Sequence alignment. Genomes change over time
|
|
|
- Shanna Hodges
- 5 years ago
- Views:
Transcription
1 Sequence alignment Genomes change over time 1
2 Goal of alignment: Infer edit operations What is sequence alignment? 2
3 Align biological sequences Statement of the problem Given 2 sequences Scoring system for evaluating match (or mismatch) of two characters Penalty function for gaps in sequences Produce: Optimal pairing of sequences that Retains the order of the sequences Introduces gaps Maximizes total score 3
4 Enumeration of all possible alignments Number of possible alignments of 2 sequences with length n and m For 2 sequences of length n n enumeration , E E+58 Naïve Z box algorithm Exact string matching Boyer Moore algorithm Knuth Morris Pratt algorithm Dan Gusfield: Algorithms on Strings,Trees, and Sequences 4
5 = ordered tree data structure Suffix (prefix) tries (retrieval) Sequence ACACGT$ Suffix tree T $ 1 Suffix array G C A C T $ G $ T 6 $ A C G T $ 4 ACACGT$ CACGT$ ACGT$ CGT$ GT$ T$ $ A C G T $ 2 G O(m) preproc. time O(n+k) search time T $ 0 Burrows Wheeler transform (BWT) Append character (not part of alphabet) Cyclic Permutations ACACGT$ CACGT$A ACGT$AC CGT$ACA GT$ACAC T$ACACG $ACACGT sort lexicographic $ACACGT ACACGT$ ACGT$AC CACGT$A CGT$ACA GT$ACAC T$ACACG Index of suffix array = S(i) B[i] (6,0,2,1,3,4,5) T$CAACC Compression by move to front encoding on BWT, run length encoding, and variable length prefix code 5
6 Backward search and FM index FM index (Full text index in Minute space) Based on Ferragina and Manzini, FOCS, 2000 Suffix array can be derived from BWT in linear time Idea is to take full advantage of the structure of suffix array for fast searching and BWT to reduce overall space occupancy Count number of occurrences and find all positions of P in text T by looking at only a small portion of the compressed text. Time: O(p), Space: O(n H k (T)) + o(n) Used for sequence alignment in high throughput sequencing applications (e.g. Bowtie) Backward search algorithm First Last BWT $ACACGT ACACGT$ ACGT$AC CACGT$A CGT$ACA GT$ACAC T$ACACG C(c) $ A C G T Occ(c,q) rank $ A C G T
7 Backward search (exact string matching) First CAC Last i=3 CAC i=3 CAC i=2 $ACACGT ACACGT$ ACGT$AC CACGT$A CGT$ACA GT$ACAC T$ACACG F=4 L=5 $ACACGT ACACGT$ ACGT$AC CACGT$A CGT$ACA GT$ACAC T$ACACG F=2 L=3 CAC $ACACGT ACACGT$ ACGT$AC CACGT$A CGT$ACA GT$ACAC T$ACACG F=4 L=4 i=1 CAC $ACACGT ACACGT$ ACGT$AC CACGT$A CGT$ACA GT$ACAC T$ACACG First ACG Last i=3 i=3 ACG i=2 ACG $ACACGT ACACGT$ ACGT$AC CACGT$A CGT$ACA GT$ACAC T$ACACG F=6 L=6 $ACACGT ACACGT$ ACGT$AC CACGT$A CGT$ACA GT$ACAC T$ACACG F=5 L=5 ACG $ACACGT ACACGT$ ACGT$AC CACGT$A CGT$ACA GT$ACAC T$ACACG F=3 L=3 i=1 ACG $ACACGT ACACGT$ ACGT$AC CACGT$A CGT$ACA GT$ACAC T$ACACG Hash table based alignment strategy 7
8 How to map billions of short reads onto genomes Maq Tophat Trapnell C, Salzberg S. Nature Biotech Dot matrix Put one sequence along the top row of a matrix. Put the other sequence along the left column of the matrix. Plot a dot every time there is a match between an element of row sequence and an element of the column sequence. Diagonal lines indicate areas of match. 8
9 Dot matrix Problems with dot matrices Rely on visual analysis Difficult to find optimal alignments Need scoring schemes more sophisticated than identical match Difficult to estimate significance of alignments 9
10 Biology of gaps Gap penalties We expect to penalize gaps the standard cost associated with a gap of length g: Linear gap penalty function (g) (g) = g*d g Convex gap penalty function (more realistic) Affine score: (g) = d (g 1)*e (g) d e gap open penalty gap extend penalty g 10
11 Distant scoring matrices Distances can be calculated between sequences: The higher the distance the smaller the similarity Distances fulfill the properties of a metric: - d(s,t) 0 d(s,t) d(s,u)+d(u,t) d(s,t) = d(t,s) - d(t,s) = 0 <=> t=s Distant scoring matrices Hamming distance: Number of letters in which sequences differ (not valid if the sequences have different length) s AAT AGCAA AGCACACA t TAA ACATA A CACACTA HD(s,t) Levenshtein distance: w(a,a)=0 w(a,b)=1 for a b w(,a)=w(b, )=1 deletion insertion s AGCACAC-A t A CACACTA d(s,t) 2 For two sequences, the distance is unique, but the optimal alignment (the one with minimal cost or distance) is not unique 11
12 Substitutions matrices Unrelated or random model assumes that letter a occurs independently with some frequency qa. P(x,y R) = qxi qxj The alternative match model of aligned pairs of residues occurs with a joint probability pab. P(x,y M) = pxi yi Odds ratio P(x,y M) pxi yi pxi yi = = P(x,y R) qxi qyj qxi qyj Substitution matrices Log odds ratio (score matrix or substitution matrix) S = s(xi,yi) where s(a,b) =log for aligned pair(a,b) s>0 more likely than random, s<0 less likely than random Physical properties of amino acids (e.g. hydrophob vs. hydrophil) are the reason that there are differences in the substitution scores pab qa qb Manually align protein structures (or, more risky, sequences) Look for frequency of amino acid substitutions at structurally nearly constant sites. 12
13 PAM matrices Margaret Dayhoff, 1978 Point Accepted Mutation (PAM) Look at patterns of substitutions in related proteins The new side chain must function the same way as the old one ( acceptance ) On average, 1 PAM corresponds to 1 amino acid change per 100 residues 1 PAM ~ 1% divergence Extrapolate to predict patterns at longer distances Assumptions PAM matrices Replacement is independent of surrounding residues Sequences being compared are of average composition All sites are equally mutable Sources of error Small, globular proteins used to derive matrices (departure from average composition) Errors in PAM 1 are magnified up to PAM 250 Does not account for conserved blocks or motifs 13
14 Henikoff and Henikoff, 1992 BLOSUM matrices Blocks Substitution Matrix (BLOSUM) Look only for differences in conserved, ungapped regions of a protein family More sensitive to structural or functional substitutions BLOSUM n Contribution of sequences > n% identical weighted to 1 Substitution frequencies are more heavily influenced by sequences that are more divergent than this cutoff Clustering reduces contribution of closely related sequences Reducing n yields more distantly related sequences BLOSUM62 14
15 Summary of substitutions matrices Triple PAM strategy (Altschul, 1991) PAM 40 short alignments, highly similar PAM 120 PAM 250 longer, weaker local alignments BLOSUM (Henikoff, 1993) BLOSUM 90 short alignments, highly similar BLOSUM 62 most effective in detecting known members of a protein family BLOSUM 30 longer, weaker local alignments No single matrix is the complete answer for all sequence comparisons Programs like BLAST usually have default matrices! Dynamic programing: Fibonacci numbers function fib(n) fib_table[0] = 1 fib_table[1] = 1 for i in range(3,n): fib_table[i]= fib_table[i-1]+ fib_table[i-2] return fib_table[n] Run in linear time O(n) and constant space O(1) 15
16 Dynamic programing for sequence alignment Global alignment Sequence alignment Needleman Wunsch algorithm Local alignment Smith Waterman algorithm 16
17 Global alignment: Needleman Wunsch algorithm Construct a matrix F(i,j) where i is index from sequence 1 and j is the index from sequence 2 Starting with F(0,0)=0 F(i,j)= max F(i 1,j 1)+s(x i,y j ) F(i 1,j) d F(i,j 1) d substitution matrix gap penalty F(i-1,j-1) s(x i,y j ) F(i-1,j) -d F(i,j-1) -d F(i,j) Example with S=BLOSUM50 and d=8 Global sequence alignment start H E A G A W G H E E P A W H E A E HEAGAWGHE-E --P-AW-HEAE best score 17
18 Backtracing to get optimal alignment Pointer to the choice made at each step Remember all pointers and trace back Time needed O(m*n) Space needed O(m*n) How can this improved? Linear space alignment Do calculate the score for column j only column j 1 is needed H E A G A W G H E E P A W H E A E
19 Local alignment: Smith Waterman algorithm Look for best alignments between subsequences E.g. two proteins sharing a common domain Algorithm is similar to global alignment F(0,j) = F(i,0)=0 F(i,j) = max 0 F(i 1,j 1)+s(x i,y j ) F(i 1,j) d F(i,j 1) d Local alignment H E A G A W G H E E stop P A W H E best score A E AWGHE AW-HE 19
20 Issues for local alignment Time needed O(m*n) Space needed O(m*n) can be brought to O(m+n) Local similarities may occur in sequences with different structure or function that share common substructure/subfunction (domains, motifs) Database search for sequences 20
21 Database search How to answer the query We could just scan the whole database But: Query must be very fast Most sequences will be completely unrelated to query Individual alignment needs not be perfect. Can finetune Exploit nature of the problem If you re going to reject any match with idperc < 90%, then why bother even looking at sequences which don t have a fairly long stretch of matching a.a. in a row. 21
22 W mer indexing Preprocessing: For every W mer (e.g., W=3) store every location in the database where it occurs (can use hashing if W is large) Query: Generate W mers and look them up in the database. Process the results Running time benefit: For W=3, if the sequences are random, then roughly one W mer in 23 3 will match, i.e., one in a ten thousand We hit only a small fraction of all sequences FASTA Use hash table of short words of the database (DB) sequence and query sequence (2 6 chars) For words in query sequence, find similar words in DB using (fast) hash table lookup, and compute R = position(query) position (DB). Areas of long match will show same R for many words. Score matching segments based on content of these matches. Extend the good matches empirically. 22
23 BLAST Finds inexact, ungapped seeds using a hashing technique (like FASTA) and then extends the seed to maximum length possible. Based on strong statistical/significance framework What is a significantly high score of two segments of length N and M? Most commonly used for fast searches and alignments. New versions now do gapped segments. High scoring segment pairs 23
24 High scoring segment pairs Receive query Split query into overlapping words of length W Find neighborhood words for each word until threshold T Look into the table where these neighbor words occur: seeds Extend seeds until score drops off under X Evaluate statistical significance of score Report scores and alignments Significance of scores The number of unrelated matches with score greater than S is approximately Poisson distributed with mean E(S)=Kmne λs where λ is a scaling factor m and n are the length of the sequences The probability that there is a match of score greater than S follows a extreme value distribution: P(x>S)=1 e E(S) Karlin S, Altschul S. Proc Natl Acad Sci (1990) 24
25 NCBI Blast Program Query sequence Subject sequence BLASTN Nucleotide Nucleotide BLASTP Protein Protein BLASTX TBLASTN TBLASTX Nucleotide six frame translation Protein Nucleotide six frame translation Protein Nucleotide six frame translation Nucleotide six frame translation NCBI Blast Example 25
26 Blast Results conserved domain database (CDD) graphical visualization Best hit description E value Score (S) alignment MegaBlast MegaBLAST uses a greedy algorithm for the nucleotide sequence alignment search. This program is optimized for aligning sequences that differ slightly as a result of sequencing or other similar "errors". When larger word size is used (see explanation below), it is up to 10 times faster than more common sequence similarity programs. Mega BLAST is also able to efficiently handle much longer DNA sequences than the blastn program of traditional BLAST algorithm. 26
27 BLAT BLAST like alignment tool UCSC genome browser ( bin/hgblat) Designed to rapidly align longer nucleotide sequences (L 40) having >95% sequence similarity 500 times faster than BLAST for mrna/cdna searches On DNA, Blat works by keeping an index of an entire genome (kmers) in memory. Thus, the target database of BLAT is not a set of GenBank sequences, but instead an index derived from the assembly of the entire genome. It may miss more divergent or short sequence alignments. Can be used also for protein sequences Multiple sequence alignment Often simple extension of pairwise alignment: Given: Set of sequences Match matrix Gap penalties Find: Alignment of sequences such that optimal score is achieved. 27
28 Goals of multiple sequence alignment Determine Consensus Sequences Prosite, emotif ClustalW, MACAW, Pileup, T Coffee Building Gene Families Blocks, Prints, ProDom, pfam, DOMO, eblocks Develop Relationships & Phylogenies Clusters Relationships Evolutionary Models Phylip, GrowTree, MACAW, PAUP Model Protein Structures for Threading and Fold Prediction Profiles, Templates, HSSP, FSSP Hidden Markov Models, pfam, SAM Network Models, Neural Nets, Belief Nets Statistical Models, Generalized Linear Models Exhaustive search using Dynamic Programming Why not just use same technique as for pairwise alignment? Instead of 2 dimensional SCORE matrix, use N dimensional. Fill from one corner to diagonal corner in N dimensions. Complexity increases with number of sequences O(MN), so only N < 10 and lengths (M)~ 200 can be accommodated. 28
29 Dynamic Programming Dynamic Programming 29
30 MSA Algorithm Based on dynamic programming concept: 1. Compute optimal pairwise alignments to get upperbound on any pair of alignments. (MA can t do any better than sum of optimal pairwise alignments.) 2. Create heuristic multiple alignment in ad hoc fashion to create lowerbound on MA score (e.g. align all sequences to the first). 3. Search N dimensional scoring matrix (as in pairwise case) for optimal path, where S[i,j,k ] is the best score including ith element of sequence 1, jth of sequence 2, kth of sequence 3, etc Greedy algorithm 1. Select most similar pair of sequences 2. Join these sequences to build a profile. This reduce the number of sequences/profiles from k to k 1 3. Repeat until only one profile is left this heuristic approach is called greedy algorithm Example: s1 GATTCA s2 GTCTGA s3 GATATT s4 GTCAGC 30
31 Greedy algorithm s2 GTCTGA s4 GTCAGC (score = 2) s1 GAT-TCA s2 G-TCTGA (score = 1) s1 GAT-TCA s3 GATAT-T (score = 1) s1 GATTCA-- s4 G T-CAGC(score = 0) s2 G-TCTGA s3 GATAT-T (score = -1) s3 GAT-ATT s4 G-TCAGC (score = -1) s2 s4 GTCTGA GTCAGC s 2,4 GTCt/aGa/c s 1 s 3 s 2,4 GATTCA GATATT GTCt/aGa/c Progressive Alignment: Tree method 1. Perform hierarchical clustering (similar to arrays) 2. Merge sequences to find ancestor sequences by finding sequences with minimum edit distance to the two children sequences (see next slides). 3. Assign weights to each branch of tree, based on distance between sequences (see next slides) 4. Align sequences (starting from the closest, using a version of dynamic programming) using weights in the score function (see next slides) 31
32 ClustalW 32
33 Markov chains Markov chains: a sequence of events that occur one after another. The main restriction on a Markov chain is that the probability assigned to an event at any location in the chain can depend on only a fixed number of previous events. Scoring sequences (e.g. start codon ATG) 3 states (S1, S2, S3), p(a)=p(c)=p(g)=p(t)=0.25 S1 S2 S3 A T G p(a)=0.91 p(c)=0.03 p(g)=0.03 p(t)=0.03 p(a)=0.03 p(c)=0.03 p(g)=0.03 p(t)=0.91 p(a)=0.03 p(c)=0.03 p(g)=0.91 p(t)=0.03 Markov chain 0 th order p(atg)= =0.752 Markov chain 1 th order p(atg)=p(a)*p(t A)*p(G T) Markov chain What is the probability that we are looking at a start codon, given that the sequence is CTG? P(M CTG)= P(CTG M) * P(M)/P(CTG) (from Bayes theorem) P(CTG)= = P(CTG M)=0.03*0.91*0.91= P(M) not relevant 33
34 Hidden Markov Model (HMM) Example exon intron border 3 states: exon(e), 5 SS (5), intron (I) Emission probabilities HMM parameters (Θ) Given (S) Hidden, want to infer(π) (hidden Markov chain) Find best state path (highest score) If man possible paths than use efficient Viterbi algorithm (based on dynamic programing) G T A A G T C A log P(S,π HMM,Θ)=log(1* * *0.1*0.95*1.0*0.4*0.9*0.4*0.9*0.4*0.9*0.1*0.9*0.4*0.9*0.1*0.9*0.4*0.1) Eddy SR, Nat Biotech 2004 Profile Hidden Markov Model - For multiple alignments (e.g. DNA sequences) ACA---ATG TCAACTATC ACAC--AGC AGA---ATC ACCG--ATC P(A)=0.2 P(C)=0.4 P(G)=0.2 P(T)= Regular Expressions [AT][CG][AC][ACGT]*A[TG][GC] insertion state P(A)=0.8 P(C)=0.0 P(G)=0.0 P(T)=0.2 P(A)=0.0 P(C)=0.8 P(G)=0.2 P(T)=0.0 P(A)=0.8 P(C)=0.2 P(G)=0.0 P(T)=0.0 P(A)=1.0 P(C)=0.0 P(G)=0.0 P(T)=0.0 P(A)=0.0 P(C)=0.0 P(G)=0.2 P(T)=0.8 P(A)=0.0 P(C)=0.8 P(G)=0.2 P(T)=0.0 p(acacatc)=0.8*1*0.8*1*0.8*0.6 *0.4*0.6*1*1*0.8*1*0.8=0.047 log odds=log(p(s)/0.25 L )=log(0.047/ ) 34
35 Profile Hidden Markov Model Allows position dependent gap penalties Can be obtained from a multiple alignment (DNA or Protein) Can be used for searching a database for other members of the family Insert states Delete (silent, null) states Insert states to model highly variable regions in the alignemnt Main states (gray) Avoid overfitting by using pseudocounts (e.g. add 1 to all counts) 35
Biology 644: Bioinformatics
 Find the best alignment between 2 sequences with lengths n and m, respectively Best alignment is very dependent upon the substitution matrix and gap penalties The Global Alignment Problem tries to find
Find the best alignment between 2 sequences with lengths n and m, respectively Best alignment is very dependent upon the substitution matrix and gap penalties The Global Alignment Problem tries to find
As of August 15, 2008, GenBank contained bases from reported sequences. The search procedure should be
 48 Bioinformatics I, WS 09-10, S. Henz (script by D. Huson) November 26, 2009 4 BLAST and BLAT Outline of the chapter: 1. Heuristics for the pairwise local alignment of two sequences 2. BLAST: search and
48 Bioinformatics I, WS 09-10, S. Henz (script by D. Huson) November 26, 2009 4 BLAST and BLAT Outline of the chapter: 1. Heuristics for the pairwise local alignment of two sequences 2. BLAST: search and
An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST
 An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST Alexander Chan 5075504 Biochemistry 218 Final Project An Analysis of Pairwise
An Analysis of Pairwise Sequence Alignment Algorithm Complexities: Needleman-Wunsch, Smith-Waterman, FASTA, BLAST and Gapped BLAST Alexander Chan 5075504 Biochemistry 218 Final Project An Analysis of Pairwise
BLAST & Genome assembly
 BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies May 15, 2014 1 BLAST What is BLAST? The algorithm 2 Genome assembly De novo assembly Mapping assembly 3
BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies May 15, 2014 1 BLAST What is BLAST? The algorithm 2 Genome assembly De novo assembly Mapping assembly 3
CISC 636 Computational Biology & Bioinformatics (Fall 2016)
 CISC 636 Computational Biology & Bioinformatics (Fall 2016) Sequence pairwise alignment Score statistics: E-value and p-value Heuristic algorithms: BLAST and FASTA Database search: gene finding and annotations
CISC 636 Computational Biology & Bioinformatics (Fall 2016) Sequence pairwise alignment Score statistics: E-value and p-value Heuristic algorithms: BLAST and FASTA Database search: gene finding and annotations
BLAST MCDB 187. Friday, February 8, 13
 BLAST MCDB 187 BLAST Basic Local Alignment Sequence Tool Uses shortcut to compute alignments of a sequence against a database very quickly Typically takes about a minute to align a sequence against a database
BLAST MCDB 187 BLAST Basic Local Alignment Sequence Tool Uses shortcut to compute alignments of a sequence against a database very quickly Typically takes about a minute to align a sequence against a database
24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, This lecture is based on the following papers, which are all recommended reading:
 24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, 2010 3 BLAST and FASTA This lecture is based on the following papers, which are all recommended reading: D.J. Lipman and W.R. Pearson, Rapid
24 Grundlagen der Bioinformatik, SS 10, D. Huson, April 26, 2010 3 BLAST and FASTA This lecture is based on the following papers, which are all recommended reading: D.J. Lipman and W.R. Pearson, Rapid
Bioinformatics for Biologists
 Bioinformatics for Biologists Sequence Analysis: Part I. Pairwise alignment and database searching Fran Lewitter, Ph.D. Director Bioinformatics & Research Computing Whitehead Institute Topics to Cover
Bioinformatics for Biologists Sequence Analysis: Part I. Pairwise alignment and database searching Fran Lewitter, Ph.D. Director Bioinformatics & Research Computing Whitehead Institute Topics to Cover
Basic Local Alignment Search Tool (BLAST)
 BLAST 26.04.2018 Basic Local Alignment Search Tool (BLAST) BLAST (Altshul-1990) is an heuristic Pairwise Alignment composed by six-steps that search for local similarities. The most used access point to
BLAST 26.04.2018 Basic Local Alignment Search Tool (BLAST) BLAST (Altshul-1990) is an heuristic Pairwise Alignment composed by six-steps that search for local similarities. The most used access point to
.. Fall 2011 CSC 570: Bioinformatics Alexander Dekhtyar..
 .. Fall 2011 CSC 570: Bioinformatics Alexander Dekhtyar.. PAM and BLOSUM Matrices Prepared by: Jason Banich and Chris Hoover Background As DNA sequences change and evolve, certain amino acids are more
.. Fall 2011 CSC 570: Bioinformatics Alexander Dekhtyar.. PAM and BLOSUM Matrices Prepared by: Jason Banich and Chris Hoover Background As DNA sequences change and evolve, certain amino acids are more
Bioinformatics explained: BLAST. March 8, 2007
 Bioinformatics Explained Bioinformatics explained: BLAST March 8, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com Bioinformatics
Bioinformatics Explained Bioinformatics explained: BLAST March 8, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com Bioinformatics
BLAST, Profile, and PSI-BLAST
 BLAST, Profile, and PSI-BLAST Jianlin Cheng, PhD School of Electrical Engineering and Computer Science University of Central Florida 26 Free for academic use Copyright @ Jianlin Cheng & original sources
BLAST, Profile, and PSI-BLAST Jianlin Cheng, PhD School of Electrical Engineering and Computer Science University of Central Florida 26 Free for academic use Copyright @ Jianlin Cheng & original sources
Computational Molecular Biology
 Computational Molecular Biology Erwin M. Bakker Lecture 3, mainly from material by R. Shamir [2] and H.J. Hoogeboom [4]. 1 Pairwise Sequence Alignment Biological Motivation Algorithmic Aspect Recursive
Computational Molecular Biology Erwin M. Bakker Lecture 3, mainly from material by R. Shamir [2] and H.J. Hoogeboom [4]. 1 Pairwise Sequence Alignment Biological Motivation Algorithmic Aspect Recursive
Lecture Overview. Sequence search & alignment. Searching sequence databases. Sequence Alignment & Search. Goals: Motivations:
 Lecture Overview Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating
Lecture Overview Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating
Sequence Alignment & Search
 Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating the first version
Sequence Alignment & Search Karin Verspoor, Ph.D. Faculty, Computational Bioscience Program University of Colorado School of Medicine With credit and thanks to Larry Hunter for creating the first version
Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence or library of DNA.
 Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence or library of DNA. Fasta is used to compare a protein or DNA sequence to all of the
Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence or library of DNA. Fasta is used to compare a protein or DNA sequence to all of the
PROTEIN MULTIPLE ALIGNMENT MOTIVATION: BACKGROUND: Marina Sirota
 Marina Sirota MOTIVATION: PROTEIN MULTIPLE ALIGNMENT To study evolution on the genetic level across a wide range of organisms, biologists need accurate tools for multiple sequence alignment of protein
Marina Sirota MOTIVATION: PROTEIN MULTIPLE ALIGNMENT To study evolution on the genetic level across a wide range of organisms, biologists need accurate tools for multiple sequence alignment of protein
Lecture 10: Local Alignments
 Lecture 10: Local Alignments Study Chapter 6.8-6.10 1 Outline Edit Distances Longest Common Subsequence Global Sequence Alignment Scoring Matrices Local Sequence Alignment Alignment with Affine Gap Penalties
Lecture 10: Local Alignments Study Chapter 6.8-6.10 1 Outline Edit Distances Longest Common Subsequence Global Sequence Alignment Scoring Matrices Local Sequence Alignment Alignment with Affine Gap Penalties
Scoring and heuristic methods for sequence alignment CG 17
 Scoring and heuristic methods for sequence alignment CG 17 Amino Acid Substitution Matrices Used to score alignments. Reflect evolution of sequences. Unitary Matrix: M ij = 1 i=j { 0 o/w Genetic Code Matrix:
Scoring and heuristic methods for sequence alignment CG 17 Amino Acid Substitution Matrices Used to score alignments. Reflect evolution of sequences. Unitary Matrix: M ij = 1 i=j { 0 o/w Genetic Code Matrix:
Similarity searches in biological sequence databases
 Similarity searches in biological sequence databases Volker Flegel september 2004 Page 1 Outline Keyword search in databases General concept Examples SRS Entrez Expasy Similarity searches in databases
Similarity searches in biological sequence databases Volker Flegel september 2004 Page 1 Outline Keyword search in databases General concept Examples SRS Entrez Expasy Similarity searches in databases
MULTIPLE SEQUENCE ALIGNMENT
 MULTIPLE SEQUENCE ALIGNMENT Multiple Alignment versus Pairwise Alignment Up until now we have only tried to align two sequences. What about more than two? A faint similarity between two sequences becomes
MULTIPLE SEQUENCE ALIGNMENT Multiple Alignment versus Pairwise Alignment Up until now we have only tried to align two sequences. What about more than two? A faint similarity between two sequences becomes
Database Searching Using BLAST
 Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
Mahidol University Objectives SCMI512 Molecular Sequence Analysis Database Searching Using BLAST Lecture 2B After class, students should be able to: explain the FASTA algorithm for database searching explain
TCCAGGTG-GAT TGCAAGTGCG-T. Local Sequence Alignment & Heuristic Local Aligners. Review: Probabilistic Interpretation. Chance or true homology?
 Local Sequence Alignment & Heuristic Local Aligners Lectures 18 Nov 28, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 12:00-1:20 Johnson Hall
Local Sequence Alignment & Heuristic Local Aligners Lectures 18 Nov 28, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 12:00-1:20 Johnson Hall
Computational Genomics and Molecular Biology, Fall
 Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
Computational Genomics and Molecular Biology, Fall 2015 1 Sequence Alignment Dannie Durand Pairwise Sequence Alignment The goal of pairwise sequence alignment is to establish a correspondence between the
Bioinformatics. Sequence alignment BLAST Significance. Next time Protein Structure
 Bioinformatics Sequence alignment BLAST Significance Next time Protein Structure 1 Experimental origins of sequence data The Sanger dideoxynucleotide method F Each color is one lane of an electrophoresis
Bioinformatics Sequence alignment BLAST Significance Next time Protein Structure 1 Experimental origins of sequence data The Sanger dideoxynucleotide method F Each color is one lane of an electrophoresis
B L A S T! BLAST: Basic local alignment search tool. Copyright notice. February 6, Pairwise alignment: key points. Outline of tonight s lecture
 February 6, 2008 BLAST: Basic local alignment search tool B L A S T! Jonathan Pevsner, Ph.D. Introduction to Bioinformatics pevsner@jhmi.edu 4.633.0 Copyright notice Many of the images in this powerpoint
February 6, 2008 BLAST: Basic local alignment search tool B L A S T! Jonathan Pevsner, Ph.D. Introduction to Bioinformatics pevsner@jhmi.edu 4.633.0 Copyright notice Many of the images in this powerpoint
Introduction to Computational Molecular Biology
 18.417 Introduction to Computational Molecular Biology Lecture 13: October 21, 2004 Scribe: Eitan Reich Lecturer: Ross Lippert Editor: Peter Lee 13.1 Introduction We have been looking at algorithms to
18.417 Introduction to Computational Molecular Biology Lecture 13: October 21, 2004 Scribe: Eitan Reich Lecturer: Ross Lippert Editor: Peter Lee 13.1 Introduction We have been looking at algorithms to
FASTA. Besides that, FASTA package provides SSEARCH, an implementation of the optimal Smith- Waterman algorithm.
 FASTA INTRODUCTION Definition (by David J. Lipman and William R. Pearson in 1985) - Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence
FASTA INTRODUCTION Definition (by David J. Lipman and William R. Pearson in 1985) - Compares a sequence of protein to another sequence or database of a protein, or a sequence of DNA to another sequence
Sequence analysis Pairwise sequence alignment
 UMF11 Introduction to bioinformatics, 25 Sequence analysis Pairwise sequence alignment 1. Sequence alignment Lecturer: Marina lexandersson 12 September, 25 here are two types of sequence alignments, global
UMF11 Introduction to bioinformatics, 25 Sequence analysis Pairwise sequence alignment 1. Sequence alignment Lecturer: Marina lexandersson 12 September, 25 here are two types of sequence alignments, global
CS 284A: Algorithms for Computational Biology Notes on Lecture: BLAST. The statistics of alignment scores.
 CS 284A: Algorithms for Computational Biology Notes on Lecture: BLAST. The statistics of alignment scores. prepared by Oleksii Kuchaiev, based on presentation by Xiaohui Xie on February 20th. 1 Introduction
CS 284A: Algorithms for Computational Biology Notes on Lecture: BLAST. The statistics of alignment scores. prepared by Oleksii Kuchaiev, based on presentation by Xiaohui Xie on February 20th. 1 Introduction
Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014
 Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014 Dynamic programming is a group of mathematical methods used to sequentially split a complicated problem into
Dynamic Programming User Manual v1.0 Anton E. Weisstein, Truman State University Aug. 19, 2014 Dynamic programming is a group of mathematical methods used to sequentially split a complicated problem into
BLAST & Genome assembly
 BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies November 17, 2012 1 Introduction Introduction 2 BLAST What is BLAST? The algorithm 3 Genome assembly De
BLAST & Genome assembly Solon P. Pissis Tomáš Flouri Heidelberg Institute for Theoretical Studies November 17, 2012 1 Introduction Introduction 2 BLAST What is BLAST? The algorithm 3 Genome assembly De
Sequence alignment theory and applications Session 3: BLAST algorithm
 Sequence alignment theory and applications Session 3: BLAST algorithm Introduction to Bioinformatics online course : IBT Sonal Henson Learning Objectives Understand the principles of the BLAST algorithm
Sequence alignment theory and applications Session 3: BLAST algorithm Introduction to Bioinformatics online course : IBT Sonal Henson Learning Objectives Understand the principles of the BLAST algorithm
From Smith-Waterman to BLAST
 From Smith-Waterman to BLAST Jeremy Buhler July 23, 2015 Smith-Waterman is the fundamental tool that we use to decide how similar two sequences are. Isn t that all that BLAST does? In principle, it is
From Smith-Waterman to BLAST Jeremy Buhler July 23, 2015 Smith-Waterman is the fundamental tool that we use to decide how similar two sequences are. Isn t that all that BLAST does? In principle, it is
Dynamic Programming in 3-D Progressive Alignment Profile Progressive Alignment (ClustalW) Scoring Multiple Alignments Entropy Sum of Pairs Alignment
 Dynamic Programming in 3-D Progressive Alignment Profile Progressive Alignment (ClustalW) Scoring Multiple Alignments Entropy Sum of Pairs Alignment Partial Order Alignment (POA) A-Bruijin (ABA) Approach
Dynamic Programming in 3-D Progressive Alignment Profile Progressive Alignment (ClustalW) Scoring Multiple Alignments Entropy Sum of Pairs Alignment Partial Order Alignment (POA) A-Bruijin (ABA) Approach
Short Read Alignment. Mapping Reads to a Reference
 Short Read Alignment Mapping Reads to a Reference Brandi Cantarel, Ph.D. & Daehwan Kim, Ph.D. BICF 05/2018 Introduction to Mapping Short Read Aligners DNA vs RNA Alignment Quality Pitfalls and Improvements
Short Read Alignment Mapping Reads to a Reference Brandi Cantarel, Ph.D. & Daehwan Kim, Ph.D. BICF 05/2018 Introduction to Mapping Short Read Aligners DNA vs RNA Alignment Quality Pitfalls and Improvements
Heuristic methods for pairwise alignment:
 Bi03c_1 Unit 03c: Heuristic methods for pairwise alignment: k-tuple-methods k-tuple-methods for alignment of pairs of sequences Bi03c_2 dynamic programming is too slow for large databases Use heuristic
Bi03c_1 Unit 03c: Heuristic methods for pairwise alignment: k-tuple-methods k-tuple-methods for alignment of pairs of sequences Bi03c_2 dynamic programming is too slow for large databases Use heuristic
Sequence Alignment. GBIO0002 Archana Bhardwaj University of Liege
 Sequence Alignment GBIO0002 Archana Bhardwaj University of Liege 1 What is Sequence Alignment? A sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity.
Sequence Alignment GBIO0002 Archana Bhardwaj University of Liege 1 What is Sequence Alignment? A sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity.
Profiles and Multiple Alignments. COMP 571 Luay Nakhleh, Rice University
 Profiles and Multiple Alignments COMP 571 Luay Nakhleh, Rice University Outline Profiles and sequence logos Profile hidden Markov models Aligning profiles Multiple sequence alignment by gradual sequence
Profiles and Multiple Alignments COMP 571 Luay Nakhleh, Rice University Outline Profiles and sequence logos Profile hidden Markov models Aligning profiles Multiple sequence alignment by gradual sequence
Pairwise Sequence Alignment. Zhongming Zhao, PhD
 Pairwise Sequence Alignment Zhongming Zhao, PhD Email: zhongming.zhao@vanderbilt.edu http://bioinfo.mc.vanderbilt.edu/ Sequence Similarity match mismatch A T T A C G C G T A C C A T A T T A T G C G A T
Pairwise Sequence Alignment Zhongming Zhao, PhD Email: zhongming.zhao@vanderbilt.edu http://bioinfo.mc.vanderbilt.edu/ Sequence Similarity match mismatch A T T A C G C G T A C C A T A T T A T G C G A T
Principles of Bioinformatics. BIO540/STA569/CSI660 Fall 2010
 Principles of Bioinformatics BIO540/STA569/CSI660 Fall 2010 Lecture 11 Multiple Sequence Alignment I Administrivia Administrivia The midterm examination will be Monday, October 18 th, in class. Closed
Principles of Bioinformatics BIO540/STA569/CSI660 Fall 2010 Lecture 11 Multiple Sequence Alignment I Administrivia Administrivia The midterm examination will be Monday, October 18 th, in class. Closed
BGGN 213 Foundations of Bioinformatics Barry Grant
 BGGN 213 Foundations of Bioinformatics Barry Grant http://thegrantlab.org/bggn213 Recap From Last Time: 25 Responses: https://tinyurl.com/bggn213-02-f17 Why ALIGNMENT FOUNDATIONS Why compare biological
BGGN 213 Foundations of Bioinformatics Barry Grant http://thegrantlab.org/bggn213 Recap From Last Time: 25 Responses: https://tinyurl.com/bggn213-02-f17 Why ALIGNMENT FOUNDATIONS Why compare biological
Sequence alignment is an essential concept for bioinformatics, as most of our data analysis and interpretation techniques make use of it.
 Sequence Alignments Overview Sequence alignment is an essential concept for bioinformatics, as most of our data analysis and interpretation techniques make use of it. Sequence alignment means arranging
Sequence Alignments Overview Sequence alignment is an essential concept for bioinformatics, as most of our data analysis and interpretation techniques make use of it. Sequence alignment means arranging
Multiple Sequence Alignment. Mark Whitsitt - NCSA
 Multiple Sequence Alignment Mark Whitsitt - NCSA What is a Multiple Sequence Alignment (MA)? GMHGTVYANYAVDSSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKQPHV GMHGTVYANYAVEHSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKTPHV
Multiple Sequence Alignment Mark Whitsitt - NCSA What is a Multiple Sequence Alignment (MA)? GMHGTVYANYAVDSSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKQPHV GMHGTVYANYAVEHSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKTPHV
Read Mapping. Slides by Carl Kingsford
 Read Mapping Slides by Carl Kingsford Bowtie Ultrafast and memory-efficient alignment of short DNA sequences to the human genome Ben Langmead, Cole Trapnell, Mihai Pop and Steven L Salzberg, Genome Biology
Read Mapping Slides by Carl Kingsford Bowtie Ultrafast and memory-efficient alignment of short DNA sequences to the human genome Ben Langmead, Cole Trapnell, Mihai Pop and Steven L Salzberg, Genome Biology
Lecture 5 Advanced BLAST
 Introduction to Bioinformatics for Medical Research Gideon Greenspan gdg@cs.technion.ac.il Lecture 5 Advanced BLAST BLAST Recap Sequence Alignment Complexity and indexing BLASTN and BLASTP Basic parameters
Introduction to Bioinformatics for Medical Research Gideon Greenspan gdg@cs.technion.ac.il Lecture 5 Advanced BLAST BLAST Recap Sequence Alignment Complexity and indexing BLASTN and BLASTP Basic parameters
Similarity Searches on Sequence Databases
 Similarity Searches on Sequence Databases Lorenza Bordoli Swiss Institute of Bioinformatics EMBnet Course, Zürich, October 2004 Swiss Institute of Bioinformatics Swiss EMBnet node Outline Importance of
Similarity Searches on Sequence Databases Lorenza Bordoli Swiss Institute of Bioinformatics EMBnet Course, Zürich, October 2004 Swiss Institute of Bioinformatics Swiss EMBnet node Outline Importance of
Wilson Leung 01/03/2018 An Introduction to NCBI BLAST. Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment
 An Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at https://blast.ncbi.nlm.nih.gov/blast.cgi
An Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at https://blast.ncbi.nlm.nih.gov/blast.cgi
Lecture 10. Sequence alignments
 Lecture 10 Sequence alignments Alignment algorithms: Overview Given a scoring system, we need to have an algorithm for finding an optimal alignment for a pair of sequences. We want to maximize the score
Lecture 10 Sequence alignments Alignment algorithms: Overview Given a scoring system, we need to have an algorithm for finding an optimal alignment for a pair of sequences. We want to maximize the score
6.047 / Computational Biology: Genomes, Networks, Evolution Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, Networks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, Networks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
EECS730: Introduction to Bioinformatics
 EECS730: Introduction to Bioinformatics Lecture 06: Multiple Sequence Alignment https://upload.wikimedia.org/wikipedia/commons/thumb/7/79/rplp0_90_clustalw_aln.gif/575px-rplp0_90_clustalw_aln.gif Slides
EECS730: Introduction to Bioinformatics Lecture 06: Multiple Sequence Alignment https://upload.wikimedia.org/wikipedia/commons/thumb/7/79/rplp0_90_clustalw_aln.gif/575px-rplp0_90_clustalw_aln.gif Slides
Algorithmic Approaches for Biological Data, Lecture #20
 Algorithmic Approaches for Biological Data, Lecture #20 Katherine St. John City University of New York American Museum of Natural History 20 April 2016 Outline Aligning with Gaps and Substitution Matrices
Algorithmic Approaches for Biological Data, Lecture #20 Katherine St. John City University of New York American Museum of Natural History 20 April 2016 Outline Aligning with Gaps and Substitution Matrices
BLAST: Basic Local Alignment Search Tool Altschul et al. J. Mol Bio CS 466 Saurabh Sinha
 BLAST: Basic Local Alignment Search Tool Altschul et al. J. Mol Bio. 1990. CS 466 Saurabh Sinha Motivation Sequence homology to a known protein suggest function of newly sequenced protein Bioinformatics
BLAST: Basic Local Alignment Search Tool Altschul et al. J. Mol Bio. 1990. CS 466 Saurabh Sinha Motivation Sequence homology to a known protein suggest function of newly sequenced protein Bioinformatics
CISC 889 Bioinformatics (Spring 2003) Multiple Sequence Alignment
 CISC 889 Bioinformatics (Spring 2003) Multiple Sequence Alignment Courtesy of jalview 1 Motivations Collective statistic Protein families Identification and representation of conserved sequence features
CISC 889 Bioinformatics (Spring 2003) Multiple Sequence Alignment Courtesy of jalview 1 Motivations Collective statistic Protein families Identification and representation of conserved sequence features
Algorithms in Bioinformatics: A Practical Introduction. Database Search
 Algorithms in Bioinformatics: A Practical Introduction Database Search Biological databases Biological data is double in size every 15 or 16 months Increasing in number of queries: 40,000 queries per day
Algorithms in Bioinformatics: A Practical Introduction Database Search Biological databases Biological data is double in size every 15 or 16 months Increasing in number of queries: 40,000 queries per day
COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1. Database Searching
 COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1 Database Searching In database search, we typically have a large sequence database
COS 551: Introduction to Computational Molecular Biology Lecture: Oct 17, 2000 Lecturer: Mona Singh Scribe: Jacob Brenner 1 Database Searching In database search, we typically have a large sequence database
NGS Data and Sequence Alignment
 Applications and Servers SERVER/REMOTE Compute DB WEB Data files NGS Data and Sequence Alignment SSH WEB SCP Manpreet S. Katari App Aug 11, 2016 Service Terminal IGV Data files Window Personal Computer/Local
Applications and Servers SERVER/REMOTE Compute DB WEB Data files NGS Data and Sequence Alignment SSH WEB SCP Manpreet S. Katari App Aug 11, 2016 Service Terminal IGV Data files Window Personal Computer/Local
Biologically significant sequence alignments using Boltzmann probabilities
 Biologically significant sequence alignments using Boltzmann probabilities P Clote Department of Biology, Boston College Gasson Hall 16, Chestnut Hill MA 0267 clote@bcedu Abstract In this paper, we give
Biologically significant sequence alignments using Boltzmann probabilities P Clote Department of Biology, Boston College Gasson Hall 16, Chestnut Hill MA 0267 clote@bcedu Abstract In this paper, we give
ICB Fall G4120: Introduction to Computational Biology. Oliver Jovanovic, Ph.D. Columbia University Department of Microbiology
 ICB Fall 2008 G4120: Computational Biology Oliver Jovanovic, Ph.D. Columbia University Department of Microbiology Copyright 2008 Oliver Jovanovic, All Rights Reserved. The Digital Language of Computers
ICB Fall 2008 G4120: Computational Biology Oliver Jovanovic, Ph.D. Columbia University Department of Microbiology Copyright 2008 Oliver Jovanovic, All Rights Reserved. The Digital Language of Computers
Giri Narasimhan. CAP 5510: Introduction to Bioinformatics. ECS 254; Phone: x3748
 CAP 5510: Introduction to Bioinformatics Giri Narasimhan ECS 254; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs07.html 1/30/07 CAP5510 1 BLAST & FASTA FASTA [Lipman, Pearson 85, 88]
CAP 5510: Introduction to Bioinformatics Giri Narasimhan ECS 254; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs07.html 1/30/07 CAP5510 1 BLAST & FASTA FASTA [Lipman, Pearson 85, 88]
Lecture 4: January 1, Biological Databases and Retrieval Systems
 Algorithms for Molecular Biology Fall Semester, 1998 Lecture 4: January 1, 1999 Lecturer: Irit Orr Scribe: Irit Gat and Tal Kohen 4.1 Biological Databases and Retrieval Systems In recent years, biological
Algorithms for Molecular Biology Fall Semester, 1998 Lecture 4: January 1, 1999 Lecturer: Irit Orr Scribe: Irit Gat and Tal Kohen 4.1 Biological Databases and Retrieval Systems In recent years, biological
Chapter 6. Multiple sequence alignment (week 10)
 Course organization Introduction ( Week 1,2) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 3)» Algorithm complexity analysis
Course organization Introduction ( Week 1,2) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 3)» Algorithm complexity analysis
Machine Learning. Computational biology: Sequence alignment and profile HMMs
 10-601 Machine Learning Computational biology: Sequence alignment and profile HMMs Central dogma DNA CCTGAGCCAACTATTGATGAA transcription mrna CCUGAGCCAACUAUUGAUGAA translation Protein PEPTIDE 2 Growth
10-601 Machine Learning Computational biology: Sequence alignment and profile HMMs Central dogma DNA CCTGAGCCAACTATTGATGAA transcription mrna CCUGAGCCAACUAUUGAUGAA translation Protein PEPTIDE 2 Growth
BLAST - Basic Local Alignment Search Tool
 Lecture for ic Bioinformatics (DD2450) April 11, 2013 Searching 1. Input: Query Sequence 2. Database of sequences 3. Subject Sequence(s) 4. Output: High Segment Pairs (HSPs) Sequence Similarity Measures:
Lecture for ic Bioinformatics (DD2450) April 11, 2013 Searching 1. Input: Query Sequence 2. Database of sequences 3. Subject Sequence(s) 4. Output: High Segment Pairs (HSPs) Sequence Similarity Measures:
Chapter 8 Multiple sequence alignment. Chaochun Wei Spring 2018
 1896 1920 1987 2006 Chapter 8 Multiple sequence alignment Chaochun Wei Spring 2018 Contents 1. Reading materials 2. Multiple sequence alignment basic algorithms and tools how to improve multiple alignment
1896 1920 1987 2006 Chapter 8 Multiple sequence alignment Chaochun Wei Spring 2018 Contents 1. Reading materials 2. Multiple sequence alignment basic algorithms and tools how to improve multiple alignment
BLAST. Basic Local Alignment Search Tool. Used to quickly compare a protein or DNA sequence to a database.
 BLAST Basic Local Alignment Search Tool Used to quickly compare a protein or DNA sequence to a database. There is no such thing as a free lunch BLAST is fast and highly sensitive compared to competitors.
BLAST Basic Local Alignment Search Tool Used to quickly compare a protein or DNA sequence to a database. There is no such thing as a free lunch BLAST is fast and highly sensitive compared to competitors.
Lecture 2 Pairwise sequence alignment. Principles Computational Biology Teresa Przytycka, PhD
 Lecture 2 Pairwise sequence alignment. Principles Computational Biology Teresa Przytycka, PhD Assumptions: Biological sequences evolved by evolution. Micro scale changes: For short sequences (e.g. one
Lecture 2 Pairwise sequence alignment. Principles Computational Biology Teresa Przytycka, PhD Assumptions: Biological sequences evolved by evolution. Micro scale changes: For short sequences (e.g. one
Alignment of Pairs of Sequences
 Bi03a_1 Unit 03a: Alignment of Pairs of Sequences Partners for alignment Bi03a_2 Protein 1 Protein 2 =amino-acid sequences (20 letter alphabeth + gap) LGPSSKQTGKGS-SRIWDN LN-ITKSAGKGAIMRLGDA -------TGKG--------
Bi03a_1 Unit 03a: Alignment of Pairs of Sequences Partners for alignment Bi03a_2 Protein 1 Protein 2 =amino-acid sequences (20 letter alphabeth + gap) LGPSSKQTGKGS-SRIWDN LN-ITKSAGKGAIMRLGDA -------TGKG--------
Sequence Alignment Heuristics
 Sequence Alignment Heuristics Some slides from: Iosif Vaisman, GMU mason.gmu.edu/~mmasso/binf630alignment.ppt Serafim Batzoglu, Stanford http://ai.stanford.edu/~serafim/ Geoffrey J. Barton, Oxford Protein
Sequence Alignment Heuristics Some slides from: Iosif Vaisman, GMU mason.gmu.edu/~mmasso/binf630alignment.ppt Serafim Batzoglu, Stanford http://ai.stanford.edu/~serafim/ Geoffrey J. Barton, Oxford Protein
Efficient Implementation of a Generalized Pair HMM for Comparative Gene Finding. B. Majoros M. Pertea S.L. Salzberg
 Efficient Implementation of a Generalized Pair HMM for Comparative Gene Finding B. Majoros M. Pertea S.L. Salzberg ab initio gene finder genome 1 MUMmer Whole-genome alignment (optional) ROSE Region-Of-Synteny
Efficient Implementation of a Generalized Pair HMM for Comparative Gene Finding B. Majoros M. Pertea S.L. Salzberg ab initio gene finder genome 1 MUMmer Whole-genome alignment (optional) ROSE Region-Of-Synteny
Distributed Protein Sequence Alignment
 Distributed Protein Sequence Alignment ABSTRACT J. Michael Meehan meehan@wwu.edu James Hearne hearne@wwu.edu Given the explosive growth of biological sequence databases and the computational complexity
Distributed Protein Sequence Alignment ABSTRACT J. Michael Meehan meehan@wwu.edu James Hearne hearne@wwu.edu Given the explosive growth of biological sequence databases and the computational complexity
Chapter 4: Blast. Chaochun Wei Fall 2014
 Course organization Introduction ( Week 1-2) Course introduction A brief introduction to molecular biology A brief introduction to sequence comparison Part I: Algorithms for Sequence Analysis (Week 3-11)
Course organization Introduction ( Week 1-2) Course introduction A brief introduction to molecular biology A brief introduction to sequence comparison Part I: Algorithms for Sequence Analysis (Week 3-11)
3.4 Multiple sequence alignment
 3.4 Multiple sequence alignment Why produce a multiple sequence alignment? Using more than two sequences results in a more convincing alignment by revealing conserved regions in ALL of the sequences Aligned
3.4 Multiple sequence alignment Why produce a multiple sequence alignment? Using more than two sequences results in a more convincing alignment by revealing conserved regions in ALL of the sequences Aligned
Divya R. Singh. Faster Sequence Alignment using Suffix Tree and Data-Mining Techniques. February A Thesis Presented by
 Faster Sequence Alignment using Suffix Tree and Data-Mining Techniques A Thesis Presented by Divya R. Singh to The Faculty of the Graduate College of the University of Vermont In Partial Fulfillment of
Faster Sequence Alignment using Suffix Tree and Data-Mining Techniques A Thesis Presented by Divya R. Singh to The Faculty of the Graduate College of the University of Vermont In Partial Fulfillment of
Wilson Leung 05/27/2008 A Simple Introduction to NCBI BLAST
 A Simple Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at http://www.ncbi.nih.gov/blast/
A Simple Introduction to NCBI BLAST Prerequisites: Detecting and Interpreting Genetic Homology: Lecture Notes on Alignment Resources: The BLAST web server is available at http://www.ncbi.nih.gov/blast/
Alignment of Long Sequences
 Alignment of Long Sequences BMI/CS 776 www.biostat.wisc.edu/bmi776/ Spring 2009 Mark Craven craven@biostat.wisc.edu Pairwise Whole Genome Alignment: Task Definition Given a pair of genomes (or other large-scale
Alignment of Long Sequences BMI/CS 776 www.biostat.wisc.edu/bmi776/ Spring 2009 Mark Craven craven@biostat.wisc.edu Pairwise Whole Genome Alignment: Task Definition Given a pair of genomes (or other large-scale
15-780: Graduate Artificial Intelligence. Computational biology: Sequence alignment and profile HMMs
 5-78: Graduate rtificial Intelligence omputational biology: Sequence alignment and profile HMMs entral dogma DN GGGG transcription mrn UGGUUUGUG translation Protein PEPIDE 2 omparison of Different Organisms
5-78: Graduate rtificial Intelligence omputational biology: Sequence alignment and profile HMMs entral dogma DN GGGG transcription mrn UGGUUUGUG translation Protein PEPIDE 2 omparison of Different Organisms
C E N T R. Introduction to bioinformatics 2007 E B I O I N F O R M A T I C S V U F O R I N T. Lecture 13 G R A T I V. Iterative homology searching,
 C E N T R E F O R I N T E G R A T I V E B I O I N F O R M A T I C S V U Introduction to bioinformatics 2007 Lecture 13 Iterative homology searching, PSI (Position Specific Iterated) BLAST basic idea use
C E N T R E F O R I N T E G R A T I V E B I O I N F O R M A T I C S V U Introduction to bioinformatics 2007 Lecture 13 Iterative homology searching, PSI (Position Specific Iterated) BLAST basic idea use
COMBINATORIAL PATTERN MATCHING
 COMBINATORIAL PATTERN MATCHING OUTLINE: EXACT MATCHING Tabulating patterns in long texts Short patterns (direct indexing) Longer patterns (hash tables) Finding exact patterns in a text Brute force (run
COMBINATORIAL PATTERN MATCHING OUTLINE: EXACT MATCHING Tabulating patterns in long texts Short patterns (direct indexing) Longer patterns (hash tables) Finding exact patterns in a text Brute force (run
FINDING APPROXIMATE REPEATS WITH MULTIPLE SPACED SEEDS
 FINDING APPROXIMATE REPEATS WITH MULTIPLE SPACED SEEDS FINDING APPROXIMATE REPEATS IN DNA SEQUENCES USING MULTIPLE SPACED SEEDS By SARAH BANYASSADY, B.S. A Thesis Submitted to the School of Graduate Studies
FINDING APPROXIMATE REPEATS WITH MULTIPLE SPACED SEEDS FINDING APPROXIMATE REPEATS IN DNA SEQUENCES USING MULTIPLE SPACED SEEDS By SARAH BANYASSADY, B.S. A Thesis Submitted to the School of Graduate Studies
Today s Lecture. Multiple sequence alignment. Improved scoring of pairwise alignments. Affine gap penalties Profiles
 Today s Lecture Multiple sequence alignment Improved scoring of pairwise alignments Affine gap penalties Profiles 1 The Edit Graph for a Pair of Sequences G A C G T T G A A T G A C C C A C A T G A C G
Today s Lecture Multiple sequence alignment Improved scoring of pairwise alignments Affine gap penalties Profiles 1 The Edit Graph for a Pair of Sequences G A C G T T G A A T G A C C C A C A T G A C G
Alignment ABC. Most slides are modified from Serafim s lectures
 Alignment ABC Most slides are modified from Serafim s lectures Complete genomes Evolution Evolution at the DNA level C ACGGTGCAGTCACCA ACGTTGCAGTCCACCA SEQUENCE EDITS REARRANGEMENTS Sequence conservation
Alignment ABC Most slides are modified from Serafim s lectures Complete genomes Evolution Evolution at the DNA level C ACGGTGCAGTCACCA ACGTTGCAGTCCACCA SEQUENCE EDITS REARRANGEMENTS Sequence conservation
Brief review from last class
 Sequence Alignment Brief review from last class DNA is has direction, we will use only one (5 -> 3 ) and generate the opposite strand as needed. DNA is a 3D object (see lecture 1) but we will model it
Sequence Alignment Brief review from last class DNA is has direction, we will use only one (5 -> 3 ) and generate the opposite strand as needed. DNA is a 3D object (see lecture 1) but we will model it
GLOBEX Bioinformatics (Summer 2015) Multiple Sequence Alignment
 GLOBEX Bioinformatics (Summer 2015) Multiple Sequence Alignment Scoring Dynamic Programming algorithms Heuristic algorithms CLUSTAL W Courtesy of jalview Motivations Collective (or aggregate) statistic
GLOBEX Bioinformatics (Summer 2015) Multiple Sequence Alignment Scoring Dynamic Programming algorithms Heuristic algorithms CLUSTAL W Courtesy of jalview Motivations Collective (or aggregate) statistic
Pairwise Sequence Alignment: Dynamic Programming Algorithms. COMP Spring 2015 Luay Nakhleh, Rice University
 Pairwise Sequence Alignment: Dynamic Programming Algorithms COMP 571 - Spring 2015 Luay Nakhleh, Rice University DP Algorithms for Pairwise Alignment The number of all possible pairwise alignments (if
Pairwise Sequence Alignment: Dynamic Programming Algorithms COMP 571 - Spring 2015 Luay Nakhleh, Rice University DP Algorithms for Pairwise Alignment The number of all possible pairwise alignments (if
Short Read Alignment Algorithms
 Short Read Alignment Algorithms Raluca Gordân Department of Biostatistics and Bioinformatics Department of Computer Science Department of Molecular Genetics and Microbiology Center for Genomic and Computational
Short Read Alignment Algorithms Raluca Gordân Department of Biostatistics and Bioinformatics Department of Computer Science Department of Molecular Genetics and Microbiology Center for Genomic and Computational
Bioinformatics explained: Smith-Waterman
 Bioinformatics Explained Bioinformatics explained: Smith-Waterman May 1, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com
Bioinformatics Explained Bioinformatics explained: Smith-Waterman May 1, 2007 CLC bio Gustav Wieds Vej 10 8000 Aarhus C Denmark Telephone: +45 70 22 55 09 Fax: +45 70 22 55 19 www.clcbio.com info@clcbio.com
Stephen Scott.
 1 / 33 sscott@cse.unl.edu 2 / 33 Start with a set of sequences In each column, residues are homolgous Residues occupy similar positions in 3D structure Residues diverge from a common ancestral residue
1 / 33 sscott@cse.unl.edu 2 / 33 Start with a set of sequences In each column, residues are homolgous Residues occupy similar positions in 3D structure Residues diverge from a common ancestral residue
Lecture 5: Multiple sequence alignment
 Lecture 5: Multiple sequence alignment Introduction to Computational Biology Teresa Przytycka, PhD (with some additions by Martin Vingron) Why do we need multiple sequence alignment Pairwise sequence alignment
Lecture 5: Multiple sequence alignment Introduction to Computational Biology Teresa Przytycka, PhD (with some additions by Martin Vingron) Why do we need multiple sequence alignment Pairwise sequence alignment
Sequence Alignment AGGCTATCACCTGACCTCCAGGCCGATGCCC TAGCTATCACGACCGCGGTCGATTTGCCCGAC -AGGCTATCACCTGACCTCCAGGCCGA--TGCCC--
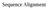 Sequence Alignment Sequence Alignment AGGCTATCACCTGACCTCCAGGCCGATGCCC TAGCTATCACGACCGCGGTCGATTTGCCCGAC -AGGCTATCACCTGACCTCCAGGCCGA--TGCCC-- TAG-CTATCAC--GACCGC--GGTCGATTTGCCCGAC Distance from sequences
Sequence Alignment Sequence Alignment AGGCTATCACCTGACCTCCAGGCCGATGCCC TAGCTATCACGACCGCGGTCGATTTGCCCGAC -AGGCTATCACCTGACCTCCAGGCCGA--TGCCC-- TAG-CTATCAC--GACCGC--GGTCGATTTGCCCGAC Distance from sequences
Proceedings of the 11 th International Conference for Informatics and Information Technology
 Proceedings of the 11 th International Conference for Informatics and Information Technology Held at Hotel Molika, Bitola, Macedonia 11-13th April, 2014 Editors: Vangel V. Ajanovski Gjorgji Madjarov ISBN
Proceedings of the 11 th International Conference for Informatics and Information Technology Held at Hotel Molika, Bitola, Macedonia 11-13th April, 2014 Editors: Vangel V. Ajanovski Gjorgji Madjarov ISBN
Multiple sequence alignment. November 20, 2018
 Multiple sequence alignment November 20, 2018 Why do multiple alignment? Gain insight into evolutionary history Can assess time of divergence by looking at the number of mutations needed to change one
Multiple sequence alignment November 20, 2018 Why do multiple alignment? Gain insight into evolutionary history Can assess time of divergence by looking at the number of mutations needed to change one
INTRODUCTION TO BIOINFORMATICS
 Molecular Biology-2019 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
Molecular Biology-2019 1 INTRODUCTION TO BIOINFORMATICS In this section, we want to provide a simple introduction to using the web site of the National Center for Biotechnology Information NCBI) to obtain
Pairwise alignment II
 Pairwise alignment II Agenda - Previous Lesson: Minhala + Introduction - Review Dynamic Programming - Pariwise Alignment Biological Motivation Today: - Quick Review: Sequence Alignment (Global, Local,
Pairwise alignment II Agenda - Previous Lesson: Minhala + Introduction - Review Dynamic Programming - Pariwise Alignment Biological Motivation Today: - Quick Review: Sequence Alignment (Global, Local,
Biochemistry 324 Bioinformatics. Multiple Sequence Alignment (MSA)
 Biochemistry 324 Bioinformatics Multiple Sequence Alignment (MSA) Big- Οh notation Greek omicron symbol Ο The Big-Oh notation indicates the complexity of an algorithm in terms of execution speed and storage
Biochemistry 324 Bioinformatics Multiple Sequence Alignment (MSA) Big- Οh notation Greek omicron symbol Ο The Big-Oh notation indicates the complexity of an algorithm in terms of execution speed and storage
Database Similarity Searching
 An Introduction to Bioinformatics BSC4933/ISC5224 Florida State University Feb. 23, 2009 Database Similarity Searching Steven M. Thompson Florida State University of Department Scientific Computing How
An Introduction to Bioinformatics BSC4933/ISC5224 Florida State University Feb. 23, 2009 Database Similarity Searching Steven M. Thompson Florida State University of Department Scientific Computing How
Outline. Sequence Alignment. Types of Sequence Alignment. Genomics & Computational Biology. Section 2. How Computers Store Information
 enomics & omputational Biology Section Lan Zhang Sep. th, Outline How omputers Store Information Sequence lignment Dot Matrix nalysis Dynamic programming lobal: NeedlemanWunsch lgorithm Local: SmithWaterman
enomics & omputational Biology Section Lan Zhang Sep. th, Outline How omputers Store Information Sequence lignment Dot Matrix nalysis Dynamic programming lobal: NeedlemanWunsch lgorithm Local: SmithWaterman
FastA and the chaining problem, Gunnar Klau, December 1, 2005, 10:
 FastA and the chaining problem, Gunnar Klau, December 1, 2005, 10:56 4001 4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem
FastA and the chaining problem, Gunnar Klau, December 1, 2005, 10:56 4001 4 FastA and the chaining problem We will discuss: Heuristics used by the FastA program for sequence alignment Chaining problem
Multiple Sequence Alignment II
 Multiple Sequence Alignment II Lectures 20 Dec 5, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 12:00-1:20 Johnson Hall (JHN) 022 1 Outline
Multiple Sequence Alignment II Lectures 20 Dec 5, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 12:00-1:20 Johnson Hall (JHN) 022 1 Outline
Reconstructing long sequences from overlapping sequence fragment. Searching databases for related sequences and subsequences
 SEQUENCE ALIGNMENT ALGORITHMS 1 Why compare sequences? Reconstructing long sequences from overlapping sequence fragment Searching databases for related sequences and subsequences Storing, retrieving and
SEQUENCE ALIGNMENT ALGORITHMS 1 Why compare sequences? Reconstructing long sequences from overlapping sequence fragment Searching databases for related sequences and subsequences Storing, retrieving and
