Visualization strategies for major white matter tracts for intraoperative use
|
|
|
- Winfred Daniel
- 5 years ago
- Views:
Transcription
1 Int J CARS (2006) 1:13 22 DOI /s ORIGINAL ARTICLE Visualization strategies for major white matter tracts for intraoperative use Christopher Nimsky Oliver Ganslandt Frank Enders Dorit Merhof Thilo Hammen Michael Buchfelder Published online: 21 February 2006 CARS 2006 Abstract Streamline representation of major fiber tract systems along with high-resolution anatomical data provides a reliable orientation for the neurosurgeon. For intraoperative visualization of these data either on navigation screens near the surgical field or directly in the surgical field applying heads-up displays of operating microscopes, wrapping of all streamlines of interest to render an individual object representing the whole fiber bundle is the most suitable representation. Integration of fiber tract data into a neuronavigation setup allows removal of tumors adjacent to eloquent brain areas with low morbidity. Keywords Diffusion tensor imaging Glyph representation Fiber tracking Functional neuronavigation visualization strategies Major white matter tracts Introduction In white matter of the brain, diffusion is anisotropic, i.e. the water diffusion is not equal in all three orthogonal directions. C. Nimsky (B) O. Ganslandt M. Buchfelder Department of Neurosurgery, University Erlangen Nuremberg, Erlangen, Germany nimsky@nch.imed.uni-erlangen.de URL: Phone: Fax: C. Nimsky O. Ganslandt F. Enders D. Merhof T. Hammen M. Buchfelder Neurocenter, University Erlangen Nuremberg, Erlangen, Germany T. Hammen Department of Neurology, University Erlangen Nuremberg, Erlangen, Germany This is due to the strong aligned microstructure, including cell membranes and the myelin sheath surrounding myelinated white matter, causing impediment of the water motion. Isotropic diffusion can be graphically represented as a sphere, whereas anisotropic diffusion can be graphically expressed as an ellipsoid, with water molecules moving farther along the long axis of a fiber bundle and less movement perpendicularly. To estimate the nine tensor matrix elements required for a Gaussian description of water mobility, the diffusion gradient must be applied to at least six noncollinear directions. The eigenvalues represent the three principal diffusion coefficients measured along the three coordinate directions of the ellipsoid. The eigenvectors represent the directions of the tensor. Diffusion tensor imaging (DTI) can resolve the dominant fiber orientation in each voxel element. The direction of greatest diffusion measured by DTI parallels the dominant orientation of the tissue structure in each voxel, representing the mean longitudinal direction of axons in white matter tracts. DTI can be applied to identify major white matter tracts, such as the pyramidal tract or the visual pathway [1,6,17]. Avoiding postoperative neurological deficits by preserving eloquent brain areas is a major principle in neurosurgery. Cortical eloquent brain areas can be preserved successfully by identification of these areas by methods such as magnetoencephalography (MEG) and functional magnetic resonance imaging (fmri). Registration of MEG and fmri data with three-dimensional (3D) anatomical MR datasets allows representation of the spatial information of the identified cortical eloquent brain areas in the surgical field by using neuronavigation systems [9,15,22]. Heads-up displays visualizing segmented data in the surgical field are routinely used in socalled microscope-based neuronavigation. While MEG and fmri only identify cortical eloquent brain areas, DTI can be used to assess the major white matter tracts connecting to
2 14 Int J CARS (2006) 1:13 22 these cortical brain areas [4,10,11]. These major white matter tracts have also to be preserved during surgery to avoid postoperative deficits. Different visualization techniques of DTI data, such as glyph representation and fiber tracking have evolved in the recent years. The aim of this study is to find a suitable method to visualize the course of major white matter tracts during neurosurgical procedures. Methods Single-shot spin-echo diffusion weighted echo planar imaging on a 1.5 T MR scanner (Magnetom Sonata, Siemens Medical Solutions, Erlangen, Germany) was used for DTI. The sequence parameters were: TE 86 ms, TR 9,200 ms, matrix size , FOV 240 mm, slice thickness 1.9 mm, bandwidth 1,502 Hz/pixel. A diffusion weighting of 1,000 s/mm 2 (high b value) was used. One null image (low b value: 0s/mm 2 ) and six diffusion weighted images were obtained with the diffusion-encoding gradients directed along the following axes (±1,1,0), (±1,0,1), and (1,±1,0). The voxel size was mm 3 ; 60 slices with no intersection gap were measured. Applying five averages, the total DTI measurement required 5 min and 31 s. Directly after image acquisition, the DTI data could be evaluated with the DTI task card version 1.7 (Magnetic Resonance Center, Massachusetts General Hospital, Boston) on a Siemens scanner using MR software MRease N4_VA21B under syngo VB10I. The diffusion tensor information could then be represented as color-encoded fractional anisotropy (FA) maps, which were generated by mapping the principal eigenvector components into red, green, and blue color channels, weighted by fractional anisotropy. Assuming the patient is lying in supine position and the head is not tilted, then the color mapping defines white matter tracts oriented in an anterior/posterior direction in green, a left/right direction in red, and a superior/inferior direction in blue (Fig. 1). Furthermore, 3-D fibertracking was generated applying a knowledge-based multiple-roi (region of interest) approach with user-defined seed regions. Tracking was initiated in both retro- and orthograde directions according to the direction of the principal eigenvector in each voxel of the ROI. These data could be displayed on a screen in the operating theater. Fiber tracts were visualized as streamtubes along the B0 images in coronal, sagittal or axial orientation (Fig. 2). In parallel DTI data were displayed using an in-house visualizing platform, allowing generating different glyph and fibertract representations, such as evenly spaced streamlines (Fig. 3). Additionally standard anatomical image data could be registered, so that glyph or streamline fiber tract visualization could be displayed along standard anatomical image data (Fig. 4). To integrate fibertracking in a navigation system (iplan 2.5, BrainLab, Heimstetten, Germany) the b = 0 images were rigidly registered with the T1-weighted MPRAGE data (magnetization prepared rapid acquisition gradient echo sequence; TE 4.38 ms, TR 2020 ms, matrix size , FOV 250 mm, slice thickness 1 mm, slab 16 cm, measurement time 8 min 39 s). For fiber tracking, we implemented a tracking algorithm based on a local diffusion approach. The algorithm is based on a tensor deflection algorithm where the trend of the current generated fibers is also considered, which was first described by Lazar, Weinstein, and Westin [16,28,29]. In contrast to the syngo approach (see above) tract seeding is performed by defining a rectangular volume of interest (VOI) either in the FA maps or in the co-registered standard anatomical datasets. The definition of the VOI depends on the fiber structures to be displayed. A raster of a third of the voxel size is applied in the VOI to define the starting points. The tract is propagated both in the forward and reverse directions of the major eigenvector, so that each fiber consists of two iterative calculated segments beginning at the starting point. After termination of the iteration process, the calculated fibers must meet all general conditions regarding threshold, local angulation, and total fiber length. Before tracking is initiated, the user can adjust the FA threshold and the minimum fiber length. The final result of the tracking calculations is a parametric display of fibers, which are represented as streamlines, using the standard direction color encoding: left right oriented fibers are displayed in red, anterior posterior in green, and cranio caudal in blue (Fig. 5a, b). For further selecting the fiber bundle of interest, it is possible to define a VOI in the initially tracked fibers, so that among these, VOI fibers can be retained, excluded, and deleted. This selection of fibers of interest can be repeated as often as necessary, allowing an interactive selection of complex volumes of interest to distinguish single fiber bundles, disconnect branching fibers, or to remove entire areas (Fig. 5c, d). For representation in the navigation software, the maximum outline of the fiber tracts was segmented automatically (Fig. 6), so that this object wrapping all fibers is displayed as a single object and could be displayed on the navigation screen (Fig. 6d), as well as in the surgical field. Results In over 50 patients, most of them undergoing glioma surgery, DTI data were visualized for intraoperative use. Figs. 1, 2, 3, 4, 5, and 6 depict typical examples of the different tools and visualization strategies in the same patient. Table 1 summarizes the clinical suitability for the different visualization techniques.
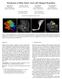
3 Int J CARS (2006) 1: Fig. 1 Diffusion tensor imaging visualization on a 28-year-old female patient with a lesion (white arrow) close to the left pyramidal tract, that was biopsied: a b = 0 image; b fractional anisotropy (FA) map; c color-encoded FA map; d glyph representation of the same FA map by ellipsoids; e glyph representation of the same slice as quadrics; f enlarged view of (e) Fig. 2 Fiber tract visualization in the same patient applying a streamtube representation displayed along the b = 0 images in standard projection (a coronal; b sagittal; c axial) The DTI task card integrated in the MR scanner syngo interface allowed, besides standard display of FA maps (Fig. 1), an immediate visualization of the major fiber tracts (Fig. 2), so that this information could be displayed during neurosurgical procedures, even if these data were acquired intraoperatively. However, this visualization lacked the parallel display of high-resolution anatomy, so that the direct mental transfer of the course of the fiber tracts to the surgical field depended much on the clinical experience of the individual surgeon. Fractional anisotropy map representation (Fig. 1) and glyph representation (Fig. 3) of the data was only used to define the seed regions for the tracking algorithm. This visualization method is of value and interest when displaying the tumor border and comparing this information to other modalities such as MR spectroscopy or PET. Furthermore, FA maps are of great value, when DTI data are used to investigate invasion of major white matter tracts by a tumor by analysis of the various DTI parameters.
4 16 Int J CARS (2006) 1:13 22 Fig. 3 Whole head DTI representation, the lesion is segmented in red; a glyph visualization; b d streamline visualization, applying different techniques. b Standard tracking with filtering; c without filtering; d applying evenly spaced streamlines Fig. 4 Simultaneous display of DTI data and standard image data, glyphs or streamlines are displayed along a three-dimensional (3D) rendering of a T1-weighted 3D dataset (a glyphs; b streamlines along the lesion segmented in red).
5 Int J CARS (2006) 1: Fig. 5 Implementation of fiber tracking in the navigation software; a initial tracking starting from seed points in a large volume of interest (VOI) covering the left hemisphere; b enlarged view of the streamline representation of (a); c after excluding some fibers, the fibers belonging to the left pyramidal tract are visualized; d enlarged view of the streamline representation of (c) (note: the axial slice are visualized viewing from above and the coronal slice is visualized viewing from behind) Fiber tract representation of DTI data was the preferred display mode (Fig. 2). However, the standard display with smoothed streamlines, suggested a high accuracy and resolution of the data, which effectively is not provided by the voxel size of mm 3 of the actually measured data. What was most important was the simultaneous display of fiber tract data along with standard anatomical images, allowing a clear spatial orientation (Figs. 4 and 5). Integration of DTI information into navigational systems, either by registration of the color-encoded FA maps and manual segmentation of the respective fiber tracts by an expert (applied in 16 glioma patients) or by integrating the reconstructed streamlines in the navigational setup (n = 20) allowed a direct visualization of the pyramidal tract or the optic radiation in the surgical field. The fiber tract generation and integration into the navigation system needed about min in each case. Integrating the reconstructed streamlines in the navigational setup was less time consuming and less user dependent, i.e. less biased than the approach applying the direct registration of color-encoded FA maps. Final visualization of a 3-D representing the major white matter tract of interest proved to be most suitable for intraoperative visuali-
6 18 Int J CARS (2006) 1:13 22 Fig. 6 Generation of a 3D object representing the left pyramidal tract; a final tracking result with streamline representation; b automaticcomputation of a hull wrapping all fibers simultaneous display; c display of the 3D object without any streamlines; d actual intraoperative screenshot from the navigation screen depicting the 3D object visualization during surgery (note: the axial slice are visualized viewing from above and the coronal slice is visualized viewing from behind) Table 1 Different visualization techniques of diffusion tensor imaging (DTI) data Detailed analysis Spatial analysis Preoperative planning Intraoperative visualization Fractional anisotropy maps + O Glyphs + O Volume growing O Fiber tracking + + Evenly spaced streamlines + + Hulls Three-dimensional (3D) objects
7 Int J CARS (2006) 1: zation (Fig. 6). For the visualization in the surgical field, using the heads-up display systems of navigation microscopes a smooth wrapping of all fibers of interest proved to be of major importance, otherwise the displayed structures (contours) in the surgical field could not be related easily to the structures of interest. Diffusion tensor imaging data navigation allowed extended resections with low new postoperative deficits; only in one patient (2.8%; 1 of 36) we observed a permanent new postoperative neurological deterioration. Discussion Diffusion tensor imaging data are of increasing interest for neurosurgeons since DTI provides information about the normal course, the displacement, or interruption of white matter tracts around a tumor, as well as a widening of fiber bundles due to edema or tumor infiltration can be detected [3,4,10, 18,25,30]. Various visualization approaches have emerged in the recent years, however, upto now there is no common standard how DTI data are presented during neurosurgical procedures. In the following the major different visualization strategies are discussed. Glyphs The simplification of a tensor to a scalar metric reduces the amount of information contained in the tensor. A possibility to show the entire information of the second order tensors is to use glyphs. Several shapes have been presented for representing tensors using glyphs [24]. A popular approach is to use ellipsoids whereas the axes of the ellipsoid correlate with the directions of the eigenvectors and are scaled according to the corresponding eigenvalues. An advantage of tensor ellipsoids is that they can be rendered in real-time using graphics hardware [7]. An even more satisfying shape for tensors are superquadric tensor glyphs which provide a better and less ambiguous spatial impression [14]. However, superquadrics are computationally much more expensive and are hard to achieve in real-time. The advantage of glyph-based techniques is that the entire tensor information is visualized. Therefore, it is especially appropriate for a detailed examination of the data. However, the results of this visualization approach are difficult to interpret in terms of underlying major white matter structures since no global connectivity information is provided. Consequently, a combination with other visualization techniques such as fiber tracing is recommended to obtain a meaningful representation. In addition, clipping or slice views of glyphs are necessary to avoid an overloading and cluttering of information in 3D [7,14]. Fiber tracking Fiber tracking which is maybe the most appealing and understandable technique for representing white matter has been investigated by several groups [2,8,20,26,27,31]. All these approaches are based on streamline techniques known from flow visualization. Thereby, the respective vector field is derived by taking the major eigenvector of each tensor. Fiber tracking algorithms often utilize thresholds, angle criterions, regularization techniques and local filters to improve tracking results. In standard fiber tracking approaches, the streamlines extracted by tracking algorithms are assumed to represent the most likely pathways through the tensor field. Note that the term fibers is used for streamlines which do not represent real anatomical fibers but provide an abstract model of neural structures. Starting from seed voxels, the tracking is performed in forward and backward direction with sub-voxel precision. For the selection of seed voxels and for aborting the streamline propagation, FA is used as a threshold. FA represents the degree of anisotropic diffusion and therefore is a proper measure for the probability of white matter. Following this assumption, voxels with high FA are used as seed voxels. If FA falls below a certain threshold, the tracking stops. Accordingly, a single tracking step of the streamline propagation looks as follows: The tensor at the current end point of the fiber is computed using trilinear interpolation which is separately performed for each tensor entry. The eigensystem of the tensor is then determined. The eigenvector belonging to the highest eigenvalue, in the following referred to as principal eigenvector, correlates with the direction of highest diffusion. In case of Euler integration, the next streamline propagation step would be in direction of this principal eigenvector. For reasons of numerical accuracy, we apply a higher order integration scheme (Runge-Kutta of order 4) which needs repeated tensor interpolation and principal eigenvector computations until the direction of streamline propagation is determined. The step size is set to a fixed value which is a quarter of the voxel size. Since the field of the principal eigenvectors does not correspond to a flow field, we found it more convenient to choose a sufficiently low fixed step size instead of adaptive adjustments. Contrary to real fibers, particle flow poses physical properties such as inertia which ensure that sudden changes of direction do not occur. For fiber tracking, a fixed step size is safer to prevent missing turnoffs. Apart from the FA threshold for aborting fiber tracking, streamline propagation is aborted in the subsequently described cases. If a streamline has reached a maximum length or if the angle between the last two steps is above a certain threshold, tracking stops. A further criterion for accepting a fiber is its length. Streamlines are rejected if they are below a minimum length. Beside these thresholds, the user may choose between a tracking encompassing the whole brain and a tracking extracting
8 20 Int J CARS (2006) 1:13 22 fibers that run through user-defined regions of interest. The latter approach enables the reconstruction of separate tract systems which is of special interest to delineate major white matter tracts. Meanwhile standard fiber tracking techniques provide a straightforward possibility to visualize major white matter tracts and their spatial relation to intracerebral lesions [6, 13,21]. However, a lot of software tools allow only tracking combined with the display of the diffusion data; co-registration with standard anatomical data is a must for an intuitive analysis in space-occupying lesions. Evenly spaced streamlines Due to the diverging nature of tract systems, the density of streamlines varies over the domain without control resulting in sparse areas as well as cramped regions. To overcome this problem, the concept of evenly spaced streamlines [12] was adapted to fiber tracking providing streamlines equally distributed over the domain. An adaptive control of the distance between separate streamlines depending on the magnitude of anisotropic diffusion provides a mechanism to emphasize dominant tract systems. The basic principle of evenly spaced streamlines is to calculate streamlines until a user-defined density level is reached. Thereby, a regular distribution of the streamlines is achieved and areas with a sparse distribution of streamlines are filled. In the context of fiber tracking, this is a highly desirable property to capture all features, i.e. the location of white matter, of the diffusion tensor field. Upto now, this technique does not provide further advantages compared to standard techniques, looked at from the intraoperative visualization viewpoint. Evenly based streamlines are an interesting tool to further investigate tracking techniques per se and may be interesting to more user-independent extract major white matter tracts. Streamtubes The visualization of streamlines is solved by rendering lines or more significant tubes. However, utilizing simple line rendering has several disadvantages. In case of curved fibers, lines thicker than one pixel suffer from gaps between neighboring segments. Furthermore, a constant width independent of the zoom restricts the quality of visualization. Tubes, on the other hand, enable a significantly improved visualization, but rendering becomes much more time-consuming due to the high geometric complexity restricting their interactive application. Alternatively, streamline visualizations which are very similar to tubes are available providing interactive rendering times since no complex geometry is necessary. Hulls So far, all presented approaches are motivated by the exploration of the diffusion tensor data. Unfortunately, they do not satisfy the needs of neurosurgical planning. In this case, the problem is the exact determination of the border of a specific major white tract. Line representation lacks the ability to provide a border. Instead, it is the job of the user to interpret the visualization as a model for the tract. The generation of hulls is a possibility to overcome this issue [7]. Thereby, a surface is generated which wraps a particular subset of previously computed streamlines, representing a certain nerve tract. The wrapping itself is based on the determination of a centerline of the bundle and the subsequent construction of bounding curves around the set of corresponding lines. Finally, a mesh is generated by connecting the curves, representing the surface of the nerve tract model. The major advantage of this method is the intuitive visualization. The results look like flexible tubes which are closely related to the expected appearance of a major white matter bundle. The borders are depicted directly and the combination with volume, rendering of anatomical MR data provides a good spatial orientation. It must be noted that the surface is entirely dependent on the previously performed fiber tracking. Errors in the streamline calculation have immediate influence on the resulting hull. Volume growing In general, volume growing algorithms start from a predefined seed region and spread out within the volume until some terminating criterion is reached. Similarly to ROI-based fiber tracking, volume growing needs an initial ROI for starting the growing procedure. The algorithm starts from each voxel within the ROI and proceeds to neighboring voxels. In the same way as for fiber tracking, the growing process stops as soon as a FA value below a specified threshold is reached. In addition, directional volume growing takes into account the shape of the local tensor which controls the direction in which the process expands [19]. The advantage of volume growing approaches over isosurface-based approaches is that a connected volume is generated. The fragmented nature of an isosurface based on the FA volume makes it difficult to clearly identify connectivity information which is important for surgery. In contrast, volume growing produces a connected volume without any detached parts. Illumination A general problem of all 3D visualization approaches is the projection into two-dimensional (2D) screen space. To overcome this limitation, some further dodges are necessary.
9 Int J CARS (2006) 1: Beside the perspective projection, the appliance of light is an important factor to receive a good 3D impression. Thereby, the color of a structure is modified dependent on the orientation with respect to the light source. Further on, specular highlights and shadows significantly contribute to the visual perception of 3D structures. In general, this is also valid for the case of DTI visualization. Hulls and glyphs highly benefit from the use of illumination since they have a spatial structure and a surface. In particular, detailed surface structures, as they appear on hulls, can hardly be distinguished by the user without the application of light. In contrast to that, the use of light in combination with lines is problematic. Since lines do not have an explicit surface, illumination can only be achieved by the simulation of infinitesimal small tubes. For correlating lines, this works satisfactorily and the highlights look expectedly. In case of turbulent lines or line bundles which fan out, the results look extremely scattered and the illumination is counterproductive [32]. Clinical application and intraoperative use Integration of DTI data into navigation systems allowing an immediate visualization in the surgical field has resulted in resections with low morbidity [5,13,23]. We have successfully implemented a fiber tracking algorithm in a common navigation software environment, allowing broad availability and providing a common platform to compare the results of different groups. Our current study showed that for intraoperative visualization, a single object representing the major white matter tract of interest, such as e.g. the pyramidal tract, proved to be most efficient for a reliable intraoperative orientation. This 3D object can be displayed along with other objects representing the tumor outline on the navigation screen, as well as it can be superimposed as contour in the surgical field. Direct visualization of fiber tract data as sophisticated 3D renderings applying illuminated streamtubes may be an impressive view for a layperson, however, superimposition of these detailed fiber tract information, suggesting an accuracy of the data, that in fact is not present, would obscure the view for the surgeon and prevent concentrating on the most critical structures. This is the area where a tumor is reaching the nearest point of a major white matter tract system, because entering major white matter tracts has to be avoided during surgery to prevent postoperative new neurological deficits. This explains why the representation as a 3D object is sufficient for intraoperative use. A promising approach for an even more sophisticated visualization is adding layers to the hulls of the 3D object with a thickness defined by safety margins. These additional hulls would allow visualizing the quality of the data, as well as representing safety corridors. So a thick hull in case of noisy unreliable data, or a thin hull in the area of high reliability would be added. This would allow that the neurosurgeon has an immediate impression to what extent the displayed data can be trusted during surgery. The technical, as well as clinical definition of the extent of these safety margins has yet to be established. When integrating DTI data in a neuronavigational setup, the brain shift has to be compensated for. Intraoperative fiber tracking revealed a marked shifting of major white matter tracts during tumor resection [21]. These intraoperative DTI data can be visualized during surgery with the techniques described above; the integration of fiber tracking in standard neuronavigation system makes it even feasible that fiber tract data are update during surgery. The ideal platform would combine the different features: allowing basic display and analysis of FA maps, including numeric evaluation of the different diffusion parameters, which may be of special interest when looking at the relation of a major white matter tract system and a tumor in respect to edema and tumor invasion; upto integration and co-registration of data from various other modalities, allowing generation of streamtubes and 3D glyph representations, as well as features defining hulls wrapping major white matter tracts of interest. Furthermore, automatic clustering techniques, facilitating the identification of the white matter tract of interest, should be established. All these data should be integrated in a navigational setup environment, so that the different information can be displayed during surgery in adapted detail and representation. The visualization strategy has to avoid disturbing the neurosurgeon form the surgical field, i.e. preventing an information overflow; however, complex data have to be presented in a hierarchical fashion, so that they can be easily used during surgery. Conclusion Streamline representation of major fiber tract systems along with high-resolution anatomical data provides a reliable orientation for the neurosurgeon. For direct visualization of these data in the surgical field applying heads-up displays of operating microscopes, wrapping of all streamlines of interest to render an individual object representing the whole fiber bundle is mandatory. Integration of fiber tract data into a neuronavigation setup allows removal of tumors adjacent to eloquent brain areas with low morbidity. Acknowledgements We express our special thanks to Greg Sorensen (Dept. Radiology, MGH, Boston, USA) for providing the DTI syngo taskcard software, as well as to G. Ogrezeanu, U. Mezger, and T. Seiler (BrainLab, Heimstetten, Germany) for implementation of the fiber tracking software in the navigation environment; Theodor Vetter, Ph.D., and Michael Zwanger, Ph.D. (Siemens Medical Solutions, Erlangen, Germany) for their continuous technical advice, as well as to Stefanie Kreckel, RT, for her technical support in MR imaging. This work was supported in part by the Deutsche Forschungsgemeinschaft in the context of project C9 of SFB603.
10 22 Int J CARS (2006) 1:13 22 References 1. Basser PJ, Mattiello J, LeBihan D (1994) MR diffusion tensor spectroscopy and imaging. Biophys J 66: Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A (2000) In vivo fiber tractography using DT-MRI data. Magn Reson Med 44: Berman JI, Berger MS, Mukherjee P, Henry RG (2004) Diffusiontensor imaging-guided tracking of fibers of the pyramidal tract combined with intraoperative cortical stimulation mapping in patients with gliomas. J Neurosurg 101: Clark CA, Barrick TR, Murphy MM, Bell BA (2003) White matter fiber tracking in patients with space-occupying lesions of the brain: a new technique for neurosurgical planning? Neuroimage 20: Coenen VA, Krings T, Mayfrank L, Polin RS, Reinges MH, Thron A, Gilsbach JM (2001) Three-dimensional visualization of the pyramidal tract in a neuronavigation system during brain tumor surgery: first experiences and technical note. Neurosurgery 49: Cruz Junior LC, Sorensen AG (2005) Diffusion tensor magnetic resonance imaging of brain tumors. Neurosurg Clin N Am 16: Enders F, Iserhardt-Bauer S, Hastreiter P, Nimsky C, Ertl T (2005) Hardware-accelerated glyph based visualization of major white matter tracts for analysis of brain tumors. In: Galloway RJ, Cleary K (eds) Medical imaging 2005: visualization, image-guided procedures, and display, Proceedings of SPIE, vol 5744, Bellingham, WA, pp Fillard P, Gilmore J, Lin W, Gerig G (2003) Quantitative analysis of white matter fiber properties along geodesic paths. In: Ellis RE, Peters TM (eds) medical image computing and computer-assisted intervention MICCAI Springer, Berlin Heidelberg New York, pp Ganslandt O, Fahlbusch R, Nimsky C, Kober H, Möller M, Steinmeier R, Romstöck J, Vieth J (1999) Functional neuronavigation with magnetoencephalography: outcome in 50 patients with lesions around the motor cortex. J Neurosurg 91: Hendler T, Pianka P, Sigal M, Kafri M, Ben-Bashat D, Constantini S, Graif M, Fried I, Assaf Y (2003) Delineating gray and white matter involvement in brain lesions: three-dimensional alignment of functional magnetic resonance and diffusion-tensor imaging. J Neurosurg 99: Holodny AI, Schwartz TH, Ollenschleger M, Liu WC, Schulder M (2001) Tumor involvement of the corticospinal tract: diffusion magnetic resonance tractography with intraoperative correlation. J Neurosurg 95: Jobard B, Lefer W (1997) Creating evenlyspaced streamlines of arbitrary density. Visualization in scientific computing 97. In: Proceedings of the Eurographics workshop in Boulogne-sur-Mer, France. Springer, Berlin Heidelberg New York, pp Kamada K, Todo T, Masutani Y, Aoki S, Ino K, Takano T, Kirino T, Kawahara N, Morita A (2005) Combined use of tractographyintegrated functional neuronavigation and direct fiber stimulation. J Neurosurg 102: Kindlmann G (2004) Superquadric tensor glyphs. In: Proceedings of Eurographics - IEEE TCVG symposium on visualization, pp Kober H, Nimsky C, Möller M, Hastreiter P, Fahlbusch R, Ganslandt O (2001) Correlation of sensorimotor activation with functional magnetic resonance imaging and magnetoencephalography in presurgical functional imaging: a spatial analysis. Neuroimage 14: Lazar M, Weinstein DM, Tsuruda JS, Hasan KM, Arfanakis K, Meyerand ME, Badie B, Rowley HA, Haughton V, Field A, Alexander AL (2003) White matter tractography using diffusion tensor deflection. Hum Brain Mapp 18: Le Bihan D, Van Zijl P (2002) From the diffusion coefficient to the diffusion tensor. NMR Biomed 15: Lu S, Ahn D, Johnson G, Cha S (2003) Peritumoral diffusion tensor imaging of high-grade gliomas and metastatic brain tumors. AJNR Am J Neuroradiol 24: Merhof D, Hastreiter P, Nimsky C, Fahlbusch R, Greiner G (2005) Directional volume growing for the extraction of white matter tracts from diffusion tensor data. In: Galloway RJ, Cleary K (eds) Medical imaging 2005: visualization, image-guided procedures, and display, Proceedings of SPIE, vol 5744, Bellingham, WA, USA, pp Mori S, Van Zijl PC (2002) Fiber tracking: principles and strategies a technical review. NMR Biomed 15: Nimsky C, Ganslandt O, Hastreiter P, Wang R, Benner T, Sorensen AG, Fahlbusch R (2005) Preoperative and intraoperative diffusion tensor imaging-based fiber tracking in glioma surgery. Neurosurgery 56: Nimsky C, Ganslandt O, Kober H, Möller M, Ulmer S, Tomandl B, Fahlbusch R (1999) Integration of functional magnetic resonance imaging supported by magnetoencephalography in functional neuronavigation. Neurosurgery 44: Nimsky C, Grummich P, Sorensen AG, Fahlbusch R, Ganslandt O (2005) Visualization of the pyramidal tract in glioma surgery by integrating diffusion tensor imaging in functional neuronavigation. Zentralbl Neurochir 66: Pierpaoli C, Jezzard P, Basser PJ, Barnett A, Di Chiro G (1996) Diffusion tensor MR imaging of the human brain. Radiology 201: Price S, Burnet N, Donovan T, Green H, Pena A, Antoun N, Pickard J, Carpenter T, Gillard J (2003) Diffusion tensor imaging of brain tumours at 3T: a potential tool for assessing white matter tract invasion? Clin Radiol 58: Stieltjes B, Kaufmann WE, van Zijl PC, Fredericksen K, Pearlson GD, Solaiyappan M, Mori S (2001) Diffusion tensor imaging and axonal tracking in the human brainstem. Neuroimage 14: Vilanova A, Berenschot G, Pul van C (2004) DTI Visualization with streamsurfaces and evenly-spaced volume seeding. In: Proceedings of Joint EG/IEEE TCVG visual Symposium pp Weinstein DM, Kindlman GL, Lundberg EC (1999) Tensorlines: advection-diffusion based propagation through diffusion tensor fields. In: Ebert DS, Gross MH, Hamann B (eds) IEEE visualization IEEE Computer Society and ACM, San Francisco, pp Westin CF, Maier SE, Mamata H, Nabavi A, Jolesz FA, Kikinis R (2002) Processing and visualization for diffusion tensor MRI. Med Image Anal 6: Yamada K, Kizu O, Mori S, Ito H, Nakamura H, Yuen S, Kubota T, Tanaka O, Akada W, Sasajima H, Mineura K, Nishimura T (2003) Brain fiber tracking with clinically feasible diffusion-tensor MR imaging: initial experience. Radiology 227: Zhukov L, Barr A (2002) Oriented tensor reconstruction: tracing neural pathways from diffusion tensor MRI. Visualization. In: Proceedings of the conference on visualization 02. IEEE Computer Society, Washington, DC, USA, pp Zöckler M, Stalling D, Hege H-C (1996) Interactive visualization of 3D-vector fields using illuminated stream lines. IEEE visualization. In: Proceedings of the 7th conference on visualization 96. IEEE Computer Society Press, Los Alamitos, CA, USA, pp
Fiber Selection from Diffusion Tensor Data based on Boolean Operators
 Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhof 1, G. Greiner 2, M. Buchfelder 3, C. Nimsky 4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Evaluation of Local Filter Approaches for Diffusion Tensor based Fiber Tracking
 Evaluation of Local Filter Approaches for Diffusion Tensor based Fiber Tracking D. Merhof 1, M. Buchfelder 2, C. Nimsky 3 1 Visual Computing, University of Konstanz, Konstanz 2 Department of Neurosurgery,
Evaluation of Local Filter Approaches for Diffusion Tensor based Fiber Tracking D. Merhof 1, M. Buchfelder 2, C. Nimsky 3 1 Visual Computing, University of Konstanz, Konstanz 2 Department of Neurosurgery,
Fiber Selection from Diffusion Tensor Data based on Boolean Operators
 Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhofl, G. Greiner 2, M. Buchfelder 3, C. Nimsky4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Fiber Selection from Diffusion Tensor Data based on Boolean Operators D. Merhofl, G. Greiner 2, M. Buchfelder 3, C. Nimsky4 1 Visual Computing, University of Konstanz, Konstanz, Germany 2 Computer Graphics
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes
 Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
Generation of Hulls Encompassing Neuronal Pathways Based on Tetrahedralization and 3D Alpha Shapes Dorit Merhof 1,2, Martin Meister 1, Ezgi Bingöl 1, Peter Hastreiter 1,2, Christopher Nimsky 2,3, Günther
Automatic Quantification of DTI Parameters along Fiber Bundles
 Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein 1, Simon Hermann 1, Olaf Konrad 1, Horst K. Hahn 1, and Heinz-Otto Peitgen 1 1 MeVis Research, 28359 Bremen Email: klein@mevis.de
Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein 1, Simon Hermann 1, Olaf Konrad 1, Horst K. Hahn 1, and Heinz-Otto Peitgen 1 1 MeVis Research, 28359 Bremen Email: klein@mevis.de
Automatic Quantification of DTI Parameters along Fiber Bundles
 Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein, Simon Hermann, Olaf Konrad, Horst K. Hahn, Heinz-Otto Peitgen MeVis Research, 28359 Bremen Email: klein@mevis.de Abstract. We introduce
Automatic Quantification of DTI Parameters along Fiber Bundles Jan Klein, Simon Hermann, Olaf Konrad, Horst K. Hahn, Heinz-Otto Peitgen MeVis Research, 28359 Bremen Email: klein@mevis.de Abstract. We introduce
A Ray-based Approach for Boundary Estimation of Fiber Bundles Derived from Diffusion Tensor Imaging
 A Ray-based Approach for Boundary Estimation of Fiber Bundles Derived from Diffusion Tensor Imaging M. H. A. Bauer 1,3, S. Barbieri 2, J. Klein 2, J. Egger 1,3, D. Kuhnt 1, B. Freisleben 3, H.-K. Hahn
A Ray-based Approach for Boundary Estimation of Fiber Bundles Derived from Diffusion Tensor Imaging M. H. A. Bauer 1,3, S. Barbieri 2, J. Klein 2, J. Egger 1,3, D. Kuhnt 1, B. Freisleben 3, H.-K. Hahn
HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets
 syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets Julien Gervais; Lisa Chuah Siemens Healthcare, Magnetic
syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets Julien Gervais; Lisa Chuah Siemens Healthcare, Magnetic
The Application of GPU Particle Tracing to Diffusion Tensor Field Visualization
 The Application of GPU Particle Tracing to Diffusion Tensor Field Visualization Polina Kondratieva, Jens Krüger, Rüdiger Westermann Computer Graphics and Visualization Group Technische Universität München
The Application of GPU Particle Tracing to Diffusion Tensor Field Visualization Polina Kondratieva, Jens Krüger, Rüdiger Westermann Computer Graphics and Visualization Group Technische Universität München
MITK-DI. A new Diffusion Imaging Component for MITK. Klaus Fritzsche, Hans-Peter Meinzer
 MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
Advanced Visual Medicine: Techniques for Visual Exploration & Analysis
 Advanced Visual Medicine: Techniques for Visual Exploration & Analysis Interactive Visualization of Multimodal Volume Data for Neurosurgical Planning Felix Ritter, MeVis Research Bremen Multimodal Neurosurgical
Advanced Visual Medicine: Techniques for Visual Exploration & Analysis Interactive Visualization of Multimodal Volume Data for Neurosurgical Planning Felix Ritter, MeVis Research Bremen Multimodal Neurosurgical
Diffusion Imaging Visualization
 Diffusion Imaging Visualization Thomas Schultz URL: http://cg.cs.uni-bonn.de/schultz/ E-Mail: schultz@cs.uni-bonn.de 1 Outline Introduction to Diffusion Imaging Basic techniques Glyph-based Visualization
Diffusion Imaging Visualization Thomas Schultz URL: http://cg.cs.uni-bonn.de/schultz/ E-Mail: schultz@cs.uni-bonn.de 1 Outline Introduction to Diffusion Imaging Basic techniques Glyph-based Visualization
8. Tensor Field Visualization
 8. Tensor Field Visualization Tensor: extension of concept of scalar and vector Tensor data for a tensor of level k is given by t i1,i2,,ik (x 1,,x n ) Second-order tensor often represented by matrix Examples:
8. Tensor Field Visualization Tensor: extension of concept of scalar and vector Tensor data for a tensor of level k is given by t i1,i2,,ik (x 1,,x n ) Second-order tensor often represented by matrix Examples:
Correction of susceptibility artifacts in diffusion tensor data using non-linear registration
 Correction of susceptibility artifacts in diffusion tensor data using non-linear registration D. Merhof a,b,, G. Soza c, A. Stadlbauer b, G. Greiner a, C. Nimsky b a Computer Graphics Group, University
Correction of susceptibility artifacts in diffusion tensor data using non-linear registration D. Merhof a,b,, G. Soza c, A. Stadlbauer b, G. Greiner a, C. Nimsky b a Computer Graphics Group, University
Correction of susceptibility artifacts in diffusion tensor data using non-linear registration
 Available online at www.sciencedirect.com Medical Image Analysis 11 (2007) 588 603 www.elsevier.com/locate/media Correction of susceptibility artifacts in diffusion tensor data using non-linear registration
Available online at www.sciencedirect.com Medical Image Analysis 11 (2007) 588 603 www.elsevier.com/locate/media Correction of susceptibility artifacts in diffusion tensor data using non-linear registration
Multimodal Visualization of DTI and fmri Data using Illustrative Methods
 Multimodal Visualization of DTI and fmri Data using Illustrative Methods Silvia Born 1, Werner Jainek 1, Mario Hlawitschka 2, Gerik Scheuermann 3, Christos Trantakis 4, Jürgen Meixensberger 4, Dirk Bartz
Multimodal Visualization of DTI and fmri Data using Illustrative Methods Silvia Born 1, Werner Jainek 1, Mario Hlawitschka 2, Gerik Scheuermann 3, Christos Trantakis 4, Jürgen Meixensberger 4, Dirk Bartz
Complex Fiber Visualization
 Annales Mathematicae et Informaticae 34 (2007) pp. 103 109 http://www.ektf.hu/tanszek/matematika/ami Complex Fiber Visualization Henrietta Tomán a, Róbert Tornai b, Marianna Zichar c a Department of Computer
Annales Mathematicae et Informaticae 34 (2007) pp. 103 109 http://www.ektf.hu/tanszek/matematika/ami Complex Fiber Visualization Henrietta Tomán a, Róbert Tornai b, Marianna Zichar c a Department of Computer
MITK-DI. A new Diffusion Imaging Component for MITK. Klaus Fritzsche, Hans-Peter Meinzer
 MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
MITK-DI A new Diffusion Imaging Component for MITK Klaus Fritzsche, Hans-Peter Meinzer Division of Medical and Biological Informatics, DKFZ Heidelberg k.fritzsche@dkfz-heidelberg.de Abstract. Diffusion-MRI
NEURO M203 & BIOMED M263 WINTER 2014
 NEURO M203 & BIOMED M263 WINTER 2014 MRI Lab 2: Neuroimaging Connectivity Lab In today s lab we will work with sample diffusion imaging data and the group averaged fmri data collected during your scanning
NEURO M203 & BIOMED M263 WINTER 2014 MRI Lab 2: Neuroimaging Connectivity Lab In today s lab we will work with sample diffusion imaging data and the group averaged fmri data collected during your scanning
A Method for Registering Diffusion Weighted Magnetic Resonance Images
 A Method for Registering Diffusion Weighted Magnetic Resonance Images Xiaodong Tao and James V. Miller GE Research, Niskayuna, New York, USA Abstract. Diffusion weighted magnetic resonance (DWMR or DW)
A Method for Registering Diffusion Weighted Magnetic Resonance Images Xiaodong Tao and James V. Miller GE Research, Niskayuna, New York, USA Abstract. Diffusion weighted magnetic resonance (DWMR or DW)
Diffusion Tensor Imaging and Reading Development
 Diffusion Tensor Imaging and Reading Development Bob Dougherty Stanford Institute for Reading and Learning Reading and Anatomy Every brain is different... Not all brains optimized for highly proficient
Diffusion Tensor Imaging and Reading Development Bob Dougherty Stanford Institute for Reading and Learning Reading and Anatomy Every brain is different... Not all brains optimized for highly proficient
Visualization of White Matter Tracts with Wrapped Streamlines
 Visualization of White Matter Tracts with Wrapped Streamlines Frank Enders Natascha Sauber Dorit Merhof Peter Hastreiter Neurocenter, Dept. of Neurosurgery Neurocenter, Dept. of Neurosurgery Neurocenter,
Visualization of White Matter Tracts with Wrapped Streamlines Frank Enders Natascha Sauber Dorit Merhof Peter Hastreiter Neurocenter, Dept. of Neurosurgery Neurocenter, Dept. of Neurosurgery Neurocenter,
ECE1778 Final Report MRI Visualizer
 ECE1778 Final Report MRI Visualizer David Qixiang Chen Alex Rodionov Word Count: 2408 Introduction We aim to develop a mobile phone/tablet based neurosurgical MRI visualization application with the goal
ECE1778 Final Report MRI Visualizer David Qixiang Chen Alex Rodionov Word Count: 2408 Introduction We aim to develop a mobile phone/tablet based neurosurgical MRI visualization application with the goal
Saturn User Manual. Rubén Cárdenes. 29th January 2010 Image Processing Laboratory, University of Valladolid. Abstract
 Saturn User Manual Rubén Cárdenes 29th January 2010 Image Processing Laboratory, University of Valladolid Abstract Saturn is a software package for DTI processing and visualization, provided with a graphic
Saturn User Manual Rubén Cárdenes 29th January 2010 Image Processing Laboratory, University of Valladolid Abstract Saturn is a software package for DTI processing and visualization, provided with a graphic
A Workflow Optimized Software Platform for Multimodal Neurosurgical Planning and Monitoring
 A Workflow Optimized Software Platform for Multimodal Neurosurgical Planning and Monitoring Eine Workflow Optimierte Software Umgebung für Multimodale Neurochirurgische Planung und Verlaufskontrolle A
A Workflow Optimized Software Platform for Multimodal Neurosurgical Planning and Monitoring Eine Workflow Optimierte Software Umgebung für Multimodale Neurochirurgische Planung und Verlaufskontrolle A
n o r d i c B r a i n E x Tutorial DTI Module
 m a k i n g f u n c t i o n a l M R I e a s y n o r d i c B r a i n E x Tutorial DTI Module Please note that this tutorial is for the latest released nordicbrainex. If you are using an older version please
m a k i n g f u n c t i o n a l M R I e a s y n o r d i c B r a i n E x Tutorial DTI Module Please note that this tutorial is for the latest released nordicbrainex. If you are using an older version please
Computational Medical Imaging Analysis Chapter 4: Image Visualization
 Computational Medical Imaging Analysis Chapter 4: Image Visualization Jun Zhang Laboratory for Computational Medical Imaging & Data Analysis Department of Computer Science University of Kentucky Lexington,
Computational Medical Imaging Analysis Chapter 4: Image Visualization Jun Zhang Laboratory for Computational Medical Imaging & Data Analysis Department of Computer Science University of Kentucky Lexington,
Quantitative MRI of the Brain: Investigation of Cerebral Gray and White Matter Diseases
 Quantities Measured by MR - Quantitative MRI of the Brain: Investigation of Cerebral Gray and White Matter Diseases Static parameters (influenced by molecular environment): T, T* (transverse relaxation)
Quantities Measured by MR - Quantitative MRI of the Brain: Investigation of Cerebral Gray and White Matter Diseases Static parameters (influenced by molecular environment): T, T* (transverse relaxation)
HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu HST.583 Functional Magnetic Resonance Imaging: Data Acquisition and Analysis Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
A method of visualization of a brain neural pathway by using critical points and target regions
 Modelling in Medicine and Biology VII 309 A method of visualization of a brain neural pathway by using critical points and target regions A. Doi 1, H. Fujimura 2, M. Nagano 1, T. Inoue 3 & A. Ogawa 3 1
Modelling in Medicine and Biology VII 309 A method of visualization of a brain neural pathway by using critical points and target regions A. Doi 1, H. Fujimura 2, M. Nagano 1, T. Inoue 3 & A. Ogawa 3 1
An independent component analysis based tool for exploring functional connections in the brain
 An independent component analysis based tool for exploring functional connections in the brain S. M. Rolfe a, L. Finney b, R. F. Tungaraza b, J. Guan b, L.G. Shapiro b, J. F. Brinkely b, A. Poliakov c,
An independent component analysis based tool for exploring functional connections in the brain S. M. Rolfe a, L. Finney b, R. F. Tungaraza b, J. Guan b, L.G. Shapiro b, J. F. Brinkely b, A. Poliakov c,
Visualization and Analysis of White Matter Structural Asymmetry in Diffusion Tensor MRI Data
 Magnetic Resonance in Medicine 51:140 147 (2004) Visualization and Analysis of White Matter Structural Asymmetry in Diffusion Tensor MRI Data Song Zhang, 1 Mark E. Bastin, 2 * David H. Laidlaw, 1 Saurabh
Magnetic Resonance in Medicine 51:140 147 (2004) Visualization and Analysis of White Matter Structural Asymmetry in Diffusion Tensor MRI Data Song Zhang, 1 Mark E. Bastin, 2 * David H. Laidlaw, 1 Saurabh
An Immersive Virtual Environment for DT-MRI Volume Visualization Applications: a Case Study
 An Immersive Virtual Environment for DT-MRI Volume Visualization Applications: a Case Study S. Zhang, Ç. Demiralp,D.F.Keefe M. DaSilva, D. H. Laidlaw, B. D. Greenberg Brown University P.J. Basser C. Pierpaoli
An Immersive Virtual Environment for DT-MRI Volume Visualization Applications: a Case Study S. Zhang, Ç. Demiralp,D.F.Keefe M. DaSilva, D. H. Laidlaw, B. D. Greenberg Brown University P.J. Basser C. Pierpaoli
On Classifying Disease-Induced Patterns in the Brain Using Diffusion Tensor Images
 On Classifying Disease-Induced Patterns in the Brain Using Diffusion Tensor Images Peng Wang 1,2 and Ragini Verma 1 1 Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania,
On Classifying Disease-Induced Patterns in the Brain Using Diffusion Tensor Images Peng Wang 1,2 and Ragini Verma 1 1 Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania,
NeuroQLab A Software Assistant for Neurosurgical Planning and Quantitative Image Analysis
 NeuroQLab A Software Assistant for Neurosurgical Planning and Quantitative Image Analysis Florian Weiler 1, Jan Rexilius 2, Jan Klein 1, Horst K. Hahn 1 1 Fraunhofer MEVIS, Universitätsallee 29, 28359
NeuroQLab A Software Assistant for Neurosurgical Planning and Quantitative Image Analysis Florian Weiler 1, Jan Rexilius 2, Jan Klein 1, Horst K. Hahn 1 1 Fraunhofer MEVIS, Universitätsallee 29, 28359
Qualitative and quantitative analysis of probabilistic and deterministic fiber tracking
 Qualitative and quantitative analysis of probabilistic and deterministic fiber tracking Jan Klein a, Adrian Grötsch a, Daniel Betz a, Sebastiano Barbieri a, Ola Friman a, Bram Stieltjes b, Helmut Hildebrandt
Qualitative and quantitative analysis of probabilistic and deterministic fiber tracking Jan Klein a, Adrian Grötsch a, Daniel Betz a, Sebastiano Barbieri a, Ola Friman a, Bram Stieltjes b, Helmut Hildebrandt
Diffusion model fitting and tractography: A primer
 Diffusion model fitting and tractography: A primer Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 03/18/10 Why n how Diffusion model fitting and tractography 0/18 Why
Diffusion model fitting and tractography: A primer Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 03/18/10 Why n how Diffusion model fitting and tractography 0/18 Why
Intraoperative Prostate Tracking with Slice-to-Volume Registration in MR
 Intraoperative Prostate Tracking with Slice-to-Volume Registration in MR Sean Gill a, Purang Abolmaesumi a,b, Siddharth Vikal a, Parvin Mousavi a and Gabor Fichtinger a,b,* (a) School of Computing, Queen
Intraoperative Prostate Tracking with Slice-to-Volume Registration in MR Sean Gill a, Purang Abolmaesumi a,b, Siddharth Vikal a, Parvin Mousavi a and Gabor Fichtinger a,b,* (a) School of Computing, Queen
FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING. Francesca Pizzorni Ferrarese
 FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING Francesca Pizzorni Ferrarese Pipeline overview WM and GM Segmentation Registration Data reconstruction Tractography
FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING Francesca Pizzorni Ferrarese Pipeline overview WM and GM Segmentation Registration Data reconstruction Tractography
Advanced MRI Techniques (and Applications)
 Advanced MRI Techniques (and Applications) Jeffry R. Alger, PhD Department of Neurology Ahmanson-Lovelace Brain Mapping Center Brain Research Institute Jonsson Comprehensive Cancer Center University of
Advanced MRI Techniques (and Applications) Jeffry R. Alger, PhD Department of Neurology Ahmanson-Lovelace Brain Mapping Center Brain Research Institute Jonsson Comprehensive Cancer Center University of
NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2010 November 10.
 NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2010 November 10. Published in final edited form as: Med Image Comput Comput Assist Interv.
NIH Public Access Author Manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2010 November 10. Published in final edited form as: Med Image Comput Comput Assist Interv.
Direct Registration of White Matter Tractographies with Application to Atlas Construction
 Direct Registration of White Matter Tractographies with Application to Atlas Construction Arnaldo Mayer, Hayit Greenspan Medical image processing lab, Tel-Aviv University, Ramat-Aviv, Israel arnakdom@post.tau.ac.il,
Direct Registration of White Matter Tractographies with Application to Atlas Construction Arnaldo Mayer, Hayit Greenspan Medical image processing lab, Tel-Aviv University, Ramat-Aviv, Israel arnakdom@post.tau.ac.il,
Analysis of White Matter Fiber Tracts via Fiber Clustering and Parametrization
 Analysis of White Matter Fiber Tracts via Fiber Clustering and Parametrization Sylvain Gouttard 2,4, Isabelle Corouge 1,2, Weili Lin 3, John Gilmore 2, and Guido Gerig 1,2 1 Departments of Computer Science,
Analysis of White Matter Fiber Tracts via Fiber Clustering and Parametrization Sylvain Gouttard 2,4, Isabelle Corouge 1,2, Weili Lin 3, John Gilmore 2, and Guido Gerig 1,2 1 Departments of Computer Science,
iplan RT Image Advanced Contouring Workstation - Driving Physician Collaboration
 iplan RT Image Advanced Contouring Workstation - Driving Physician Collaboration The iplan Contouring Workstation offers unique and innovative capabilities for faster contouring and consistent segmentation
iplan RT Image Advanced Contouring Workstation - Driving Physician Collaboration The iplan Contouring Workstation offers unique and innovative capabilities for faster contouring and consistent segmentation
Automatic Subthalamic Nucleus Targeting for Deep Brain Stimulation. A Validation Study
 Automatic Subthalamic Nucleus Targeting for Deep Brain Stimulation. A Validation Study F. Javier Sánchez Castro a, Claudio Pollo a,b, Jean-Guy Villemure b, Jean-Philippe Thiran a a École Polytechnique
Automatic Subthalamic Nucleus Targeting for Deep Brain Stimulation. A Validation Study F. Javier Sánchez Castro a, Claudio Pollo a,b, Jean-Guy Villemure b, Jean-Philippe Thiran a a École Polytechnique
Generating Fiber Crossing Phantoms Out of Experimental DWIs
 Generating Fiber Crossing Phantoms Out of Experimental DWIs Matthan Caan 1,2, Anne Willem de Vries 2, Ganesh Khedoe 2,ErikAkkerman 1, Lucas van Vliet 2, Kees Grimbergen 1, and Frans Vos 1,2 1 Department
Generating Fiber Crossing Phantoms Out of Experimental DWIs Matthan Caan 1,2, Anne Willem de Vries 2, Ganesh Khedoe 2,ErikAkkerman 1, Lucas van Vliet 2, Kees Grimbergen 1, and Frans Vos 1,2 1 Department
Neural Network-Assisted Fiber Tracking of Synthetic and White Matter DT-MR Images
 Neural Network-Assisted Fiber Tracking of Synthetic and White Matter DT-MR Images L.M. San-José-Revuelta, M. Martín-Fernández and C. Alberola-López Abstract In this paper, a recently developed fiber tracking
Neural Network-Assisted Fiber Tracking of Synthetic and White Matter DT-MR Images L.M. San-José-Revuelta, M. Martín-Fernández and C. Alberola-López Abstract In this paper, a recently developed fiber tracking
Whole Body MRI Intensity Standardization
 Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
2 Michael E. Leventon and Sarah F. F. Gibson a b c d Fig. 1. (a, b) Two MR scans of a person's knee. Both images have high resolution in-plane, but ha
 Model Generation from Multiple Volumes using Constrained Elastic SurfaceNets Michael E. Leventon and Sarah F. F. Gibson 1 MIT Artificial Intelligence Laboratory, Cambridge, MA 02139, USA leventon@ai.mit.edu
Model Generation from Multiple Volumes using Constrained Elastic SurfaceNets Michael E. Leventon and Sarah F. F. Gibson 1 MIT Artificial Intelligence Laboratory, Cambridge, MA 02139, USA leventon@ai.mit.edu
Slide 1. Technical Aspects of Quality Control in Magnetic Resonance Imaging. Slide 2. Annual Compliance Testing. of MRI Systems.
 Slide 1 Technical Aspects of Quality Control in Magnetic Resonance Imaging Slide 2 Compliance Testing of MRI Systems, Ph.D. Department of Radiology Henry Ford Hospital, Detroit, MI Slide 3 Compliance Testing
Slide 1 Technical Aspects of Quality Control in Magnetic Resonance Imaging Slide 2 Compliance Testing of MRI Systems, Ph.D. Department of Radiology Henry Ford Hospital, Detroit, MI Slide 3 Compliance Testing
Blood Particle Trajectories in Phase-Contrast-MRI as Minimal Paths Computed with Anisotropic Fast Marching
 Blood Particle Trajectories in Phase-Contrast-MRI as Minimal Paths Computed with Anisotropic Fast Marching Michael Schwenke 1, Anja Hennemuth 1, Bernd Fischer 2, Ola Friman 1 1 Fraunhofer MEVIS, Institute
Blood Particle Trajectories in Phase-Contrast-MRI as Minimal Paths Computed with Anisotropic Fast Marching Michael Schwenke 1, Anja Hennemuth 1, Bernd Fischer 2, Ola Friman 1 1 Fraunhofer MEVIS, Institute
Visualizing Diffusion Tensor Imaging Data with Merging Ellipsoids
 Visualizing Diffusion Tensor Imaging Data with Merging Ellipsoids Wei Chen Song Zhang Stephen Correia David F. Tate State Key Lab of CAD&CG, ZJU, China Mississippi State University Brown University Brigham
Visualizing Diffusion Tensor Imaging Data with Merging Ellipsoids Wei Chen Song Zhang Stephen Correia David F. Tate State Key Lab of CAD&CG, ZJU, China Mississippi State University Brown University Brigham
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy
 The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
Appendix E1. Supplementary Methods. MR Image Acquisition. MR Image Analysis
 RSNA, 2015 10.1148/radiol.2015150532 Appendix E1 Supplementary Methods MR Image Acquisition By using a 1.5-T system (Avanto, Siemens Medical, Erlangen, Germany) under a program of regular maintenance (no
RSNA, 2015 10.1148/radiol.2015150532 Appendix E1 Supplementary Methods MR Image Acquisition By using a 1.5-T system (Avanto, Siemens Medical, Erlangen, Germany) under a program of regular maintenance (no
Scene-Based Segmentation of Multiple Muscles from MRI in MITK
 Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
Scene-Based Segmentation of Multiple Muscles from MRI in MITK Yan Geng 1, Sebastian Ullrich 2, Oliver Grottke 3, Rolf Rossaint 3, Torsten Kuhlen 2, Thomas M. Deserno 1 1 Department of Medical Informatics,
CHAPTER 9: Magnetic Susceptibility Effects in High Field MRI
 Figure 1. In the brain, the gray matter has substantially more blood vessels and capillaries than white matter. The magnified image on the right displays the rich vasculature in gray matter forming porous,
Figure 1. In the brain, the gray matter has substantially more blood vessels and capillaries than white matter. The magnified image on the right displays the rich vasculature in gray matter forming porous,
Constrained Reconstruction of Sparse Cardiac MR DTI Data
 Constrained Reconstruction of Sparse Cardiac MR DTI Data Ganesh Adluru 1,3, Edward Hsu, and Edward V.R. DiBella,3 1 Electrical and Computer Engineering department, 50 S. Central Campus Dr., MEB, University
Constrained Reconstruction of Sparse Cardiac MR DTI Data Ganesh Adluru 1,3, Edward Hsu, and Edward V.R. DiBella,3 1 Electrical and Computer Engineering department, 50 S. Central Campus Dr., MEB, University
Quantitative Analysis of White Matter Fiber Properties along Geodesic Paths
 Quantitative Analysis of White Matter Fiber Properties along Geodesic Paths Pierre Fillard 1,4, John Gilmore 2, Joseph Piven 2, Weili Lin 3, and Guido Gerig 1,2 1 Department of Computer Science, 2 Department
Quantitative Analysis of White Matter Fiber Properties along Geodesic Paths Pierre Fillard 1,4, John Gilmore 2, Joseph Piven 2, Weili Lin 3, and Guido Gerig 1,2 1 Department of Computer Science, 2 Department
Metrics for Uncertainty Analysis and Visualization of Diffusion Tensor Images
 Metrics for Uncertainty Analysis and Visualization of Diffusion Tensor Images Fangxiang Jiao 1, Jeff M. Phillips 2, Jeroen Stinstra 3, Jens Krger 4, Raj Varma 2, Edward HSU 5, Julie Korenberg 6, and Chris
Metrics for Uncertainty Analysis and Visualization of Diffusion Tensor Images Fangxiang Jiao 1, Jeff M. Phillips 2, Jeroen Stinstra 3, Jens Krger 4, Raj Varma 2, Edward HSU 5, Julie Korenberg 6, and Chris
NIH Public Access Author Manuscript Proc Soc Photo Opt Instrum Eng. Author manuscript; available in PMC 2014 October 07.
 NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
Lilla Zöllei A.A. Martinos Center, MGH; Boston, MA
 Lilla Zöllei lzollei@nmr.mgh.harvard.edu A.A. Martinos Center, MGH; Boston, MA Bruce Fischl Gheorghe Postelnicu Jean Augustinack Anastasia Yendiki Allison Stevens Kristen Huber Sita Kakonoori + the FreeSurfer
Lilla Zöllei lzollei@nmr.mgh.harvard.edu A.A. Martinos Center, MGH; Boston, MA Bruce Fischl Gheorghe Postelnicu Jean Augustinack Anastasia Yendiki Allison Stevens Kristen Huber Sita Kakonoori + the FreeSurfer
Atelier 2 : Calcul Haute Performance et Sciences du Vivant Forum er juillet, Paris, France
 From Diffusion MR Image Analysis to Whole Brain Connectivity Simulation Jean-Philippe Thiran EPFL Lausanne, Switzerland EPFL - Lausanne HPC in life sciences at EPFL The Blue Brain project: create a biologically
From Diffusion MR Image Analysis to Whole Brain Connectivity Simulation Jean-Philippe Thiran EPFL Lausanne, Switzerland EPFL - Lausanne HPC in life sciences at EPFL The Blue Brain project: create a biologically
Supplementary methods
 Supplementary methods This section provides additional technical details on the sample, the applied imaging and analysis steps and methods. Structural imaging Trained radiographers placed all participants
Supplementary methods This section provides additional technical details on the sample, the applied imaging and analysis steps and methods. Structural imaging Trained radiographers placed all participants
Technische Universiteit Eindhoven Department of Mathematics and Computer Science. Master s Thesis HIERARCHICAL VISUALIZATION USING FIBER CLUSTERING
 Technische Universiteit Eindhoven Department of Mathematics and Computer Science Master s Thesis HIERARCHICAL VISUALIZATION USING FIBER CLUSTERING by Ing. B. Moberts Supervisors: Dr. A. Vilanova Prof.dr.ir.
Technische Universiteit Eindhoven Department of Mathematics and Computer Science Master s Thesis HIERARCHICAL VISUALIZATION USING FIBER CLUSTERING by Ing. B. Moberts Supervisors: Dr. A. Vilanova Prof.dr.ir.
Video Registration Virtual Reality for Non-linkage Stereotactic Surgery
 Video Registration Virtual Reality for Non-linkage Stereotactic Surgery P.L. Gleason, Ron Kikinis, David Altobelli, William Wells, Eben Alexander III, Peter McL. Black, Ferenc Jolesz Surgical Planning
Video Registration Virtual Reality for Non-linkage Stereotactic Surgery P.L. Gleason, Ron Kikinis, David Altobelli, William Wells, Eben Alexander III, Peter McL. Black, Ferenc Jolesz Surgical Planning
Head motion in diffusion MRI
 Head motion in diffusion MRI Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 11/06/13 Head motion in diffusion MRI 0/33 Diffusion contrast Basic principle of diffusion
Head motion in diffusion MRI Anastasia Yendiki HMS/MGH/MIT Athinoula A. Martinos Center for Biomedical Imaging 11/06/13 Head motion in diffusion MRI 0/33 Diffusion contrast Basic principle of diffusion
Magnetic Resonance Elastography (MRE) of Liver Disease
 Magnetic Resonance Elastography (MRE) of Liver Disease Authored by: Jennifer Dolan Fox, PhD VirtualScopics Inc. jennifer_fox@virtualscopics.com 1-585-249-6231 1. Overview of MRE Imaging MRE is a magnetic
Magnetic Resonance Elastography (MRE) of Liver Disease Authored by: Jennifer Dolan Fox, PhD VirtualScopics Inc. jennifer_fox@virtualscopics.com 1-585-249-6231 1. Overview of MRE Imaging MRE is a magnetic
Lab Location: MRI, B2, Cardinal Carter Wing, St. Michael s Hospital, 30 Bond Street
 Lab Location: MRI, B2, Cardinal Carter Wing, St. Michael s Hospital, 30 Bond Street MRI is located in the sub basement of CC wing. From Queen or Victoria, follow the baby blue arrows and ride the CC south
Lab Location: MRI, B2, Cardinal Carter Wing, St. Michael s Hospital, 30 Bond Street MRI is located in the sub basement of CC wing. From Queen or Victoria, follow the baby blue arrows and ride the CC south
The organization of the human cerebral cortex estimated by intrinsic functional connectivity
 1 The organization of the human cerebral cortex estimated by intrinsic functional connectivity Journal: Journal of Neurophysiology Author: B. T. Thomas Yeo, et al Link: https://www.ncbi.nlm.nih.gov/pubmed/21653723
1 The organization of the human cerebral cortex estimated by intrinsic functional connectivity Journal: Journal of Neurophysiology Author: B. T. Thomas Yeo, et al Link: https://www.ncbi.nlm.nih.gov/pubmed/21653723
RT_Image v0.2β User s Guide
 RT_Image v0.2β User s Guide RT_Image is a three-dimensional image display and analysis suite developed in IDL (ITT, Boulder, CO). It offers a range of flexible tools for the visualization and quantitation
RT_Image v0.2β User s Guide RT_Image is a three-dimensional image display and analysis suite developed in IDL (ITT, Boulder, CO). It offers a range of flexible tools for the visualization and quantitation
Network connectivity via inference over curvature-regularizing line graphs
 Network connectivity via inference over curvature-regularizing line graphs Asian Conference on Computer Vision Maxwell D. Collins 1,2, Vikas Singh 2,1, Andrew L. Alexander 3 1 Department of Computer Sciences
Network connectivity via inference over curvature-regularizing line graphs Asian Conference on Computer Vision Maxwell D. Collins 1,2, Vikas Singh 2,1, Andrew L. Alexander 3 1 Department of Computer Sciences
Chapter 6 Visualization Techniques for Vector Fields
 Chapter 6 Visualization Techniques for Vector Fields 6.1 Introduction 6.2 Vector Glyphs 6.3 Particle Advection 6.4 Streamlines 6.5 Line Integral Convolution 6.6 Vector Topology 6.7 References 2006 Burkhard
Chapter 6 Visualization Techniques for Vector Fields 6.1 Introduction 6.2 Vector Glyphs 6.3 Particle Advection 6.4 Streamlines 6.5 Line Integral Convolution 6.6 Vector Topology 6.7 References 2006 Burkhard
Quantifying Three-Dimensional Deformations of Migrating Fibroblasts
 45 Chapter 4 Quantifying Three-Dimensional Deformations of Migrating Fibroblasts This chapter presents the full-field displacements and tractions of 3T3 fibroblast cells during migration on polyacrylamide
45 Chapter 4 Quantifying Three-Dimensional Deformations of Migrating Fibroblasts This chapter presents the full-field displacements and tractions of 3T3 fibroblast cells during migration on polyacrylamide
Vector Visualization
 Vector Visualization Vector Visulization Divergence and Vorticity Vector Glyphs Vector Color Coding Displacement Plots Stream Objects Texture-Based Vector Visualization Simplified Representation of Vector
Vector Visualization Vector Visulization Divergence and Vorticity Vector Glyphs Vector Color Coding Displacement Plots Stream Objects Texture-Based Vector Visualization Simplified Representation of Vector
Super-Resolution Reconstruction of Diffusion-Weighted Images from Distortion Compensated Orthogonal Anisotropic Acquisitions.
 Super-Resolution Reconstruction of Diffusion-Weighted Images from Distortion Compensated Orthogonal Anisotropic Acquisitions. Benoit Scherrer Ali Gholipour Simon K. Warfield Children s Hospital Boston,
Super-Resolution Reconstruction of Diffusion-Weighted Images from Distortion Compensated Orthogonal Anisotropic Acquisitions. Benoit Scherrer Ali Gholipour Simon K. Warfield Children s Hospital Boston,
A Generic Lie Group Model for Computer Vision
 A Generic Lie Group Model for Computer Vision Within this research track we follow a generic Lie group approach to computer vision based on recent physiological research on how the primary visual cortex
A Generic Lie Group Model for Computer Vision Within this research track we follow a generic Lie group approach to computer vision based on recent physiological research on how the primary visual cortex
Spatial Normalization of Diffusion Tensor Fields
 Magnetic Resonance in Medicine 50:175 182 (2003) Spatial Normalization of Diffusion Tensor Fields Dongrong Xu, 1 * Susumu Mori, 2 Dinggang Shen, 1 Peter C.M. van Zijl, 2 and Christos Davatzikos 1 A method
Magnetic Resonance in Medicine 50:175 182 (2003) Spatial Normalization of Diffusion Tensor Fields Dongrong Xu, 1 * Susumu Mori, 2 Dinggang Shen, 1 Peter C.M. van Zijl, 2 and Christos Davatzikos 1 A method
Generation of Triangle Meshes from Time-of-Flight Data for Surface Registration
 Generation of Triangle Meshes from Time-of-Flight Data for Surface Registration Thomas Kilgus, Thiago R. dos Santos, Alexander Seitel, Kwong Yung, Alfred M. Franz, Anja Groch, Ivo Wolf, Hans-Peter Meinzer,
Generation of Triangle Meshes from Time-of-Flight Data for Surface Registration Thomas Kilgus, Thiago R. dos Santos, Alexander Seitel, Kwong Yung, Alfred M. Franz, Anja Groch, Ivo Wolf, Hans-Peter Meinzer,
Supplementary Information
 Supplementary Information Magnetic resonance imaging reveals functional anatomy and biomechanics of a living dragon tree Linnea Hesse 1,2,*, Tom Masselter 1,2,3, Jochen Leupold 4, Nils Spengler 5, Thomas
Supplementary Information Magnetic resonance imaging reveals functional anatomy and biomechanics of a living dragon tree Linnea Hesse 1,2,*, Tom Masselter 1,2,3, Jochen Leupold 4, Nils Spengler 5, Thomas
COBRE Scan Information
 COBRE Scan Information Below is more information on the directory structure for the COBRE imaging data. Also below are the imaging parameters for each series. Directory structure: var/www/html/dropbox/1139_anonymized/human:
COBRE Scan Information Below is more information on the directory structure for the COBRE imaging data. Also below are the imaging parameters for each series. Directory structure: var/www/html/dropbox/1139_anonymized/human:
Coloring of DT-MRI Fiber Traces using Laplacian Eigenmaps
 Coloring of DT-MRI Fiber Traces using Laplacian Eigenmaps Anders Brun 1, Hae-Jeong Park 2, Hans Knutsson 3, and Carl-Fredrik Westin 1 1 Laboratory of Mathematics in Imaging, Brigham and Women s Hospital,
Coloring of DT-MRI Fiber Traces using Laplacian Eigenmaps Anders Brun 1, Hae-Jeong Park 2, Hans Knutsson 3, and Carl-Fredrik Westin 1 1 Laboratory of Mathematics in Imaging, Brigham and Women s Hospital,
Vector Visualization. CSC 7443: Scientific Information Visualization
 Vector Visualization Vector data A vector is an object with direction and length v = (v x,v y,v z ) A vector field is a field which associates a vector with each point in space The vector data is 3D representation
Vector Visualization Vector data A vector is an object with direction and length v = (v x,v y,v z ) A vector field is a field which associates a vector with each point in space The vector data is 3D representation
Open Topology: A Toolkit for Brain Isosurface Correction
 Open Topology: A Toolkit for Brain Isosurface Correction Sylvain Jaume 1, Patrice Rondao 2, and Benoît Macq 2 1 National Institute of Research in Computer Science and Control, INRIA, France, sylvain@mit.edu,
Open Topology: A Toolkit for Brain Isosurface Correction Sylvain Jaume 1, Patrice Rondao 2, and Benoît Macq 2 1 National Institute of Research in Computer Science and Control, INRIA, France, sylvain@mit.edu,
Vector Field Visualisation
 Vector Field Visualisation Computer Animation and Visualization Lecture 14 Institute for Perception, Action & Behaviour School of Informatics Visualising Vectors Examples of vector data: meteorological
Vector Field Visualisation Computer Animation and Visualization Lecture 14 Institute for Perception, Action & Behaviour School of Informatics Visualising Vectors Examples of vector data: meteorological
Bone registration with 3D CT and ultrasound data sets
 International Congress Series 1256 (2003) 426 432 Bone registration with 3D CT and ultrasound data sets B. Brendel a, *,1, S. Winter b,1, A. Rick c,1, M. Stockheim d,1, H. Ermert a,1 a Institute of High
International Congress Series 1256 (2003) 426 432 Bone registration with 3D CT and ultrasound data sets B. Brendel a, *,1, S. Winter b,1, A. Rick c,1, M. Stockheim d,1, H. Ermert a,1 a Institute of High
DIFFUSION TENSOR IMAGING ANALYSIS. Using Analyze
 DIFFUSION TENSOR IMAGING ANALYSIS Using Analyze 2 Table of Contents 1. Introduction page 3 2. Loading DTI Data page 4 3. Computing DTI Maps page 5 4. Defining ROIs for Fiber Tracking page 6 5. Visualizing
DIFFUSION TENSOR IMAGING ANALYSIS Using Analyze 2 Table of Contents 1. Introduction page 3 2. Loading DTI Data page 4 3. Computing DTI Maps page 5 4. Defining ROIs for Fiber Tracking page 6 5. Visualizing
Landmark-based 3D Elastic Registration of Pre- and Postoperative Liver CT Data
 Landmark-based 3D Elastic Registration of Pre- and Postoperative Liver CT Data An Experimental Comparison Thomas Lange 1, Stefan Wörz 2, Karl Rohr 2, Peter M. Schlag 3 1 Experimental and Clinical Research
Landmark-based 3D Elastic Registration of Pre- and Postoperative Liver CT Data An Experimental Comparison Thomas Lange 1, Stefan Wörz 2, Karl Rohr 2, Peter M. Schlag 3 1 Experimental and Clinical Research
MITK Global Tractography
 MITK Global Tractography Peter F. Neher a, Bram Stieltjes b, Marco Reisert c, Ignaz Reicht a, Hans-Peter Meinzer a, Klaus H. Fritzsche a,b a German Cancer Research Center, Medical and Biological Informatics,
MITK Global Tractography Peter F. Neher a, Bram Stieltjes b, Marco Reisert c, Ignaz Reicht a, Hans-Peter Meinzer a, Klaus H. Fritzsche a,b a German Cancer Research Center, Medical and Biological Informatics,
SURFACE RECONSTRUCTION OF EX-VIVO HUMAN V1 THROUGH IDENTIFICATION OF THE STRIA OF GENNARI USING MRI AT 7T
 SURFACE RECONSTRUCTION OF EX-VIVO HUMAN V1 THROUGH IDENTIFICATION OF THE STRIA OF GENNARI USING MRI AT 7T Oliver P. Hinds 1, Jonathan R. Polimeni 2, Megan L. Blackwell 3, Christopher J. Wiggins 3, Graham
SURFACE RECONSTRUCTION OF EX-VIVO HUMAN V1 THROUGH IDENTIFICATION OF THE STRIA OF GENNARI USING MRI AT 7T Oliver P. Hinds 1, Jonathan R. Polimeni 2, Megan L. Blackwell 3, Christopher J. Wiggins 3, Graham
Fmri Spatial Processing
 Educational Course: Fmri Spatial Processing Ray Razlighi Jun. 8, 2014 Spatial Processing Spatial Re-alignment Geometric distortion correction Spatial Normalization Smoothing Why, When, How, Which Why is
Educational Course: Fmri Spatial Processing Ray Razlighi Jun. 8, 2014 Spatial Processing Spatial Re-alignment Geometric distortion correction Spatial Normalization Smoothing Why, When, How, Which Why is
A validation of the biexponential model in diffusion MRI signal attenuation using diffusion Monte Carlo simulator
 A validation of the biexponential model in diffusion MRI signal attenuation using diffusion Monte Carlo simulator Poster No.: C-0331 Congress: ECR 2014 Type: Scientific Exhibit Authors: D. Nishigake, S.
A validation of the biexponential model in diffusion MRI signal attenuation using diffusion Monte Carlo simulator Poster No.: C-0331 Congress: ECR 2014 Type: Scientific Exhibit Authors: D. Nishigake, S.
Reconstruction of Fiber Trajectories via Population-Based Estimation of Local Orientations
 IDEA Reconstruction of Fiber Trajectories via Population-Based Estimation of Local Orientations Pew-Thian Yap, John H. Gilmore, Weili Lin, Dinggang Shen Email: ptyap@med.unc.edu 2011-09-21 Poster: P2-46-
IDEA Reconstruction of Fiber Trajectories via Population-Based Estimation of Local Orientations Pew-Thian Yap, John H. Gilmore, Weili Lin, Dinggang Shen Email: ptyap@med.unc.edu 2011-09-21 Poster: P2-46-
Navigation System for ACL Reconstruction Using Registration between Multi-Viewpoint X-ray Images and CT Images
 Navigation System for ACL Reconstruction Using Registration between Multi-Viewpoint X-ray Images and CT Images Mamoru Kuga a*, Kazunori Yasuda b, Nobuhiko Hata a, Takeyoshi Dohi a a Graduate School of
Navigation System for ACL Reconstruction Using Registration between Multi-Viewpoint X-ray Images and CT Images Mamoru Kuga a*, Kazunori Yasuda b, Nobuhiko Hata a, Takeyoshi Dohi a a Graduate School of
Machine Learning for Medical Image Analysis. A. Criminisi
 Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Diffusion Tractography of the Corticospinal Tract with Multi-fiber Orientation Filtering
 Diffusion Tractography of the Corticospinal Tract with Multi-fiber Orientation Filtering Ryan P. Cabeen, David H. Laidlaw Computer Science Department Brown University Providence, RI, USA {cabeen,dhl}@cs.brown.edu
Diffusion Tractography of the Corticospinal Tract with Multi-fiber Orientation Filtering Ryan P. Cabeen, David H. Laidlaw Computer Science Department Brown University Providence, RI, USA {cabeen,dhl}@cs.brown.edu
Fast 3D Mean Shift Filter for CT Images
 Fast 3D Mean Shift Filter for CT Images Gustavo Fernández Domínguez, Horst Bischof, and Reinhard Beichel Institute for Computer Graphics and Vision, Graz University of Technology Inffeldgasse 16/2, A-8010,
Fast 3D Mean Shift Filter for CT Images Gustavo Fernández Domínguez, Horst Bischof, and Reinhard Beichel Institute for Computer Graphics and Vision, Graz University of Technology Inffeldgasse 16/2, A-8010,
Digital Imaging and Communications in Medicine (DICOM) Supplement 189: Parametrical Blending Presentation State Storage
 5 Digital Imaging and Communications in edicine (DICO) Supplement 189: Parametrical Blending Presentation State Storage 10 15 Prepared by: DICO Standards Committee, Working Group 16 (R sub group Functional
5 Digital Imaging and Communications in edicine (DICO) Supplement 189: Parametrical Blending Presentation State Storage 10 15 Prepared by: DICO Standards Committee, Working Group 16 (R sub group Functional
Segmentation of Bony Structures with Ligament Attachment Sites
 Segmentation of Bony Structures with Ligament Attachment Sites Heiko Seim 1, Hans Lamecker 1, Markus Heller 2, Stefan Zachow 1 1 Visualisierung und Datenanalyse, Zuse-Institut Berlin (ZIB), 14195 Berlin
Segmentation of Bony Structures with Ligament Attachment Sites Heiko Seim 1, Hans Lamecker 1, Markus Heller 2, Stefan Zachow 1 1 Visualisierung und Datenanalyse, Zuse-Institut Berlin (ZIB), 14195 Berlin
Learning-based Neuroimage Registration
 Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
