
For Research Use Only. Not for use in diagnostic procedures.
 SMRT View Guide For Research Use Only. Not for use in diagnostic procedures. P/N 100-088-600-02 Copyright 2012, Pacific Biosciences of California, Inc. All rights reserved. Information in this document
SMRT View Guide For Research Use Only. Not for use in diagnostic procedures. P/N 100-088-600-02 Copyright 2012, Pacific Biosciences of California, Inc. All rights reserved. Information in this document
DOCUWARE REFERENCE MANUAL
 DOCUWARE REFERENCE MANUAL Sierra Lands, Inc 2013 LOGGING INTO DOCUWARE Open a web browser and navigate to: https://docs.cariberoyale.com/dwwebclient. DocuWare will automatically log you in based on your
DOCUWARE REFERENCE MANUAL Sierra Lands, Inc 2013 LOGGING INTO DOCUWARE Open a web browser and navigate to: https://docs.cariberoyale.com/dwwebclient. DocuWare will automatically log you in based on your
SIMS 4.0 Archived Projects Users Guide
 If you have access to other modules within SIMS, you will not automatically be directed to the Archived Projects module. If that is the case, choose Archived Projects from the menu at the top left of the
If you have access to other modules within SIMS, you will not automatically be directed to the Archived Projects module. If that is the case, choose Archived Projects from the menu at the top left of the
ChIP-seq hands-on practical using Galaxy
 ChIP-seq hands-on practical using Galaxy In this exercise we will cover some of the basic NGS analysis steps for ChIP-seq using the Galaxy framework: Quality control Mapping of reads using Bowtie2 Peak-calling
ChIP-seq hands-on practical using Galaxy In this exercise we will cover some of the basic NGS analysis steps for ChIP-seq using the Galaxy framework: Quality control Mapping of reads using Bowtie2 Peak-calling
Tutorial. De Novo Assembly of Paired Data. Sample to Insight. November 21, 2017
 De Novo Assembly of Paired Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
De Novo Assembly of Paired Data November 21, 2017 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com AdvancedGenomicsSupport@qiagen.com
4.1. Access the internet and log on to the UCSC Genome Bioinformatics Web Page (Figure 1-
 1. PURPOSE To provide instructions for finding rs Numbers (SNP database ID numbers) and increasing sequence length by utilizing the UCSC Genome Bioinformatics Database. 2. MATERIALS 2.1. Sequence Information
1. PURPOSE To provide instructions for finding rs Numbers (SNP database ID numbers) and increasing sequence length by utilizing the UCSC Genome Bioinformatics Database. 2. MATERIALS 2.1. Sequence Information
Data Walkthrough: Background
 Data Walkthrough: Background File Types FASTA Files FASTA files are text-based representations of genetic information. They can contain nucleotide or amino acid sequences. For this activity, students will
Data Walkthrough: Background File Types FASTA Files FASTA files are text-based representations of genetic information. They can contain nucleotide or amino acid sequences. For this activity, students will
For Research Use Only. Not for use in diagnostic procedures.
 SMRT View Guide For Research Use Only. Not for use in diagnostic procedures. P/N 100-088-600-03 Copyright 2012, Pacific Biosciences of California, Inc. All rights reserved. Information in this document
SMRT View Guide For Research Use Only. Not for use in diagnostic procedures. P/N 100-088-600-03 Copyright 2012, Pacific Biosciences of California, Inc. All rights reserved. Information in this document
QDA Miner. Addendum v2.0
 QDA Miner Addendum v2.0 QDA Miner is an easy-to-use qualitative analysis software for coding, annotating, retrieving and reviewing coded data and documents such as open-ended responses, customer comments,
QDA Miner Addendum v2.0 QDA Miner is an easy-to-use qualitative analysis software for coding, annotating, retrieving and reviewing coded data and documents such as open-ended responses, customer comments,
Helpful Galaxy screencasts are available at:
 This user guide serves as a simplified, graphic version of the CloudMap paper for applicationoriented end-users. For more details, please see the CloudMap paper. Video versions of these user guides and
This user guide serves as a simplified, graphic version of the CloudMap paper for applicationoriented end-users. For more details, please see the CloudMap paper. Video versions of these user guides and
Google Sheets: Spreadsheet basics
 Google Sheets: Spreadsheet basics You can find all of your spreadsheets on the Google Sheets home screen or in Google Drive. Create a spreadsheet On the Sheets home screen, click Create new spreadsheet
Google Sheets: Spreadsheet basics You can find all of your spreadsheets on the Google Sheets home screen or in Google Drive. Create a spreadsheet On the Sheets home screen, click Create new spreadsheet
Tutorial: De Novo Assembly of Paired Data
 : De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : De Novo Assembly
: De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : De Novo Assembly
Creating Reports in Access 2007 Table of Contents GUIDE TO DESIGNING REPORTS... 3 DECIDE HOW TO LAY OUT YOUR REPORT... 3 MAKE A SKETCH OF YOUR
 Creating Reports in Access 2007 Table of Contents GUIDE TO DESIGNING REPORTS... 3 DECIDE HOW TO LAY OUT YOUR REPORT... 3 MAKE A SKETCH OF YOUR REPORT... 3 DECIDE WHICH DATA TO PUT IN EACH REPORT SECTION...
Creating Reports in Access 2007 Table of Contents GUIDE TO DESIGNING REPORTS... 3 DECIDE HOW TO LAY OUT YOUR REPORT... 3 MAKE A SKETCH OF YOUR REPORT... 3 DECIDE WHICH DATA TO PUT IN EACH REPORT SECTION...
Transitions and Photos
 4 Adding Lesson 3: Transitions and Photos You ve arranged your video clips in the order you want them. But the jump from one clip to the next is very abrupt, and you d like to smooth things out. You can
4 Adding Lesson 3: Transitions and Photos You ve arranged your video clips in the order you want them. But the jump from one clip to the next is very abrupt, and you d like to smooth things out. You can
Searching for Images in v10
 Searching for Images in v10 Following are the steps to search for images in Docfinity version 10. Searching for Images: Log into Docfinity on www.controller.psu.edu/docfinity Open Searching Workspace in
Searching for Images in v10 Following are the steps to search for images in Docfinity version 10. Searching for Images: Log into Docfinity on www.controller.psu.edu/docfinity Open Searching Workspace in
Introduction to Galaxy
 Introduction to Galaxy Dr Jason Wong Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW Day 1 Thurs 28 th January 2016 Overview What is Galaxy? Description of
Introduction to Galaxy Dr Jason Wong Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW Day 1 Thurs 28 th January 2016 Overview What is Galaxy? Description of
Next generation Confirmation (NGC) module
 QUICK REFERENCE Next generation Confirmation (NGC) module Catalog Number A28221 Pub. No. MAN0015891 Rev. A.0 Product description The Applied Biosystems Next generation Confirmation (NGC) module analyzes
QUICK REFERENCE Next generation Confirmation (NGC) module Catalog Number A28221 Pub. No. MAN0015891 Rev. A.0 Product description The Applied Biosystems Next generation Confirmation (NGC) module analyzes
GSEQ Software User s Guide for AccuID TM
 GSEQ Software User s Guide for AccuID TM This protocol is majorly based on Affymetrix GeneChip Sequence Analysis Software User s Guide Version 4.1 guidebook and has modified it to fit GSEQ Software User
GSEQ Software User s Guide for AccuID TM This protocol is majorly based on Affymetrix GeneChip Sequence Analysis Software User s Guide Version 4.1 guidebook and has modified it to fit GSEQ Software User
How to use the transcription tool STATE LIBRARY GUIDE
 How to use the transcription tool STATE LIBRARY GUIDE Transcription website https://transcripts.sl.nsw.gov.au/ Overview of the transcription process The Library loads the digitised image of the document
How to use the transcription tool STATE LIBRARY GUIDE Transcription website https://transcripts.sl.nsw.gov.au/ Overview of the transcription process The Library loads the digitised image of the document
New User Orientation PARTICIPANT WORKBOOK
 New User Orientation PARTICIPANT WORKBOOK INTEGRATED SOFTWARE SERIES New User Orientation PARTICIPANT WORKBOOK Version 2.0 Copyright 2005 2009. Interactive Financial Solutions, Inc. All Rights Reserved.
New User Orientation PARTICIPANT WORKBOOK INTEGRATED SOFTWARE SERIES New User Orientation PARTICIPANT WORKBOOK Version 2.0 Copyright 2005 2009. Interactive Financial Solutions, Inc. All Rights Reserved.
Colorado State University Bioinformatics Algorithms Assignment 6: Analysis of High- Throughput Biological Data Hamidreza Chitsaz, Ali Sharifi- Zarchi
 Colorado State University Bioinformatics Algorithms Assignment 6: Analysis of High- Throughput Biological Data Hamidreza Chitsaz, Ali Sharifi- Zarchi Although a little- bit long, this is an easy exercise
Colorado State University Bioinformatics Algorithms Assignment 6: Analysis of High- Throughput Biological Data Hamidreza Chitsaz, Ali Sharifi- Zarchi Although a little- bit long, this is an easy exercise
The UCSC Gene Sorter, Table Browser & Custom Tracks
 The UCSC Gene Sorter, Table Browser & Custom Tracks Advanced searching and discovery using the UCSC Table Browser and Custom Tracks Osvaldo Graña Bioinformatics Unit, CNIO 1 Table Browser and Custom Tracks
The UCSC Gene Sorter, Table Browser & Custom Tracks Advanced searching and discovery using the UCSC Table Browser and Custom Tracks Osvaldo Graña Bioinformatics Unit, CNIO 1 Table Browser and Custom Tracks
CLC Server. End User USER MANUAL
 CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
CLC Server End User USER MANUAL Manual for CLC Server 10.0.1 Windows, macos and Linux March 8, 2018 This software is for research purposes only. QIAGEN Aarhus Silkeborgvej 2 Prismet DK-8000 Aarhus C Denmark
Agilent Genomic Workbench 7.0
 Agilent Genomic Workbench 7.0 Data Viewing User Guide Agilent Technologies Notices Agilent Technologies, Inc. 2012, 2015 No part of this manual may be reproduced in any form or by any means (including
Agilent Genomic Workbench 7.0 Data Viewing User Guide Agilent Technologies Notices Agilent Technologies, Inc. 2012, 2015 No part of this manual may be reproduced in any form or by any means (including
Océ Engineering Exec. Advanced Import and Index
 Océ Engineering Exec Advanced Import and Index Océ-Technologies B.V. Copyright 2004, Océ-Technologies B.V. Venlo, The Netherlands All rights reserved. No part of this work may be reproduced, copied, adapted,
Océ Engineering Exec Advanced Import and Index Océ-Technologies B.V. Copyright 2004, Océ-Technologies B.V. Venlo, The Netherlands All rights reserved. No part of this work may be reproduced, copied, adapted,
Adding records Pasting records Deleting records Sorting records Filtering records Inserting and deleting columns Calculated columns Working with the
 Show All About spreadsheets You can use a spreadsheet to enter and calculate data. A spreadsheet consists of columns and rows of cells. You can enter data directly into the cells of the spreadsheet and
Show All About spreadsheets You can use a spreadsheet to enter and calculate data. A spreadsheet consists of columns and rows of cells. You can enter data directly into the cells of the spreadsheet and
Introduction to Genome Browsers
 Introduction to Genome Browsers Rolando Garcia-Milian, MLS, AHIP (Rolando.milian@ufl.edu) Department of Biomedical and Health Information Services Health Sciences Center Libraries, University of Florida
Introduction to Genome Browsers Rolando Garcia-Milian, MLS, AHIP (Rolando.milian@ufl.edu) Department of Biomedical and Health Information Services Health Sciences Center Libraries, University of Florida
BRIEFCASES & TASKS ZIMBRA. Briefcase can be used to share and manage documents. Documents can be shared, edited, and created using Briefcases.
 BRIEFCASES & TASKS ZIMBRA BRIEFCASES Briefcase can be used to share and manage documents. Documents can be shared, edited, and created using Briefcases. Options Briefcase New Briefcase To create briefcases,
BRIEFCASES & TASKS ZIMBRA BRIEFCASES Briefcase can be used to share and manage documents. Documents can be shared, edited, and created using Briefcases. Options Briefcase New Briefcase To create briefcases,
Genome Browsers - The UCSC Genome Browser
 Genome Browsers - The UCSC Genome Browser Background The UCSC Genome Browser is a well-curated site that provides users with a view of gene or sequence information in genomic context for a specific species,
Genome Browsers - The UCSC Genome Browser Background The UCSC Genome Browser is a well-curated site that provides users with a view of gene or sequence information in genomic context for a specific species,
Acrobat X Professional
 Acrobat X Professional Toolbar Well Page Navigations/Page Indicator Buttons for paging through document Scroll Bar/box page indicator appears when using the scroll button to navigate. When you release
Acrobat X Professional Toolbar Well Page Navigations/Page Indicator Buttons for paging through document Scroll Bar/box page indicator appears when using the scroll button to navigate. When you release
 1 of 9 8/27/2014 10:53 AM Units: Teacher: MOExcel/Access, CORE Course: MOExcel/Access Year: 2012-13 Excel Unit A What is spreadsheet software? What are the parts of the Excel window? What are labels and
1 of 9 8/27/2014 10:53 AM Units: Teacher: MOExcel/Access, CORE Course: MOExcel/Access Year: 2012-13 Excel Unit A What is spreadsheet software? What are the parts of the Excel window? What are labels and
Exercise 2: Browser-Based Annotation and RNA-Seq Data
 Exercise 2: Browser-Based Annotation and RNA-Seq Data Jeremy Buhler July 24, 2018 This exercise continues your introduction to practical issues in comparative annotation. You ll be annotating genomic sequence
Exercise 2: Browser-Based Annotation and RNA-Seq Data Jeremy Buhler July 24, 2018 This exercise continues your introduction to practical issues in comparative annotation. You ll be annotating genomic sequence
Quick Start Guide. Table of contents. Browsing in the Navigator... 2 The Navigator makes browsing and navigation easier.
 Table of contents Browsing in the Navigator... 2 The Navigator makes browsing and navigation easier. Searching in Windchill... 3 Quick and simple searches are always available at the top of the Windchill
Table of contents Browsing in the Navigator... 2 The Navigator makes browsing and navigation easier. Searching in Windchill... 3 Quick and simple searches are always available at the top of the Windchill
Juniata County, Pennsylvania
 GIS Parcel Viewer Web Mapping Application Functional Documentation June 21, 2017 Juniata County, Pennsylvania Presented by www.worldviewsolutions.com (804) 767-1870 (phone) (804) 545-0792 (fax) 115 South
GIS Parcel Viewer Web Mapping Application Functional Documentation June 21, 2017 Juniata County, Pennsylvania Presented by www.worldviewsolutions.com (804) 767-1870 (phone) (804) 545-0792 (fax) 115 South
HPHConnect for Providers. Member Roster User Guide
 HPHConnect for Providers Member Roster User Guide March 2015 HPHCONNECT MEMBER ROSTER Table of Contents A. OVERVIEW Introduction................................................................. 1 Member
HPHConnect for Providers Member Roster User Guide March 2015 HPHCONNECT MEMBER ROSTER Table of Contents A. OVERVIEW Introduction................................................................. 1 Member
Quick Start Guide. Table of contents. Browsing in the Navigator... 2 The Navigator makes browsing and navigation easier.
 Table of contents Browsing in the Navigator... 2 The Navigator makes browsing and navigation easier. Searching in Windchill... 3 Quick and simple searches are always available at the top of the Windchill
Table of contents Browsing in the Navigator... 2 The Navigator makes browsing and navigation easier. Searching in Windchill... 3 Quick and simple searches are always available at the top of the Windchill
The tracing tool in SQL-Hero tries to deal with the following weaknesses found in the out-of-the-box SQL Profiler tool:
 Revision Description 7/21/2010 Original SQL-Hero Tracing Introduction Let s start by asking why you might want to do SQL tracing in the first place. As it turns out, this can be an extremely useful activity
Revision Description 7/21/2010 Original SQL-Hero Tracing Introduction Let s start by asking why you might want to do SQL tracing in the first place. As it turns out, this can be an extremely useful activity
Analyzing ChIP- Seq Data in Galaxy
 Analyzing ChIP- Seq Data in Galaxy Lauren Mills RISS ABSTRACT Step- by- step guide to basic ChIP- Seq analysis using the Galaxy platform. Table of Contents Introduction... 3 Links to helpful information...
Analyzing ChIP- Seq Data in Galaxy Lauren Mills RISS ABSTRACT Step- by- step guide to basic ChIP- Seq analysis using the Galaxy platform. Table of Contents Introduction... 3 Links to helpful information...
QUICK EXCEL TUTORIAL. The Very Basics
 QUICK EXCEL TUTORIAL The Very Basics You Are Here. Titles & Column Headers Merging Cells Text Alignment When we work on spread sheets we often need to have a title and/or header clearly visible. Merge
QUICK EXCEL TUTORIAL The Very Basics You Are Here. Titles & Column Headers Merging Cells Text Alignment When we work on spread sheets we often need to have a title and/or header clearly visible. Merge
Enterprise Application Systems
 INFORMATION TECHNOLOGY Enterprise Application Systems Argos Report Viewer Guide Mt. San Antonio College Information Technology 1100 North Grand Avenue Building 23 Walnut, CA 91789 Help Desk 909.274.4357
INFORMATION TECHNOLOGY Enterprise Application Systems Argos Report Viewer Guide Mt. San Antonio College Information Technology 1100 North Grand Avenue Building 23 Walnut, CA 91789 Help Desk 909.274.4357
The following instructions cover how to edit an existing report in IBM Cognos Analytics.
 IBM Cognos Analytics Edit a Report The following instructions cover how to edit an existing report in IBM Cognos Analytics. Navigate to Cognos Cognos Analytics supports all browsers with the exception
IBM Cognos Analytics Edit a Report The following instructions cover how to edit an existing report in IBM Cognos Analytics. Navigate to Cognos Cognos Analytics supports all browsers with the exception
CCRS Quick Start Guide for Program Administrators. September Bank Handlowy w Warszawie S.A.
 CCRS Quick Start Guide for Program Administrators September 2017 www.citihandlowy.pl Bank Handlowy w Warszawie S.A. CitiManager Quick Start Guide for Program Administrators Table of Contents Table of Contents
CCRS Quick Start Guide for Program Administrators September 2017 www.citihandlowy.pl Bank Handlowy w Warszawie S.A. CitiManager Quick Start Guide for Program Administrators Table of Contents Table of Contents
Tutorial: Resequencing Analysis using Tracks
 : Resequencing Analysis using Tracks September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : Resequencing
: Resequencing Analysis using Tracks September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45 86 20 12 22 www.clcbio.com support@clcbio.com : Resequencing
m6aviewer Version Documentation
 m6aviewer Version 1.6.0 Documentation Contents 1. About 2. Requirements 3. Launching m6aviewer 4. Running Time Estimates 5. Basic Peak Calling 6. Running Modes 7. Multiple Samples/Sample Replicates 8.
m6aviewer Version 1.6.0 Documentation Contents 1. About 2. Requirements 3. Launching m6aviewer 4. Running Time Estimates 5. Basic Peak Calling 6. Running Modes 7. Multiple Samples/Sample Replicates 8.
KaryoStudio v1.4 User Guide
 KaryoStudio v1.4 User Guide FOR RESEARCH USE ONLY ILLUMINA PROPRIETARY Part # 11328837 Rev. C June 2011 Notice This document and its contents are proprietary to Illumina, Inc. and its affiliates ("Illumina"),
KaryoStudio v1.4 User Guide FOR RESEARCH USE ONLY ILLUMINA PROPRIETARY Part # 11328837 Rev. C June 2011 Notice This document and its contents are proprietary to Illumina, Inc. and its affiliates ("Illumina"),
DecisionPoint For Excel
 DecisionPoint For Excel Getting Started Guide 2015 Antivia Group Ltd Notation used in this workbook Indicates where you need to click with your mouse Indicates a drag and drop path State >= N Indicates
DecisionPoint For Excel Getting Started Guide 2015 Antivia Group Ltd Notation used in this workbook Indicates where you need to click with your mouse Indicates a drag and drop path State >= N Indicates
ChIP-seq practical: peak detection and peak annotation. Mali Salmon-Divon Remco Loos Myrto Kostadima
 ChIP-seq practical: peak detection and peak annotation Mali Salmon-Divon Remco Loos Myrto Kostadima March 2012 Introduction The goal of this hands-on session is to perform some basic tasks in the analysis
ChIP-seq practical: peak detection and peak annotation Mali Salmon-Divon Remco Loos Myrto Kostadima March 2012 Introduction The goal of this hands-on session is to perform some basic tasks in the analysis
Google Sheets: Spreadsheet basics
 Google Sheets: Spreadsheet basics To view all of your Google sheets, or to create a new spreadsheet, visit docs.google.com/spreadsheets. Create a spreadsheet From the Google Sheets home screen, click the
Google Sheets: Spreadsheet basics To view all of your Google sheets, or to create a new spreadsheet, visit docs.google.com/spreadsheets. Create a spreadsheet From the Google Sheets home screen, click the
Supplementary Figure 1. Fast read-mapping algorithm of BrowserGenome.
 Supplementary Figure 1 Fast read-mapping algorithm of BrowserGenome. (a) Indexing strategy: The genome sequence of interest is divided into non-overlapping 12-mers. A Hook table is generated that contains
Supplementary Figure 1 Fast read-mapping algorithm of BrowserGenome. (a) Indexing strategy: The genome sequence of interest is divided into non-overlapping 12-mers. A Hook table is generated that contains
ChIP-seq hands-on practical using Galaxy
 ChIP-seq hands-on practical using Galaxy In this exercise we will cover some of the basic NGS analysis steps for ChIP-seq using the Galaxy framework: Quality control Mapping of reads using Bowtie2 Peak-calling
ChIP-seq hands-on practical using Galaxy In this exercise we will cover some of the basic NGS analysis steps for ChIP-seq using the Galaxy framework: Quality control Mapping of reads using Bowtie2 Peak-calling
PDF Splitter User Guide. PDF Splitter. User Guide. CoolUtils.com. CoolUtils.com Page 2
 USER GUIDE PDF Splitter User Guide CoolUtils.com CoolUtils.com Page 2 Table of Contents 1. Introduction to PDF Splitter... 5 2. How to Get Help... 6 3. How to Install PDF Splitter... 7 4. PDF Splitter
USER GUIDE PDF Splitter User Guide CoolUtils.com CoolUtils.com Page 2 Table of Contents 1. Introduction to PDF Splitter... 5 2. How to Get Help... 6 3. How to Install PDF Splitter... 7 4. PDF Splitter
Access 2013 Introduction to Forms and Reports
 Forms Overview You can create forms to present data in a more attractive and easier to use format They can be used for viewing, editing and printing data and in advanced cases, used to automate the database
Forms Overview You can create forms to present data in a more attractive and easier to use format They can be used for viewing, editing and printing data and in advanced cases, used to automate the database
An Introduction to Oracle Business Intelligence
 Table of Contents Introduction... 2 What is OBI...2 What is a Dashboard?...2 Accessing OBI... 3 Viewing Data via Dashboards... 4 Using Favorites... 5 Filtering Data... 6 Changing the way data is displayed...
Table of Contents Introduction... 2 What is OBI...2 What is a Dashboard?...2 Accessing OBI... 3 Viewing Data via Dashboards... 4 Using Favorites... 5 Filtering Data... 6 Changing the way data is displayed...
Spreadsheet definition: Starting a New Excel Worksheet: Navigating Through an Excel Worksheet
 Copyright 1 99 Spreadsheet definition: A spreadsheet stores and manipulates data that lends itself to being stored in a table type format (e.g. Accounts, Science Experiments, Mathematical Trends, Statistics,
Copyright 1 99 Spreadsheet definition: A spreadsheet stores and manipulates data that lends itself to being stored in a table type format (e.g. Accounts, Science Experiments, Mathematical Trends, Statistics,
Sage Estimating (SQL) v17.13
 Sage Estimating (SQL) v17.13 Sage 100 Contractor (SQL) Integration Guide December 2017 This is a publication of Sage Software, Inc. 2017 The Sage Group plc or its licensors. All rights reserved. Sage,
Sage Estimating (SQL) v17.13 Sage 100 Contractor (SQL) Integration Guide December 2017 This is a publication of Sage Software, Inc. 2017 The Sage Group plc or its licensors. All rights reserved. Sage,
SAS Visual Analytics 8.2: Getting Started with Reports
 SAS Visual Analytics 8.2: Getting Started with Reports Introduction Reporting The SAS Visual Analytics tools give you everything you need to produce and distribute clear and compelling reports. SAS Visual
SAS Visual Analytics 8.2: Getting Started with Reports Introduction Reporting The SAS Visual Analytics tools give you everything you need to produce and distribute clear and compelling reports. SAS Visual
Data Feedback Report Tutorial Script Data Collection Cycle
 Data Feedback Report Tutorial Script 2014-15 Data Collection Cycle The IPEDS Data Center includes a wide-range of functional options, including access to current and previous versions of your institution's
Data Feedback Report Tutorial Script 2014-15 Data Collection Cycle The IPEDS Data Center includes a wide-range of functional options, including access to current and previous versions of your institution's
Analysis of ChIP-seq data
 Before we start: 1. Log into tak (step 0 on the exercises) 2. Go to your lab space and create a folder for the class (see separate hand out) 3. Connect to your lab space through the wihtdata network and
Before we start: 1. Log into tak (step 0 on the exercises) 2. Go to your lab space and create a folder for the class (see separate hand out) 3. Connect to your lab space through the wihtdata network and
metilene - a tool for fast and sensitive detection of differential DNA methylation
 metilene - a tool for fast and sensitive detection of differential DNA methylation Frank Jühling, Helene Kretzmer, Stephan H. Bernhart, Christian Otto, Peter F. Stadler, Steve Hoffmann Version 0.23 1 Introduction
metilene - a tool for fast and sensitive detection of differential DNA methylation Frank Jühling, Helene Kretzmer, Stephan H. Bernhart, Christian Otto, Peter F. Stadler, Steve Hoffmann Version 0.23 1 Introduction
Introduction to Qualtrics Research Suite Wednesday, September 19, 2012
 Logging in to Qualtrics Introduction to Qualtrics Research Suite Wednesday, September 19, 2012 1. Open a browser and go to http://www.qualtrics.com 2. If you have a Qualtrics account, use it to login.
Logging in to Qualtrics Introduction to Qualtrics Research Suite Wednesday, September 19, 2012 1. Open a browser and go to http://www.qualtrics.com 2. If you have a Qualtrics account, use it to login.
Quick Start Guide. ARIS Architect. Version 9.7
 ARIS Architect Version 9.7 October 2014 This document applies to ARIS Version 9.7 and to all subsequent releases. Specifications contained herein are subject to change and these changes will be reported
ARIS Architect Version 9.7 October 2014 This document applies to ARIS Version 9.7 and to all subsequent releases. Specifications contained herein are subject to change and these changes will be reported
TIBCO Spotfire Lead Discovery 2.1 User s Manual
 TIBCO Spotfire Lead Discovery 2.1 User s Manual Important Information SOME TIBCO SOFTWARE EMBEDS OR BUNDLES OTHER TIBCO SOFTWARE. USE OF SUCH EMBEDDED OR BUNDLED TIBCO SOFTWARE IS SOLELY TO ENABLE THE
TIBCO Spotfire Lead Discovery 2.1 User s Manual Important Information SOME TIBCO SOFTWARE EMBEDS OR BUNDLES OTHER TIBCO SOFTWARE. USE OF SUCH EMBEDDED OR BUNDLED TIBCO SOFTWARE IS SOLELY TO ENABLE THE
Importing sequence assemblies from BAM and SAM files
 BioNumerics Tutorial: Importing sequence assemblies from BAM and SAM files 1 Aim With the BioNumerics BAM import routine, a sequence assembly in BAM or SAM format can be imported in BioNumerics. A BAM
BioNumerics Tutorial: Importing sequence assemblies from BAM and SAM files 1 Aim With the BioNumerics BAM import routine, a sequence assembly in BAM or SAM format can be imported in BioNumerics. A BAM
GO! with Microsoft Excel 2016 Comprehensive
 GO! with Microsoft Excel 2016 Comprehensive First Edition Chapter 2 Using Functions, Creating Tables, and Managing Large Workbooks Use SUM and Statistical Functions The SUM function is a predefined formula
GO! with Microsoft Excel 2016 Comprehensive First Edition Chapter 2 Using Functions, Creating Tables, and Managing Large Workbooks Use SUM and Statistical Functions The SUM function is a predefined formula
COPYRIGHTED MATERIAL. Making Excel More Efficient
 Making Excel More Efficient If you find yourself spending a major part of your day working with Excel, you can make those chores go faster and so make your overall work life more productive by making Excel
Making Excel More Efficient If you find yourself spending a major part of your day working with Excel, you can make those chores go faster and so make your overall work life more productive by making Excel
EXCEL IMPORT user guide
 18.2 user guide No Magic, Inc. 2015 All material contained herein is considered proprietary information owned by No Magic, Inc. and is not to be shared, copied, or reproduced by any means. All information
18.2 user guide No Magic, Inc. 2015 All material contained herein is considered proprietary information owned by No Magic, Inc. and is not to be shared, copied, or reproduced by any means. All information
ECSE-323 Digital System Design. Lab #1 Using the Altera Quartus II Software Fall 2008
 1 ECSE-323 Digital System Design Lab #1 Using the Altera Quartus II Software Fall 2008 2 Introduction. In this lab you will learn the basics of the Altera Quartus II FPGA design software through following
1 ECSE-323 Digital System Design Lab #1 Using the Altera Quartus II Software Fall 2008 2 Introduction. In this lab you will learn the basics of the Altera Quartus II FPGA design software through following
เพ มภาพตามเน อหาของแต ละบท. Microsoft Excel Benjamas Panyangam and Dr. Dussadee Praserttitipong. Adapted in English by Prakarn Unachak
 เพ มภาพตามเน อหาของแต ละบท Microsoft Excel 2016 Benjamas Panyangam and Dr. Dussadee Praserttitipong Adapted in English by Prakarn Unachak 204100 IT AND MODERN LIFE 1. Excel Basics 2. Calculation and Formula
เพ มภาพตามเน อหาของแต ละบท Microsoft Excel 2016 Benjamas Panyangam and Dr. Dussadee Praserttitipong Adapted in English by Prakarn Unachak 204100 IT AND MODERN LIFE 1. Excel Basics 2. Calculation and Formula
Gene Set Enrichment Analysis. GSEA User Guide
 Gene Set Enrichment Analysis GSEA User Guide 1 Software Copyright The Broad Institute SOFTWARE COPYRIGHT NOTICE AGREEMENT This software and its documentation are copyright 2009, 2010 by the Broad Institute/Massachusetts
Gene Set Enrichment Analysis GSEA User Guide 1 Software Copyright The Broad Institute SOFTWARE COPYRIGHT NOTICE AGREEMENT This software and its documentation are copyright 2009, 2010 by the Broad Institute/Massachusetts
Today's outline. Resources. Genome browser components. Genome browsers: Discovering biology through genomics. Genome browser tutorial materials
 Today's outline Genome browsers: Discovering biology through genomics BaRC Hot Topics April 2013 George Bell, Ph.D. http://jura.wi.mit.edu/bio/education/hot_topics/ Genome browser introduction Popular
Today's outline Genome browsers: Discovering biology through genomics BaRC Hot Topics April 2013 George Bell, Ph.D. http://jura.wi.mit.edu/bio/education/hot_topics/ Genome browser introduction Popular
What is New in TILOS 10.0
 Civil Engineering and Construction What is New in TILOS 10.0 Mass Haulage Add Space Above/Below the Zero Line In a mass haulage diagram, you can now add space vertically between the zero line and cut/fill
Civil Engineering and Construction What is New in TILOS 10.0 Mass Haulage Add Space Above/Below the Zero Line In a mass haulage diagram, you can now add space vertically between the zero line and cut/fill
CHAPTER 4: MICROSOFT OFFICE: EXCEL 2010
 CHAPTER 4: MICROSOFT OFFICE: EXCEL 2010 Quick Summary A workbook an Excel document that stores data contains one or more pages called a worksheet. A worksheet or spreadsheet is stored in a workbook, and
CHAPTER 4: MICROSOFT OFFICE: EXCEL 2010 Quick Summary A workbook an Excel document that stores data contains one or more pages called a worksheet. A worksheet or spreadsheet is stored in a workbook, and
BaseSpace - MiSeq Reporter Software v2.4 Release Notes
 Page 1 of 5 BaseSpace - MiSeq Reporter Software v2.4 Release Notes For MiSeq Systems Connected to BaseSpace June 2, 2014 Revision Date Description of Change A May 22, 2014 Initial Version Revision History
Page 1 of 5 BaseSpace - MiSeq Reporter Software v2.4 Release Notes For MiSeq Systems Connected to BaseSpace June 2, 2014 Revision Date Description of Change A May 22, 2014 Initial Version Revision History
Faculty Guide to Grade Center in Blackboard
 Faculty Guide to Grade Center in Blackboard Grade Center, formally known as Gradebook, is a central repository for assessment data, student information, and instructor notes. Although it includes items
Faculty Guide to Grade Center in Blackboard Grade Center, formally known as Gradebook, is a central repository for assessment data, student information, and instructor notes. Although it includes items
IBM TRIRIGA Application Platform Version 3.2. Graphics User Guide. Copyright IBM Corp i
 IBM TRIRIGA Application Platform Version 3.2 Graphics User Guide Copyright IBM Corp. 2011 i Note Before using this information and the product it supports, read the information in Notices on page 31. This
IBM TRIRIGA Application Platform Version 3.2 Graphics User Guide Copyright IBM Corp. 2011 i Note Before using this information and the product it supports, read the information in Notices on page 31. This
QIAseq Targeted RNAscan Panel Analysis Plugin USER MANUAL
 QIAseq Targeted RNAscan Panel Analysis Plugin USER MANUAL User manual for QIAseq Targeted RNAscan Panel Analysis 0.5.2 beta 1 Windows, Mac OS X and Linux February 5, 2018 This software is for research
QIAseq Targeted RNAscan Panel Analysis Plugin USER MANUAL User manual for QIAseq Targeted RNAscan Panel Analysis 0.5.2 beta 1 Windows, Mac OS X and Linux February 5, 2018 This software is for research
Tutorial. Aligning contigs manually using the Genome Finishing. Sample to Insight. February 6, 2019
 Aligning contigs manually using the Genome Finishing Module February 6, 2019 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com
Aligning contigs manually using the Genome Finishing Module February 6, 2019 Sample to Insight QIAGEN Aarhus Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.qiagenbioinformatics.com
UCSC Genome Browser Pittsburgh Workshop -- Practical Exercises
 UCSC Genome Browser Pittsburgh Workshop -- Practical Exercises We will be using human assembly hg19. These problems will take you through a variety of resources at the UCSC Genome Browser. You will learn
UCSC Genome Browser Pittsburgh Workshop -- Practical Exercises We will be using human assembly hg19. These problems will take you through a variety of resources at the UCSC Genome Browser. You will learn
Tutorial: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and Expression measures
 : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
: RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and February 24, 2014 Sample to Insight : RNA-Seq Analysis Part II (Tracks): Non-Specific Matches, Mapping Modes and : RNA-Seq Analysis
User Manual. Ver. 3.0 March 19, 2012
 User Manual Ver. 3.0 March 19, 2012 Table of Contents 1. Introduction... 2 1.1 Rationale... 2 1.2 Software Work-Flow... 3 1.3 New in GenomeGems 3.0... 4 2. Software Description... 5 2.1 Key Features...
User Manual Ver. 3.0 March 19, 2012 Table of Contents 1. Introduction... 2 1.1 Rationale... 2 1.2 Software Work-Flow... 3 1.3 New in GenomeGems 3.0... 4 2. Software Description... 5 2.1 Key Features...
IBM TRIRIGA Application Platform Version 3.3. Graphics User Guide. Copyright IBM Corp i
 IBM TRIRIGA Application Platform Version 3.3 Graphics User Guide Copyright IBM Corp. 2011 i Note Before using this information and the product it supports, read the information in Notices on page 33. This
IBM TRIRIGA Application Platform Version 3.3 Graphics User Guide Copyright IBM Corp. 2011 i Note Before using this information and the product it supports, read the information in Notices on page 33. This
Numbers Basics Website:
 Website: http://etc.usf.edu/te/ Numbers is Apple's new spreadsheet application. It is installed as part of the iwork suite, which also includes the word processing program Pages and the presentation program
Website: http://etc.usf.edu/te/ Numbers is Apple's new spreadsheet application. It is installed as part of the iwork suite, which also includes the word processing program Pages and the presentation program
Real Monitor and Real Monitor Viewer
 Real Monitor and Real Monitor Viewer This chapter describes Real Monitor and Real Monitor Viewer, two related applications which allow you to collect and review data about your system. These applications
Real Monitor and Real Monitor Viewer This chapter describes Real Monitor and Real Monitor Viewer, two related applications which allow you to collect and review data about your system. These applications
End User Guide. 2.1 Getting Started Toolbar Right-click Contextual Menu Navigation Panels... 2
 TABLE OF CONTENTS 1 OVERVIEW...1 2 WEB VIEWER DEMO ON DESKTOP...1 2.1 Getting Started... 1 2.1.1 Toolbar... 1 2.1.2 Right-click Contextual Menu... 2 2.1.3 Navigation Panels... 2 2.1.4 Floating Toolbar...
TABLE OF CONTENTS 1 OVERVIEW...1 2 WEB VIEWER DEMO ON DESKTOP...1 2.1 Getting Started... 1 2.1.1 Toolbar... 1 2.1.2 Right-click Contextual Menu... 2 2.1.3 Navigation Panels... 2 2.1.4 Floating Toolbar...
epigenomegateway.wustl.edu
 Everything can be found at epigenomegateway.wustl.edu REFERENCES 1. Zhou X, et al., Nature Methods 8, 989-990 (2011) 2. Zhou X & Wang T, Current Protocols in Bioinformatics Unit 10.10 (2012) 3. Zhou X,
Everything can be found at epigenomegateway.wustl.edu REFERENCES 1. Zhou X, et al., Nature Methods 8, 989-990 (2011) 2. Zhou X & Wang T, Current Protocols in Bioinformatics Unit 10.10 (2012) 3. Zhou X,
IBM Rational Rhapsody Gateway Add On. User Guide
 User Guide Rhapsody IBM Rational Rhapsody Gateway Add On User Guide License Agreement No part of this publication may be reproduced, transmitted, stored in a retrieval system, nor translated into any
User Guide Rhapsody IBM Rational Rhapsody Gateway Add On User Guide License Agreement No part of this publication may be reproduced, transmitted, stored in a retrieval system, nor translated into any
ChIP-seq (NGS) Data Formats
 ChIP-seq (NGS) Data Formats Biological samples Sequence reads SRA/SRF, FASTQ Quality control SAM/BAM/Pileup?? Mapping Assembly... DE Analysis Variant Detection Peak Calling...? Counts, RPKM VCF BED/narrowPeak/
ChIP-seq (NGS) Data Formats Biological samples Sequence reads SRA/SRF, FASTQ Quality control SAM/BAM/Pileup?? Mapping Assembly... DE Analysis Variant Detection Peak Calling...? Counts, RPKM VCF BED/narrowPeak/
Workbooks & Worksheets. Getting Started. Formatting. Formulas & Functions
 1 Getting Started Cells Workbooks & Worksheets Formatting Formulas & Functions Chart Printing 2 Getting Started Start a spreadsheet program Recognize the spreadsheet screen layout Use the ribbon,quick
1 Getting Started Cells Workbooks & Worksheets Formatting Formulas & Functions Chart Printing 2 Getting Started Start a spreadsheet program Recognize the spreadsheet screen layout Use the ribbon,quick
