Multi-Atlas Segmentation Using Robust Feature-Based Registration
|
|
|
- Lucy Johnson
- 5 years ago
- Views:
Transcription
1 Multi-Atlas Segmentation Using Robust Feature-Based Registration Frida Fejne, Matilda Landgren, Jennifer Alvén, Johannes Ulén, Johan Fredriksson, Viktor Larsson, Olof Enqvist, and Fredrik Kahl Abstract This paper presents a pipeline which uses a multi-atlas approach for multiorgan segmentation in whole-body CT images. In order to obtain accurate registrations between the target and the atlas images, we develop an adapted feature-based method which uses organ specific features. These features are learnt during an offline pre-processing step, and thus the algorithm still benefits from the speed of feature-based registration methods. These feature sets are then used to obtain pairwise non-rigid transformations using RANSAC, followed by a thin plate spline refinement or NIFTYREG. The fusion of the transferred atlas labels is performed using a random forest classifier, and finally the segmentation is obtained using graph cuts with a Potts model as interaction term. Our pipeline was evaluated on 20 organs in 10 whole-body CT images at the VISCERAL Anatomy Grand Challenge, in conjunction with the International Symposium on Biomedical Imaging, Brooklyn, New York, in April It performed best on a majority of the organs, with respect to the Dice index. Frida Fejne Department of Signals and Systems, Chalmers University of Technology, Sweden, fejne@chalmers.se Matilda Landgren Centre for Mathematical Sciences, Lund University, Sweden matilda@maths.lth.se Jennifer Alvén Department of Signals and Systems, Chalmers University of Technology, Sweden, alven@chalmers.se Johannes Ulén Centre for Mathematical Sciences, Lund University, Sweden ulen@maths.lth.se Johan Fredriksson Centre for Mathematical Sciences, Lund University, Sweden johanf@maths.lth.se Viktor Larsson Centre for Mathematical Sciences, Lund University, Sweden viktorl@maths.lth.se Olof Enqvist Department of Signals and Systems, Chalmers University of Technology, Sweden, olof.enqvist@chalmers.se Fredrik Kahl Department of Signals and Systems, Chalmers University of Technology, Sweden, Centre for Mathematical Sciences, Lund University, Sweden, fredrik.kahl@chalmers.se The authors assert equal contribution and joint first authorship. 1
2 2 Fejne & Landgren et al. 1 Introduction Segmentation of anatomical structures is a fundamental task in medical image analysis. It has several applications such as localization of organs, detection of tumors or other pathological structures, and the results can for example serve as input to computer aided diagnosis (CAD) systems. Multi-organ segmentation is useful e.g. in radiotherapy planning [25], where the location of the tumor is of most interest, but also the location of surrounding (vital) organs. Furthermore, it can also be used in the preparation of, and during computer-assisted surgery [30]. Automated methods are preferable due to the time consuming task to do the segmentations manually and the need of a skilled expert. In this paper we propose a pipeline that uses a multi-atlas approach for automatic multi-organ segmentation for CT images. The segmentation of each organ is independent of the others and we show that very reliable organ localization can be obtained using (i) robust optimization techniques for registration, (ii) learned feature correspondences, and (iii) refinement with a random forest classifier and graph cut segmentation. 1.1 Related Work Multi-atlas methods for segmentation, which were first introduced in [26, 17, 10], have become a very popular choice in medical image analysis due to their excellent performance. The methods have been extensively used on brain MR images [22, 5, 11], on cardiac CTA data [16], for thoracic CT segmentation [9, 32] and multi-organ segmentation in CT images [34]. Multi-atlas methods generally produce robust results but relies on multiple image registrations as each atlas image is registered to the target image. Image registration can be divided into two different approaches; feature-based and intensity-based registration, see the surveys [15, 28]. Feature-based methods are generally very fast, but may have a risk of failing due to many outlier correspondences between the images. The intensity-based methods are on the contrary capable of producing accurate registrations but may be slow and are sensitive to initialization. Multi-atlas segmentation is a further development from single-atlas segmentation [20] and works as follows. In order to capture more anatomical variations and reduce the effect of registration errors, several single-atlas segmentations are combined in the multi-atlas approach. At first, pairwise registrations are computed between each atlas image and the target image, and thereafter the single-atlas segmentation labels are transferred to the target image according to the registrations. Next, a segmentation proposal is obtained by fusing the transferred labels. The label fusion can be performed using several different methods, whereof the simplest one is majority voting for each voxel, see [26, 17, 10]. However, there exists more sophisticated methods, for instance weighted voting [31], probabilistic reasoning using, e.g. the STAPLE algorithm [33], and different types of machine learning approaches [27].
3 Multi-Atlas Segmentation Using Robust Feature-Based Registration 3 The fused segmentation proposal can be further refined into a final segmentation by using graph cut [2, 21] or random forest based methods [8]. For a comprehensive survey of multi-atlas segmentation methods and their applications, see [12]. As mentioned previously, multi-atlas methods are often used to perform multiorgan segmentation. In the work of Wolz et al. [34] a hierarchical atlas is refined at three levels; global, organ, and voxel level. Instead of utilizing all the available atlases, the authors choose the most suitable ones. Similar to our paper, their final segmentation is obtained using graph cuts, and the evaluation of the algorithm results in relatively high Dice index on the liver, kidneys, pancreas and spleen. Another example of multi-organ segmentation is the probabilistic multi-atlas used by Chu et al. [3] for abdominal segmentation. They divide the image space into N subspaces and compute weights for the probabilistic atlas at both a global level (N = 1) and for each subspace. The organ segmentation is then obtained by a maximum a posteriori estimation and graph cuts. The method is evaluated on different number of subspaces, where the best performance is obtained for N = 64. Furthermore, another variant of multi-atlas segmentation of abdominal organs was used in a recent paper by Xu et al., [35]. Atlas selection and label fusion were done using a reformulation of the selective and iterative method for performance level estimation (SIMPLE) method. The authors developed a method for atlas selection, which were regularized by a Bayesian prior, learnt from context information. When evaluating the proposed method, it outperformed the compared methods, among them [34], on 11 out of 12 organs. In [23], Okada et al. present an approach to multi-organ segmentation that uses conditional shape-location combined with unsupervised intensity priors. In their work, an organ correlation graph is used to steer the order for which the organs are segmented by utilizing spatial correlation. In addition, the authors also developed a method for modeling conditional shape-location priors. 1.2 Our approach In this paper, we propose a pipeline for segmentation of 20 different organs in whole-body CT images. The algorithm uses standard multi-atlas segmentation for initial spatial localization. For the pairwise registrations between the target image and all the atlas images, we use an adapted feature-based method that has been designed to reduce the risk of establishing incorrect point-to-point correspondences between the image pairs. The main contribution of this paper is a method to identify reliable organ specific feature points among the atlas images. The speed of general feature-based registration methods is still beneficial to the algorithm, since this identification is done in an offline pre-processing step. We fuse the transferred labels by training a random forest classifier, and the final segmentation is then obtained with graph cuts. The pipeline is described in detail in Section 2. In Section 3 we present the results from the VISCERAL Anatomy Grand Challenge [14], as well as a detailed evaluation of the different steps in the pipeline for some of the organs.
4 4 Fejne & Landgren et al. 2 Methods Our pipeline for multi-organ segmentation contains the following three main steps: 1. Pairwise Registration. For a particular organ, the atlases are registered to the target image in two steps: first, subsets of the features in each atlas image are selected and matched to the features in the target image. Next, a non-rigid transformation between the atlases and the target image is estimated using RANSAC followed by a thin plate spline (TPS) refinement, or a free-form deformation using NIFTYREG. See Section Label Fusion with a Random Forest Classifier. The pairwise registrations give us a rough estimate of the location of the target organ. However, the accuracy of the solution after the registration can be further improved by taking the local appearance surrounding the target organ into account. In order to do this, we train a random forest classifier that is used to fuse the transferred atlas labels after the registration. See Section Graph Cut Segmentation with a Potts Model. The segmentation is further refined by encouraging spatial smoothness between neighbouring pixels. For this, we formulate the labeling problem as an optimization problem and solve it using graph cuts. See Section 2.3. These steps of our pipeline will be presented in detail in the following sections. Each organ is segmented individually in our multi-atlas approach. 2.1 Pairwise Registration Determination of Organ Specific Feature Sets For each organ and atlas image I i I = {I 1,...,I n }, a subset of features that is designed to produce a reliable registration of the organ of interest is determined. The basic idea is to evaluate how well the extracted feature points in an atlas image match to other feature points in the remaining atlas images. In order to quantify the quality of a matched feature correspondence, we first establish so-called golden transformations between the two atlas images around the organ of interest, based on pre-computed (ground truth) landmark correspondences. Hence, if a feature point is matched to a feature point in another image, then this point-to-point correspondence should be consistent with the golden transformation, provided the correspondence is correct. Otherwise, it is likely to be an outlier. Feature points that always form inlier correspondences are good candidates for reliable registration, and these points determine the organ specific feature sets. Establishing golden transformations. For each atlas image, we compute golden transformations between I i and the other atlas images in I \I i. This is done by apply-
5 Multi-Atlas Segmentation Using Robust Feature-Based Registration 5 ing TPS to precomputed landmark correspondences using the method proposed in [4]. The landmark correspondences are computed through accurate non-rigid registrations between a randomly chosen reference atlas in the atlas set and each of the remaining atlases. The registration uses two channels; the image intensity and the ground truth mask of the organ of interest, and maximizes the similarity between these according to the normalized mutual information (NMI) measure using NIFTYREG, [24]. With the obtained displacement field, the mesh points of a triangular mesh of the ground truth surfaces is transformed to the coordinate system of the reference atlas. For each mesh point (landmark) of the triangulation of the reference ground truth surface, the closest point on each transformed triangulated surface is found using an algorithm based on [7], and chosen as the corresponding landmark. Feature extraction. For each atlas image, I i I, we calculate sparse features according to the method proposed by Svärm et al. in [29]. A feature point is denoted f = (i,x,d) where i is the index of the image, and x and d are the coordinates and the description vector for the point, respectively. Only the features that lie sufficiently close to the organ are considered, i.e., we keep features with distance to the organ, δ, less than a predefined threshold, D max. For the whole atlas I we thus obtain F = {F 1,...,F n }, where F i is the set of feature points for I i. For each atlas image I i, the points in F i are matched to the other feature sets in F \ F i using a symmetric neighbour approach, thus establishing point-to-point correspondences between I i and the other atlas images. Computation of organ specific feature sets. Next, we proceed by applying the golden transformations to the feature points in F \ F i that have been matched to points in F i, in order to transform them into the same coordinate system. Furthermore, we calculate the residuals between the coordinates of the feature points for I i and the corresponding feature points for the other atlas images after the transformation. If feature point f k F i is matched to f k F j, the residual is defined as r k = x k ˆT G j,i x k 2, (1) where ˆT G j,i denotes the golden transformation between I i and I j. Each feature in F i receives a score that is a weighted sum of the normalized residuals for all the corresponding points in the other atlas images and the normalized distance from the organ. More precisely, the score for f k F i is where Score[f k ] = s δ + ω r s k, (2) k R s δ = (D max δ k )/D max, s k = max(t r k,0)/t. (3) Here, ω r is the importance weight for the residuals, T is a predefined threshold, and R is the set of feature points for the atlas images I \ I i, which have been matched to f k. The features are ranked according to their score, and those with the highest
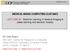
6 6 Fejne & Landgren et al. Fig. 1: Two CT slices of a target image (left) and an atlas image (right) with corresponding features after RANSAC for Lumbar Vertebra 1. scores are kept and used in the registration step. This procedure is relatively time consuming but it is an offline process, and thus only done once Pairwise Registration with RANSAC At run-time, we estimate pairwise affine transformations between the target image I t and all the images in the atlas set, using the feature sets obtained in the previous section. We apply RANSAC with the truncated l 2 norm as a cost function for outlier removal. An example of inlier feature correspondences for Lumbar Vertebra 1 is illustrated in Fig. 1. Furthermore, for about half of the organs the final coordinate transforms between I and I t are obtained by TPS interpolation between the remaining correspondences. For the rest of the organs we find that the best transformations are obtained by using NIFTYREG, with the affine transformation as initialization. These transformations are then used to transfer the labels of the atlas images into the same coordinate system as the target image, see Fig Label Fusion with a Random Forest Classifier We use the pairwise registrations to obtain a rough estimate of where the organ is located. This is done by fusing the transferred labels from each atlas into a so called voxel map, P, in which each voxel can be interpreted as a measure of the likelihood of that voxel belonging to the organ, according to the pairwise registrations. More precisely, the map P is the normalized average of the warped target masks of each of the atlas images, so if half of the atlas images think that voxel i is organ, then P(i) = 0.5. However, the map P largely ignores the local appearance around the target organ and in order to further improve the result, a random forest classifier is trained in an
7 Multi-Atlas Segmentation Using Robust Feature-Based Registration 7 Fig. 2: Example of a registration of Lumbar Vertebra 1. Left: The atlas ground truth mask. Middle: The warped target mask after the registration. Right: The masks overlaid in the same coordinate system. offline process. The classifier is then used to obtain a refined estimate of P, which will be denoted Pr. We implement this using Sherwood [6], which allows us to train and evaluate large random forest instances efficiently. The voxel map, P, and the target image, I, is used to compute a set of features for each voxel, which will be used as input to the random forest classifier. By smoothing I and P using a Gaussian kernel, we obtain two new volumes, which we refer to as Is and Ps. Furthermore, for each organ we determine a threshold level, τ, for P and use this to construct a distance map, DP, where each voxel in DP equals the (signed) distance to the boundary surface of the binary volume P > τ. For each voxel i, in each volume I, we thus obtain five features: I(i), Is (i), P(i), Ps (i) and DP (i). 2.3 Graph Cut Segmentation with a Potts Model The refined estimate, Pr, gives us a better estimation of the segmentation, but there is room for further improvement. The decision of whether voxel i should be classified as belonging to the organ or not is taken without considering the classification of neighboring voxels, which may cause noisy and inaccurate estimates along the boundaries of the target organ. Thus, we can improve the segmentation by incorporating this information into the model, and in order to do so we formulate our voxel labeling problem as an energy minimization problem, and solve it using graph cuts [1]. More precisely, let xi L = {0, 1} be a Boolean indicator variable for voxel i, that is, 1 if xi is classified as belonging to the organ (foreground), and 0 if it belongs to the background. We are now seeking the labeling x? that minimizes the energy function of the form n E(x? ) = Di (xi ) + i=1 i, j N Vi, j (xi, x j ), (4)
8 8 Fejne & Landgren et al. where the data term, D i (x i ), measures how well label x i suits voxel i, given the target image I t, and V i, j (x i,x j ) is an interaction term that regularizes the solution by assigning different costs to neighbouring voxels, which depend on the labels they take. Furthermore, n is the number of voxels in the image, and N defines the neighbourhood system of the voxels. The output from the random forest classifier is used for the data term, in which voxel i is set to take the value 1/2 P r (i) if x i = 1, and zero otherwise, i.e., D i (x i ) = x i ( 1 2 P r(i)). Thus, this model makes it more likely that voxel i is classified as foreground if P r (i) [0.5,1], and background if P r (i) [0,0.5]. As interaction term we use Potts model, which regularizes the resulting segmentation by penalizing neighboring voxels if they receive different labels. It assigns a cost to two neighbouring voxels, i and j, according to λ[x i x j ], where λ is a regularization weight, and x i and x j are the labels for voxels i and j, respectively. This interaction cost can also be expressed as V i, j (x i,x j ) = λ(x i (1 x j ) + x j (1 x i )). Thus, the final segmentation, x, is obtained by solving the following minimization problem: x = argmin n x {0,1} n i=1 ( 1 ) x i 2 P r(i) + λ n i=1 j N(i) µ i j x i (1 x j ), (5) where µ i j is a variable that compensates for anisotropic resolution [18], and N(i) is the set of voxels in the neighbourhood of voxel i, which is set to be 6-connected for all the organs, i.e. each voxel that touches a side of a voxel is a neighbour. Since the cost function in (5) is submodular, it can be minimized efficiently using graph cuts [19] with the implementation of [13]. During the minimization, we process a smaller volume, that is, a cut out around the zero level of the thresholded voxel map P > τ, which allows us to save memory and speed-up the calculations. A comparison between the resulting segmentation of the spleen with graph cuts, using the initial voxel map, P, and the refined probability map after the random forest step, P r, is illustrated in Fig Experimental Evaluation The different steps in the pipeline involve some tuning parameters and these have been set as follows. The 20 whole-body CT images that are available in the challenge are split into one training and validation (atlas) set consisting of the first 15 images, while the remaining 5 serve as a test set. For the registration, the parameters are determined by leave-one-out cross validation of the atlas images, while the remaining ones are used as validation images for the random forest classification and graph cut segmentation. For the computation of the organ specific feature sets, the same parameter settings are used for all organs. At first around 8,000-10,000 features are extracted from a whole body CT image in less than 30s. When ranking the features, the max-
9 Multi-Atlas Segmentation Using Robust Feature-Based Registration 9 GT GT GT x P r x P Fig. 3: Example of the resulting probability estimates and segmentation of the spleen for one CT slice; in each image the ground truth (GT) is indicated. Left: The initial probability, P. Middle: The probability given by random forest, P r. Right: The resulting segmentation x P using P and x P r using P r overlaid on the original image. imal distance is set to D max = 100 mm, the threshold T = 15, and the importance weight for the residuals, ω, is set to 10. We have found empirically that the 300 features with the highest score can be used to provide robust and reliable registration. RANSAC is run iterations and the truncation threshold for the l 2 cost function is set to 30 mm. The value of the standard deviation, σ, for the Gaussian kernel in the smoothing of P for the random forest classifier is 1. Table 1 lists parameters and settings for each individual organ. Note that in the segmentation of some of the organs we do not use a random forest classifier. This is either because the organ has a very large volume, which makes the computations heavy, or because the classifier does not improve the results at all. Furthermore, we do not use the learnt features for the lungs. In our experience, very simple methods yield accurate segmentations of the lungs and that is also the case when we use ordinary features. The single most time consuming online part of the algorithm is the registration and the time needed strongly depends on the size of the organ and the choice of registration method. NIFTYREG takes around s per registration compared to TPS for which a registration takes less than 10s regardless of the organ type. However, we have found empirically that NIFTYREG performs a lot better for ten of the organs, see Table 1. If more images are added to the training set, the process of determining the organ specific features, the registration of the atlas images in order to obtain the voxel map, P, and the training of the random forest classifier would have to be run again from start. The only difference for the online process is that we would have to perform one extra registration per added image. 3.1 Challenge Results In our contribution to the VISCERAL Anatomy Grand Challenge, all 20 images available for training formed the atlas set in the final submission. The algorithm
10 10 Fejne & Landgren et al. Table 1: Parameters used in the pipeline. Organ Registration method RANSAC threshold Random forest Trachea NIFTYREG 20 Yes Right lung TPS Left lung TPS Pancreas TPS 20 Yes Gallbladder TPS Urinary bladder TPS 50 Yes Sternum NIFTYREG 20 Yes Lumbar Vertebra 1 NIFTYREG 7 Yes Right kidney NIFTYREG 25 Yes Left kidney NIFTYREG 20 Yes Right adrenal gland TPS Left adrenal gland TPS Right psoas major NIFTYREG 40 Yes Left psoas major NIFTYREG 30 Yes Muscle body of right rectus abdominis NIFTYREG 40 Yes Muscle body of left rectus abdominis NIFTYREG 40 Yes Aorta NIFTYREG 30 Yes Liver TPS 50 Yes Thyroid gland TPS 20 Yes Spleen TPS 30 Yes τ λ was evaluated on a test set consisting of 10 new whole-body CT images that only were available to the organizers of the competition, and the evaluation took place at ISBI The results are measured using the dice index, which is defined as Dice(S, G) = 2 S G /( S + G ), where S and G are the computed segmentation and ground truth, respectively. Thus, a perfect segmentation would yield dice index 1, while a segmentation with no ground truth overlap would receive a dice index of 0. Our results are reported in Table 2 together with the the results from the strongest competitors: CMIV - Center for Medical Image Science and Visualization, Linköping University, HES-SO - University of Applied Sciences Western Switzerland SIAT - Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences. In summary, our algorithm provides the best results for 13 of the 20 organs. 3.2 Detailed Evaluation In this section, we evaluate what kinds of benefits that specific parts of the pipeline provide for Lumbar Vertebra 1, the left kidney, and the spleen. Dice scores after
11 Multi-Atlas Segmentation Using Robust Feature-Based Registration 11 Table 2: Final results measured in DICE index for whole-body CT images. Here * means that no segmentation was provided. Organ Our CMIV HES-SO SIAT Left Kidney * Right Kidney * Spleen Liver Left Lung Right Lung Urinary Bladder * Muscle Body of Left Rectus Abdominis * * Muscle Body of Right Rectus Abdominis * * Lumbar Vertebra * * Thyroid * * Pancreas * * Left Psoas Major Muscle * Right Psoas Major Muscle * Gallbladder * * Sternum * * Aorta * * Trachea * 0.92 * Left Adrenal Gland * * Right Adrenal Gland * * Average * * different steps in the pipeline, when using both organ specific and ordinary features sets, are presented in Table 3. We denote the segmentation that can be obtained after fusing the transferred labels from the registration with x P and it is the map P, thresholded at τ. The values in the table are the average results for the 5 images that formed the test set when using the 15 first as atlas set as described in the introduction to this section. Note that these values are not comparable with the ones in Table 2 since those were obtained with a different amount of training data and evaluated on images that are not accessible to us. The results in Table 3 should be compared among themselves in order to determine the contribution of different parts of the pipeline for the selected organs. Clearly, the organ specific features improve the results significantly for the segmentation of Lumbar Vertebra 1, which is demonstrated in Fig. 4. The left and the middle pictures in Fig. 5 show the segmentation of the spleen for CT image 19 before and after the graph cut part in the pipeline, respectively. It is clear from both the figures and the table that the random forest classifier improves the results a lot. However, note that the segmentation in the middle picture of Fig. 5 is not a valid spleen shape. The right picture of Fig. 5 illustrates the final segmentation for the left kidney. This segmentation is obtained using organ specific features, and the result is quite accurate. However, the random forest classifier and the graph cut segmentation do not seem to further improve the Dice index for the left kidney according to the
12 12 Fejne & Landgren et al. Fig. 4: Segmentation of Lumbar Vertebra 1. Here the ground truth is red and our segmentation is blue. Left: Segmentation using ordinary features. The registration fails to find the correct vertebra as it is confused by a nearby vertebra. Right: Segmentation using organ specific features. Now the correct vertebra is located. results in the table. Note that his does not necessarily mean that the segmentation is not improved. It is still possible that the produced solution is more regularized and accurate, just not according to the Dice index. Furthermore, the Dice scores for the left kidney and the spleen are more or less the same when using organ specific features compared to when using ordinary. In our experience, with a lot of training data the organ specific features generally performs a little better than the ordinary features although that may not be the case for all the organs. It is, however, worth mentioning that there are certain limitations regarding the performance of the organ specific features. It requires accurate landmark correspondences in order to establish reliable golden transformations, see Section 2.1. If this is not the case, it may not be advantageous to use organ specific features. Table 3: Results for three organs after different steps in the pipeline, measured in Dice index. Here OF indicates that ordinary feature sets were used instead of the organ specific ones. x P is the map P, thresholded at τ and x is the graph cut segmentation after the random forest step. Organ x P (OF) x (OF) x P x Lumbar Vertebra Left kidney Spleen
13 Multi-Atlas Segmentation Using Robust Feature-Based Registration 13 Fig. 5: Ground truth (red) and computed segmentation (blue) for some of the organs after different steps in the pipeline. Organ specific features are used for all the segmentations. Left: Warped ground truth mask of the spleen after the registration. Middle: Graph cut segmentation of the spleen. Right: Graph cut segmentation of the left kidney. 4 Conclusions In this paper, we have described an algorithm that uses a feature-based approach to multi-atlas segmentation of organs in whole-body CT images. The results clearly demonstrate that this method manages to locate and segment the organs with stateof-the-art results, and our approach outperforms the competitors at the VISCERAL Anatomy Grand Challenge on segmentation at ISBI 2015 for 13 out of 20 organs. However, some parts of the algorithm could benefit from further work. For instance, incorporating prior information about the organ shapes into the pipeline would help to guarantee that the algorithm produces feasible organ shapes. A more thorough evaluation of the organs specific feature sets could help determine individual parameter settings for the organs, as well as help explaining why the method improves the result for some organs but not for all. Furthermore, adding additional features to the random forest classifier would likely yield better results. Moreover, the calculations could be sped-up, e.g. by considering alternative registration methods. References 1. Boykov, Y., Veksler, O., Zabih, R.: Fast approximate energy minimization via graph cuts. IEEE Transactions on Pattern Analysis and Machine Intelligence 23(11), (2001) 2. Candemir, S., Jaeger, S., Palaniappan, K., Musco, J., Singh, R., Xue, Z., Karargyris, A., Antani, S., Thoma, G., McDonald, C.: Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Transactions on Medical Imaging 33(2), (2014)
14 14 Fejne & Landgren et al. 3. Chu, C., Oda, M., Kitasaka, T., Misawa, K., Fujiwara, M., Hayashi, Y., Nimura, Y., Rueckert, D., Mori, K.: Multi-organ segmentation based on spatially-divided probabilistic atlas from 3D abdominal CT images. In: K. Mori, I. Sakuma, Y. Sato, C. Barillot, N. Navab (eds.) Medical Image Computing and Computer-Assisted Intervention MICCAI 2013, Lecture Notes in Computer Science, vol. 8150, pp Springer Berlin Heidelberg (2013) 4. Chui, H., Rangarajan, A.: A new algorithm for non-rigid point matching. In: IEEE Conference on Computer Vision and Pattern Recognition, vol. 2, pp (2000) 5. Chupin, M., Grardin, E., Cuingnet, R., Boutet, C., Lemieux, L., Lehricy, S., Benali, H., Garnero, L., Colliot, O.: Fully automatic hippocampus segmentation and classification in Alzheimer s disease and mild cognitive impairment applied on data from adni. Hippocampus 19(6), (2009) 6. Criminisi, A., Shotton, J., Konukoglu, E.: Decision forests: A unified framework for classification, regression, density estimation, manifold learning and semi-supervised learning. Foundations and Trends R in Computer Graphics and Vision 7, (2011) 7. Eberly, D.: Distance between point and triangle in 3D (2008). Geometric Tools, LLC, Available from: 8. Han, X.: Learning-boosted label fusion for multi-atlas auto-segmentation. In: G. Wu, D. Zhang, D. Shen, P. Yan, K. Suzuki, F. Wang (eds.) Machine Learning in Medical Imaging, Lecture Notes in Computer Science, vol. 8184, pp Springer International Publishing (2013) 9. Han, X., Hoogeman, M., Levendag, P., Hibbard, L., Teguh, D., Voet, P., Cowen, A., Wolf, T.: Atlas-based auto-segmentation of head and neck CT images. In: D. Metaxas, L. Axel, G. Fichtinger, G. Szkely (eds.) Medical Image Computing and Computer-Assisted Intervention MICCAI 2008, Lecture Notes in Computer Science, vol. 5242, pp Springer Berlin Heidelberg (2008) 10. Heckemann, R.A., Hajnal, J.V., Aljabar, P., Rueckert, D., Hammers, A.: Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage 33(1), (2006) 11. Heckemann, R.A., Keihaninejad, S., Aljabar, P., Rueckert, D., Hajnal, J.V., Hammers, A.: Improving intersubject image registration using tissue-class information benefits robustness and accuracy of multi-atlas based anatomical segmentation. Neuroimage 51(1), (2010) 12. Iglesias, J.E., Sabuncu, M.R.: Multi-atlas segmentation of biomedical images: A survey. Medical Image Analysis 24(1), (2015) 13. Jamriška, O., Sýkora, D., Hornung, A.: Cache-efficient graph cuts on structured grids. IEEE Conference on Computer Vision and Pattern Recognition pp (2012) 14. Jiménez-del-Toro, O.A., Müller, H., Krenn, M., Gruenberg, K., Taha, A.A., Winterstein, M., Eggel, I., Foncubierta-Rodrguez, A., Goksel, O., Jakab, A., Kontokotsios, G., Langs, G., Menze, B., Fernandez, T.S., Schaer, R., Walleyo, A., Weber, M., Cid, Y.D., Gass, T., Heinrich, M., Jia, F., Kahl, F., Kechichian, R., Mai, D., Spanier, A.B., Vincent, G., Wang, C., Wyeth, D., Hanbury, A.: Cloudbased evaluation of anatomical structure segmentation and landmark detection algorithms: Visceral anatomy benchmarks. IEEE Transactions on Medical Imaging (2016). Accepted 15. Khalifa, F., Beache, G., Gimel farb, G., Suri, J., El-Baz, A.: State-of-the-art medical image registration methodologies: A survey. In: Multi Modality State-of-the-Art Medical Image Segmentation and Registration Methodologies, pp Springer (2011) 16. Kirisli, H.A., Schaap, M., Klein, S., Neefjes, L.A., Weustink, A.C., van Walsum, T., Niessen, W.J.: Fully automatic cardiac segmentation from 3D CTA data: A multi-atlas based approach. In: SPIE Medical Imaging. San Diego, USA (2010) 17. Klein, A., Mensh, B., Ghosh, S., Tourville, J., Hirsch, J.: Mindboggle: Automated brain labeling with multiple atlases. BMC Medical Imaging 5(1), 7 (2005) 18. Kolmogorov, V., Boykov, Y.: What metrics can be approximated by geo-cuts, or global optimization of length/area and flux. In: IEEE International Conference on Computer Vision, vol. 1, pp (2005) 19. Kolmogorov, V., Zabin, R.: What energy functions can be minimized via graph cuts? IEEE Transactions on Pattern Analysis and Machine Intelligence 26(2), (2004)
15 Multi-Atlas Segmentation Using Robust Feature-Based Registration Kurkure, U., Le, Y., Ju, T., Carson, J., Paragios, N., Kakadiaris, I.: Subdivision-based deformable model for geometric atlas fitting. In: IEEE International Conference on Computer Vision. Barcelona, Spain (2011) 21. Lee, J.G., Gumus, S., Moon, C.H., Kwoh, C.K., Bae, K.T.: Fully automated segmentation of cartilage from the MR images of knee using a multi-atlas and local structural analysis method. Medical Physics 41(9) (2014) 22. van der Lijn, F., den Heijer, T., Breteler, M.M., Niessen, W.J.: Hippocampus segmentation in MR images using atlas registration, voxel classification, and graph cuts. NeuroImage 43(4), (2008) 23. Okada, T., Linguraru, M.G., Hori, M., Summers, R.M., Tomiyama, N., Sato, Y.: Abdominal multi-organ segmentation from CT images using conditional shapelocation and unsupervised intensity priors. Medical Image Analysis 26(1), 1 18 (2015) 24. Ourselin, S., Roche, A., Subsol, G., Pennec, X., Ayache, N.: Reconstructing a 3D structure from serial histological sections. Image and Vision Computing 19(1-2), (2001) 25. Pekar, V., McNutt, T.R., Kaus, M.R.: Automated model-based organ delineation for radiotherapy planning in prostatic region. International Journal of Radiation Oncology*Biology*Physics 60(3), (2004) 26. Rohlfing, T., Brandt, R., Menzel, R., Jr., C.R.M.: Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. NeuroImage 21(4), (2004) 27. Sanroma, G., Wu, G., Gao, Y., Shen, D.: Learning-based atlas selection for multiple-atlas segmentation. In: IEEE Conference on Computer Vision and Pattern Recognition. Columbus, USA (2014) 28. Sotiras, A., Davatzikos, C., Paragios, N.: Deformable medical image registration: A survey. IEEE Transactions on Medical Imaging 32(7), (2013) 29. Svärm, L., Enqvist, O., Kahl, F., Oskarsson, M.: Improving robustness for inter-subject medical image registration using a feature-based approach. In: International Symposium on Biomedical Imaging (2015) 30. Taylor, R., Stoianovici, D.: Medical robotics in computer-integrated surgery. Robotics and Automation, IEEE Transactions on 19(5), (2003) 31. Wang, H., Suh, J., S, R.D., Pluta, J., Craige, C., Yushkevich, P.: Multi-atlas segmentation with joint label fusion. IEEE Transactions on Pattern Analysis and Machine Intelligence 35(3), (2013) 32. Wang, L., Chen, K.C., Gao, Y., Shi, F., Liao, S., Li, G., Shen, S.G.F., Yan, J., Lee, P.K.M., Chow, B., Liu, N.X., Xia, J.J., Shen, D.: Automated bone segmentation from dental CBCT images using patch-based sparse representation and convex optimization. Medical Physics 41(4), (2014) 33. Warfield, S., Zou, K., Wells, W.: Simultaneous truth and performance level estimation (STA- PLE): An algorithm for the validation of image segmentation. IEEE Transactions on Medical Imaging 27(3), (2004) 34. Wolz, R., Chu, C., Misawa, K., Fujiwara, M., Mori, K., Rueckert, D.: Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE Transactions on Medical Imaging 32(9), (2013) 35. Xu, Z., Burke, R.P., Lee, C.P., Baucom, R.B., Poulose, B.K., Abramson, R.G., Landman, B.A.: Efficient multi-atlas abdominal segmentation on clinically acquired CT with SIMPLE context learning. Medical Image Analysis 24(1), (2015)
Shape-Aware Multi-Atlas Segmentation
 Shape-Aware Multi-Atlas Segmentation Jennifer Alvén, Fredrik Kahl, Matilda Landgren, Viktor Larsson and Johannes Ulén Department of Signals and Systems, Chalmers University of Technology, Sweden Email:
Shape-Aware Multi-Atlas Segmentation Jennifer Alvén, Fredrik Kahl, Matilda Landgren, Viktor Larsson and Johannes Ulén Department of Signals and Systems, Chalmers University of Technology, Sweden Email:
Hierarchical Multi structure Segmentation Guided by Anatomical Correlations
 Hierarchical Multi structure Segmentation Guided by Anatomical Correlations Oscar Alfonso Jiménez del Toro oscar.jimenez@hevs.ch Henning Müller henningmueller@hevs.ch University of Applied Sciences Western
Hierarchical Multi structure Segmentation Guided by Anatomical Correlations Oscar Alfonso Jiménez del Toro oscar.jimenez@hevs.ch Henning Müller henningmueller@hevs.ch University of Applied Sciences Western
VISCERAL Anatomy3 Organ Segmentation Challenge
 VISCERAL@ISBI 2015 VISCERAL Anatomy3 Organ Segmentation Challenge co-located with IEEE International Symposium on Biomedical Imaging 2015 New York, NY, USA, April 16, 2015 Proceedings Orcun Goksel Oscar
VISCERAL@ISBI 2015 VISCERAL Anatomy3 Organ Segmentation Challenge co-located with IEEE International Symposium on Biomedical Imaging 2015 New York, NY, USA, April 16, 2015 Proceedings Orcun Goksel Oscar
Manifold Learning-based Data Sampling for Model Training
 Manifold Learning-based Data Sampling for Model Training Shuqing Chen 1, Sabrina Dorn 2, Michael Lell 3, Marc Kachelrieß 2,Andreas Maier 1 1 Pattern Recognition Lab, FAU Erlangen-Nürnberg 2 German Cancer
Manifold Learning-based Data Sampling for Model Training Shuqing Chen 1, Sabrina Dorn 2, Michael Lell 3, Marc Kachelrieß 2,Andreas Maier 1 1 Pattern Recognition Lab, FAU Erlangen-Nürnberg 2 German Cancer
Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search
 Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search Baochun He, Cheng Huang, Fucang Jia Shenzhen Institutes of Advanced Technology, Chinese Academy
Fully Automatic Multi-organ Segmentation based on Multi-boost Learning and Statistical Shape Model Search Baochun He, Cheng Huang, Fucang Jia Shenzhen Institutes of Advanced Technology, Chinese Academy
Supervoxel Classification Forests for Estimating Pairwise Image Correspondences
 Supervoxel Classification Forests for Estimating Pairwise Image Correspondences Fahdi Kanavati 1, Tong Tong 1, Kazunari Misawa 2, Michitaka Fujiwara 3, Kensaku Mori 4, Daniel Rueckert 1, and Ben Glocker
Supervoxel Classification Forests for Estimating Pairwise Image Correspondences Fahdi Kanavati 1, Tong Tong 1, Kazunari Misawa 2, Michitaka Fujiwara 3, Kensaku Mori 4, Daniel Rueckert 1, and Ben Glocker
Multi-Atlas Segmentation of the Cardiac MR Right Ventricle
 Multi-Atlas Segmentation of the Cardiac MR Right Ventricle Yangming Ou, Jimit Doshi, Guray Erus, and Christos Davatzikos Section of Biomedical Image Analysis (SBIA) Department of Radiology, University
Multi-Atlas Segmentation of the Cardiac MR Right Ventricle Yangming Ou, Jimit Doshi, Guray Erus, and Christos Davatzikos Section of Biomedical Image Analysis (SBIA) Department of Radiology, University
Attribute Similarity and Mutual-Saliency Weighting for Registration and Label Fusion
 Attribute Similarity and Mutual-Saliency Weighting for Registration and Label Fusion Yangming Ou, Jimit Doshi, Guray Erus, and Christos Davatzikos Section of Biomedical Image Analysis (SBIA) Department
Attribute Similarity and Mutual-Saliency Weighting for Registration and Label Fusion Yangming Ou, Jimit Doshi, Guray Erus, and Christos Davatzikos Section of Biomedical Image Analysis (SBIA) Department
Semantic Context Forests for Learning- Based Knee Cartilage Segmentation in 3D MR Images
 Semantic Context Forests for Learning- Based Knee Cartilage Segmentation in 3D MR Images MICCAI 2013: Workshop on Medical Computer Vision Authors: Quan Wang, Dijia Wu, Le Lu, Meizhu Liu, Kim L. Boyer,
Semantic Context Forests for Learning- Based Knee Cartilage Segmentation in 3D MR Images MICCAI 2013: Workshop on Medical Computer Vision Authors: Quan Wang, Dijia Wu, Le Lu, Meizhu Liu, Kim L. Boyer,
Deformable Segmentation using Sparse Shape Representation. Shaoting Zhang
 Deformable Segmentation using Sparse Shape Representation Shaoting Zhang Introduction Outline Our methods Segmentation framework Sparse shape representation Applications 2D lung localization in X-ray 3D
Deformable Segmentation using Sparse Shape Representation Shaoting Zhang Introduction Outline Our methods Segmentation framework Sparse shape representation Applications 2D lung localization in X-ray 3D
Prototype of Silver Corpus Merging Framework
 www.visceral.eu Prototype of Silver Corpus Merging Framework Deliverable number D3.3 Dissemination level Public Delivery data 30.4.2014 Status Authors Final Markus Krenn, Allan Hanbury, Georg Langs This
www.visceral.eu Prototype of Silver Corpus Merging Framework Deliverable number D3.3 Dissemination level Public Delivery data 30.4.2014 Status Authors Final Markus Krenn, Allan Hanbury, Georg Langs This
Machine Learning for Medical Image Analysis. A. Criminisi
 Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
Machine Learning for Medical Image Analysis A. Criminisi Overview Introduction to machine learning Decision forests Applications in medical image analysis Anatomy localization in CT Scans Spine Detection
PROSTATE CANCER DETECTION USING LABEL IMAGE CONSTRAINED MULTIATLAS SELECTION
 PROSTATE CANCER DETECTION USING LABEL IMAGE CONSTRAINED MULTIATLAS SELECTION Ms. Vaibhavi Nandkumar Jagtap 1, Mr. Santosh D. Kale 2 1 PG Scholar, 2 Assistant Professor, Department of Electronics and Telecommunication,
PROSTATE CANCER DETECTION USING LABEL IMAGE CONSTRAINED MULTIATLAS SELECTION Ms. Vaibhavi Nandkumar Jagtap 1, Mr. Santosh D. Kale 2 1 PG Scholar, 2 Assistant Professor, Department of Electronics and Telecommunication,
Discriminative Dictionary Learning for Abdominal Multi-Organ Segmentation
 Discriminative Dictionary Learning for Abdominal Multi-Organ Segmentation Tong Tong a,, Robin Wolz a, Zehan Wang a, Qinquan Gao a, Kazunari Misawa b, Michitaka Fujiwara c, Kensaku Mori d, Joseph V. Hajnal
Discriminative Dictionary Learning for Abdominal Multi-Organ Segmentation Tong Tong a,, Robin Wolz a, Zehan Wang a, Qinquan Gao a, Kazunari Misawa b, Michitaka Fujiwara c, Kensaku Mori d, Joseph V. Hajnal
Auto-contouring the Prostate for Online Adaptive Radiotherapy
 Auto-contouring the Prostate for Online Adaptive Radiotherapy Yan Zhou 1 and Xiao Han 1 Elekta Inc., Maryland Heights, MO, USA yan.zhou@elekta.com, xiao.han@elekta.com, Abstract. Among all the organs under
Auto-contouring the Prostate for Online Adaptive Radiotherapy Yan Zhou 1 and Xiao Han 1 Elekta Inc., Maryland Heights, MO, USA yan.zhou@elekta.com, xiao.han@elekta.com, Abstract. Among all the organs under
A Review on Label Image Constrained Multiatlas Selection
 A Review on Label Image Constrained Multiatlas Selection Ms. VAIBHAVI NANDKUMAR JAGTAP 1, Mr. SANTOSH D. KALE 2 1PG Scholar, Department of Electronics and Telecommunication, SVPM College of Engineering,
A Review on Label Image Constrained Multiatlas Selection Ms. VAIBHAVI NANDKUMAR JAGTAP 1, Mr. SANTOSH D. KALE 2 1PG Scholar, Department of Electronics and Telecommunication, SVPM College of Engineering,
Multi-atlas spectral PatchMatch: Application to cardiac image segmentation
 Multi-atlas spectral PatchMatch: Application to cardiac image segmentation W. Shi 1, H. Lombaert 3, W. Bai 1, C. Ledig 1, X. Zhuang 2, A. Marvao 1, T. Dawes 1, D. O Regan 1, and D. Rueckert 1 1 Biomedical
Multi-atlas spectral PatchMatch: Application to cardiac image segmentation W. Shi 1, H. Lombaert 3, W. Bai 1, C. Ledig 1, X. Zhuang 2, A. Marvao 1, T. Dawes 1, D. O Regan 1, and D. Rueckert 1 1 Biomedical
Adaptive Local Multi-Atlas Segmentation: Application to Heart Segmentation in Chest CT Scans
 Adaptive Local Multi-Atlas Segmentation: Application to Heart Segmentation in Chest CT Scans Eva M. van Rikxoort, Ivana Isgum, Marius Staring, Stefan Klein and Bram van Ginneken Image Sciences Institute,
Adaptive Local Multi-Atlas Segmentation: Application to Heart Segmentation in Chest CT Scans Eva M. van Rikxoort, Ivana Isgum, Marius Staring, Stefan Klein and Bram van Ginneken Image Sciences Institute,
Using Probability Maps for Multi organ Automatic Segmentation
 Using Probability Maps for Multi organ Automatic Segmentation Ranveer Joyseeree 1,2, Óscar Jiménez del Toro1, and Henning Müller 1,3 1 University of Applied Sciences Western Switzerland (HES SO), Sierre,
Using Probability Maps for Multi organ Automatic Segmentation Ranveer Joyseeree 1,2, Óscar Jiménez del Toro1, and Henning Müller 1,3 1 University of Applied Sciences Western Switzerland (HES SO), Sierre,
Überatlas: Robust Speed-Up of Feature-Based Registration and Multi-Atlas Based Segmentation. Jennifer Alvén
 Überatlas: Robust Speed-Up of Feature-Based Registration and Multi-Atlas Based Segmentation Jennifer Alvén January 2015 Abstract Registration is a key component in multi-atlas approaches to medical image
Überatlas: Robust Speed-Up of Feature-Based Registration and Multi-Atlas Based Segmentation Jennifer Alvén January 2015 Abstract Registration is a key component in multi-atlas approaches to medical image
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION
 ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
ADAPTIVE GRAPH CUTS WITH TISSUE PRIORS FOR BRAIN MRI SEGMENTATION Abstract: MIP Project Report Spring 2013 Gaurav Mittal 201232644 This is a detailed report about the course project, which was to implement
Regional Manifold Learning for Deformable Registration of Brain MR Images
 Regional Manifold Learning for Deformable Registration of Brain MR Images Dong Hye Ye, Jihun Hamm, Dongjin Kwon, Christos Davatzikos, and Kilian M. Pohl Department of Radiology, University of Pennsylvania,
Regional Manifold Learning for Deformable Registration of Brain MR Images Dong Hye Ye, Jihun Hamm, Dongjin Kwon, Christos Davatzikos, and Kilian M. Pohl Department of Radiology, University of Pennsylvania,
3D Brain Segmentation Using Active Appearance Models and Local Regressors
 3D Brain Segmentation Using Active Appearance Models and Local Regressors K.O. Babalola, T.F. Cootes, C.J. Twining, V. Petrovic, and C.J. Taylor Division of Imaging Science and Biomedical Engineering,
3D Brain Segmentation Using Active Appearance Models and Local Regressors K.O. Babalola, T.F. Cootes, C.J. Twining, V. Petrovic, and C.J. Taylor Division of Imaging Science and Biomedical Engineering,
Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion
 Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion Mattias P. Heinrich Julia A. Schnabel, Mark Jenkinson, Sir Michael Brady 2 Clinical
Methodological progress in image registration for ventilation estimation, segmentation propagation and multi-modal fusion Mattias P. Heinrich Julia A. Schnabel, Mark Jenkinson, Sir Michael Brady 2 Clinical
Chapter 11 Automatic Multiorgan Segmentation Using Hierarchically Registered Probabilistic Atlases
 Chapter 11 Automatic Multiorgan Segmentation Using Hierarchically Registered Probabilistic Atlases Razmig Kéchichian, Sébastien Valette and Michel Desvignes Abstract We propose a generic method for the
Chapter 11 Automatic Multiorgan Segmentation Using Hierarchically Registered Probabilistic Atlases Razmig Kéchichian, Sébastien Valette and Michel Desvignes Abstract We propose a generic method for the
Learning-based Neuroimage Registration
 Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
Learning-based Neuroimage Registration Leonid Teverovskiy and Yanxi Liu 1 October 2004 CMU-CALD-04-108, CMU-RI-TR-04-59 School of Computer Science Carnegie Mellon University Pittsburgh, PA 15213 Abstract
Keypoint Transfer Segmentation
 Keypoint Transfer Segmentation The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Wachinger, C.; Toews,
Keypoint Transfer Segmentation The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Wachinger, C.; Toews,
Joint Supervoxel Classification Forest for Weakly-Supervised Organ Segmentation
 Joint Supervoxel Classification Forest for Weakly-Supervised Organ Segmentation Fahdi Kanavati 1, Kazunari Misawa 2, Michitaka Fujiwara 3, Kensaku Mori 4, Daniel Rueckert 1, and Ben Glocker 1 1 Biomedical
Joint Supervoxel Classification Forest for Weakly-Supervised Organ Segmentation Fahdi Kanavati 1, Kazunari Misawa 2, Michitaka Fujiwara 3, Kensaku Mori 4, Daniel Rueckert 1, and Ben Glocker 1 1 Biomedical
Simultaneous Model-based Segmentation of Multiple Objects
 Simultaneous Model-based Segmentation of Multiple Objects Astrid Franz 1, Robin Wolz 1, Tobias Klinder 1,2, Cristian Lorenz 1, Hans Barschdorf 1, Thomas Blaffert 1, Sebastian P. M. Dries 1, Steffen Renisch
Simultaneous Model-based Segmentation of Multiple Objects Astrid Franz 1, Robin Wolz 1, Tobias Klinder 1,2, Cristian Lorenz 1, Hans Barschdorf 1, Thomas Blaffert 1, Sebastian P. M. Dries 1, Steffen Renisch
Non-rigid Image Registration
 Overview Non-rigid Image Registration Introduction to image registration - he goal of image registration - Motivation for medical image registration - Classification of image registration - Nonrigid registration
Overview Non-rigid Image Registration Introduction to image registration - he goal of image registration - Motivation for medical image registration - Classification of image registration - Nonrigid registration
MARS: Multiple Atlases Robust Segmentation
 Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Ensemble registration: Combining groupwise registration and segmentation
 PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
Rule-Based Ventral Cavity Multi-organ Automatic Segmentation in CT Scans
 Rule-Based Ventral Cavity Multi-organ Automatic Segmentation in CT Scans Assaf B. Spanier (B) and Leo Joskowicz The Rachel and Selim Benin School of Computer Science and Engineering, The Hebrew University
Rule-Based Ventral Cavity Multi-organ Automatic Segmentation in CT Scans Assaf B. Spanier (B) and Leo Joskowicz The Rachel and Selim Benin School of Computer Science and Engineering, The Hebrew University
Robust Abdominal Organ Segmentation Using Regional Convolutional Neural Networks
 Robust Abdominal Organ Segmentation Using Regional Convolutional Neural Networks Måns Larsson 1, Yuhang Zhang 1, and Fredrik Kahl 1,2 1 Chalmers University of Technology, Gothenburg, Sweden, {mans.larsson,alven,fredrik.kahl}@chalmers.se
Robust Abdominal Organ Segmentation Using Regional Convolutional Neural Networks Måns Larsson 1, Yuhang Zhang 1, and Fredrik Kahl 1,2 1 Chalmers University of Technology, Gothenburg, Sweden, {mans.larsson,alven,fredrik.kahl}@chalmers.se
Computational Radiology Lab, Children s Hospital, Harvard Medical School, Boston, MA.
 Shape prior integration in discrete optimization segmentation algorithms M. Freiman Computational Radiology Lab, Children s Hospital, Harvard Medical School, Boston, MA. Email: moti.freiman@childrens.harvard.edu
Shape prior integration in discrete optimization segmentation algorithms M. Freiman Computational Radiology Lab, Children s Hospital, Harvard Medical School, Boston, MA. Email: moti.freiman@childrens.harvard.edu
Mapping Multi-Modal Routine Imaging Data to a Single Reference via Multiple Templates
 Mapping Multi-Modal Routine Imaging Data to a Single Reference via Multiple Templates Johannes Hofmanninger 1, Bjoern Menze 2, Marc-André Weber 3,4 and Georg Langs 1 1 Department of Biomedical imaging
Mapping Multi-Modal Routine Imaging Data to a Single Reference via Multiple Templates Johannes Hofmanninger 1, Bjoern Menze 2, Marc-André Weber 3,4 and Georg Langs 1 1 Department of Biomedical imaging
Automatic Segmentation of Parotids from CT Scans Using Multiple Atlases
 Automatic Segmentation of Parotids from CT Scans Using Multiple Atlases Jinzhong Yang, Yongbin Zhang, Lifei Zhang, and Lei Dong Department of Radiation Physics, University of Texas MD Anderson Cancer Center
Automatic Segmentation of Parotids from CT Scans Using Multiple Atlases Jinzhong Yang, Yongbin Zhang, Lifei Zhang, and Lei Dong Department of Radiation Physics, University of Texas MD Anderson Cancer Center
Image Segmentation and Registration
 Image Segmentation and Registration Dr. Christine Tanner (tanner@vision.ee.ethz.ch) Computer Vision Laboratory, ETH Zürich Dr. Verena Kaynig, Machine Learning Laboratory, ETH Zürich Outline Segmentation
Image Segmentation and Registration Dr. Christine Tanner (tanner@vision.ee.ethz.ch) Computer Vision Laboratory, ETH Zürich Dr. Verena Kaynig, Machine Learning Laboratory, ETH Zürich Outline Segmentation
STIC AmSud Project. Graph cut based segmentation of cardiac ventricles in MRI: a shape-prior based approach
 STIC AmSud Project Graph cut based segmentation of cardiac ventricles in MRI: a shape-prior based approach Caroline Petitjean A joint work with Damien Grosgeorge, Pr Su Ruan, Pr JN Dacher, MD October 22,
STIC AmSud Project Graph cut based segmentation of cardiac ventricles in MRI: a shape-prior based approach Caroline Petitjean A joint work with Damien Grosgeorge, Pr Su Ruan, Pr JN Dacher, MD October 22,
Optimal MAP Parameters Estimation in STAPLE - Learning from Performance Parameters versus Image Similarity Information
 Optimal MAP Parameters Estimation in STAPLE - Learning from Performance Parameters versus Image Similarity Information Subrahmanyam Gorthi 1, Alireza Akhondi-Asl 1, Jean-Philippe Thiran 2, and Simon K.
Optimal MAP Parameters Estimation in STAPLE - Learning from Performance Parameters versus Image Similarity Information Subrahmanyam Gorthi 1, Alireza Akhondi-Asl 1, Jean-Philippe Thiran 2, and Simon K.
MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 20: Machine Learning in Medical Imaging II (deep learning and decision forests)
 SPRING 2016 1 MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 20: Machine Learning in Medical Imaging II (deep learning and decision forests) Dr. Ulas Bagci HEC 221, Center for Research in Computer Vision (CRCV),
SPRING 2016 1 MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 20: Machine Learning in Medical Imaging II (deep learning and decision forests) Dr. Ulas Bagci HEC 221, Center for Research in Computer Vision (CRCV),
ABSTRACT 1. INTRODUCTION 2. METHODS
 Finding Seeds for Segmentation Using Statistical Fusion Fangxu Xing *a, Andrew J. Asman b, Jerry L. Prince a,c, Bennett A. Landman b,c,d a Department of Electrical and Computer Engineering, Johns Hopkins
Finding Seeds for Segmentation Using Statistical Fusion Fangxu Xing *a, Andrew J. Asman b, Jerry L. Prince a,c, Bennett A. Landman b,c,d a Department of Electrical and Computer Engineering, Johns Hopkins
Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues
 Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues D. Zikic, B. Glocker, E. Konukoglu, J. Shotton, A. Criminisi, D. H. Ye, C. Demiralp 3, O. M. Thomas 4,5, T. Das 4, R. Jena
Context-sensitive Classification Forests for Segmentation of Brain Tumor Tissues D. Zikic, B. Glocker, E. Konukoglu, J. Shotton, A. Criminisi, D. H. Ye, C. Demiralp 3, O. M. Thomas 4,5, T. Das 4, R. Jena
Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information
 Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information Andreas Biesdorf 1, Stefan Wörz 1, Hans-Jürgen Kaiser 2, Karl Rohr 1 1 University of Heidelberg, BIOQUANT, IPMB,
Hybrid Spline-based Multimodal Registration using a Local Measure for Mutual Information Andreas Biesdorf 1, Stefan Wörz 1, Hans-Jürgen Kaiser 2, Karl Rohr 1 1 University of Heidelberg, BIOQUANT, IPMB,
PBSI: A symmetric probabilistic extension of the Boundary Shift Integral
 PBSI: A symmetric probabilistic extension of the Boundary Shift Integral Christian Ledig 1, Robin Wolz 1, Paul Aljabar 1,2, Jyrki Lötjönen 3, Daniel Rueckert 1 1 Department of Computing, Imperial College
PBSI: A symmetric probabilistic extension of the Boundary Shift Integral Christian Ledig 1, Robin Wolz 1, Paul Aljabar 1,2, Jyrki Lötjönen 3, Daniel Rueckert 1 1 Department of Computing, Imperial College
A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images
 A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images Jianhua Yao 1, Russell Taylor 2 1. Diagnostic Radiology Department, Clinical Center,
A Multiple-Layer Flexible Mesh Template Matching Method for Nonrigid Registration between a Pelvis Model and CT Images Jianhua Yao 1, Russell Taylor 2 1. Diagnostic Radiology Department, Clinical Center,
Multi-Organ Segmentation using Vantage Point Forests and Binary Context Features
 Multi-Organ Segmentation using Vantage Point Forests and Binary Context Features Mattias P. Heinrich and Maximilian Blendowski Institute of Medical Informatics, University of Lübeck, Germany heinrich@imi.uni-luebeck.de,
Multi-Organ Segmentation using Vantage Point Forests and Binary Context Features Mattias P. Heinrich and Maximilian Blendowski Institute of Medical Informatics, University of Lübeck, Germany heinrich@imi.uni-luebeck.de,
1 Introduction Motivation and Aims Functional Imaging Computational Neuroanatomy... 12
 Contents 1 Introduction 10 1.1 Motivation and Aims....... 10 1.1.1 Functional Imaging.... 10 1.1.2 Computational Neuroanatomy... 12 1.2 Overview of Chapters... 14 2 Rigid Body Registration 18 2.1 Introduction.....
Contents 1 Introduction 10 1.1 Motivation and Aims....... 10 1.1.1 Functional Imaging.... 10 1.1.2 Computational Neuroanatomy... 12 1.2 Overview of Chapters... 14 2 Rigid Body Registration 18 2.1 Introduction.....
Auxiliary Anatomical Labels for Joint Segmentation and Atlas Registration
 Auxiliary Anatomical Labels for Joint Segmentation and Atlas Registration Tobias Gass, Gabor Szekely and Orcun Goksel Computer Vision Lab, ETH Zurich, Switzerland. {gasst, szekely, ogoksel}@vision.ee.ethz.ch
Auxiliary Anatomical Labels for Joint Segmentation and Atlas Registration Tobias Gass, Gabor Szekely and Orcun Goksel Computer Vision Lab, ETH Zurich, Switzerland. {gasst, szekely, ogoksel}@vision.ee.ethz.ch
MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 10: Medical Image Segmentation as an Energy Minimization Problem
 SPRING 07 MEDICAL IMAGE COMPUTING (CAP 97) LECTURE 0: Medical Image Segmentation as an Energy Minimization Problem Dr. Ulas Bagci HEC, Center for Research in Computer Vision (CRCV), University of Central
SPRING 07 MEDICAL IMAGE COMPUTING (CAP 97) LECTURE 0: Medical Image Segmentation as an Energy Minimization Problem Dr. Ulas Bagci HEC, Center for Research in Computer Vision (CRCV), University of Central
Nearest Neighbor 3D Segmentation with Context Features
 Paper 10574-21 Session 4: Machine Learning, 3:30 PM - 5:30 PM, Salon B Nearest Neighbor 3D Segmentation with Context Features Evelin Hristova, Heinrich Schulz, Tom Brosch, Mattias P. Heinrich, Hannes Nickisch
Paper 10574-21 Session 4: Machine Learning, 3:30 PM - 5:30 PM, Salon B Nearest Neighbor 3D Segmentation with Context Features Evelin Hristova, Heinrich Schulz, Tom Brosch, Mattias P. Heinrich, Hannes Nickisch
Automatic Detection and Segmentation of Kidneys in 3D CT Images Using Random Forests
 Automatic Detection and Segmentation of Kidneys in 3D CT Images Using Random Forests Rémi Cuingnet 1, Raphael Prevost 1,2, David Lesage 1,LaurentD.Cohen 2, Benoît Mory 1, and Roberto Ardon 1 1 Philips
Automatic Detection and Segmentation of Kidneys in 3D CT Images Using Random Forests Rémi Cuingnet 1, Raphael Prevost 1,2, David Lesage 1,LaurentD.Cohen 2, Benoît Mory 1, and Roberto Ardon 1 1 Philips
Automatic Generation of Shape Models Using Nonrigid Registration with a Single Segmented Template Mesh
 Automatic Generation of Shape Models Using Nonrigid Registration with a Single Segmented Template Mesh Geremy Heitz, Torsten Rohlfing, and Calvin R. Maurer, Jr. Image Guidance Laboratories Department of
Automatic Generation of Shape Models Using Nonrigid Registration with a Single Segmented Template Mesh Geremy Heitz, Torsten Rohlfing, and Calvin R. Maurer, Jr. Image Guidance Laboratories Department of
Counter-Driven Regression for Label Inference in Atlas- Based Segmentation
 Counter-Driven Regression for Label Inference in Atlas- Based Segmentation The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation
Counter-Driven Regression for Label Inference in Atlas- Based Segmentation The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation
Multi-atlas Propagation Whole Heart Segmentation from MRI and CTA Using a Local Normalised Correlation Coefficient Criterion
 Multi-atlas Propagation Whole Heart Segmentation from MRI and CTA Using a Local Normalised Correlation Coefficient Criterion Maria A. Zuluaga, M. Jorge Cardoso, Marc Modat, and Sébastien Ourselin Centre
Multi-atlas Propagation Whole Heart Segmentation from MRI and CTA Using a Local Normalised Correlation Coefficient Criterion Maria A. Zuluaga, M. Jorge Cardoso, Marc Modat, and Sébastien Ourselin Centre
Multi-atlas labeling with population-specific template and non-local patch-based label fusion
 Multi-atlas labeling with population-specific template and non-local patch-based label fusion Vladimir Fonov, Pierrick Coupé, Simon Eskildsen, Jose Manjon, Louis Collins To cite this version: Vladimir
Multi-atlas labeling with population-specific template and non-local patch-based label fusion Vladimir Fonov, Pierrick Coupé, Simon Eskildsen, Jose Manjon, Louis Collins To cite this version: Vladimir
Supervoxel Classification Forests for Estimating Pairwise Image Correspondences
 Supervoxel Classification Forests for Estimating Pairwise Image Correspondences Fahdi Kanavati a,, Tong Tong a, Kazunari Misawa b, Michitaka Fujiwara c, Kensaku Mori d, Daniel Rueckert a, Ben Glocker a
Supervoxel Classification Forests for Estimating Pairwise Image Correspondences Fahdi Kanavati a,, Tong Tong a, Kazunari Misawa b, Michitaka Fujiwara c, Kensaku Mori d, Daniel Rueckert a, Ben Glocker a
Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging
 1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
Similarity Guided Feature Labeling for Lesion Detection
 Similarity Guided Feature Labeling for Lesion Detection Yang Song 1, Weidong Cai 1, Heng Huang 2, Xiaogang Wang 3, Stefan Eberl 4, Michael Fulham 4,5, Dagan Feng 1 1 BMIT Research Group, School of IT,
Similarity Guided Feature Labeling for Lesion Detection Yang Song 1, Weidong Cai 1, Heng Huang 2, Xiaogang Wang 3, Stefan Eberl 4, Michael Fulham 4,5, Dagan Feng 1 1 BMIT Research Group, School of IT,
Automated Model-Based Rib Cage Segmentation and Labeling in CT Images
 Automated Model-Based Rib Cage Segmentation and Labeling in CT Images Tobias Klinder 1,2,CristianLorenz 2,JensvonBerg 2, Sebastian P.M. Dries 2, Thomas Bülow 2,andJörn Ostermann 1 1 Institut für Informationsverarbeitung,
Automated Model-Based Rib Cage Segmentation and Labeling in CT Images Tobias Klinder 1,2,CristianLorenz 2,JensvonBerg 2, Sebastian P.M. Dries 2, Thomas Bülow 2,andJörn Ostermann 1 1 Institut für Informationsverarbeitung,
Semi-supervised Segmentation Using Multiple Segmentation Hypotheses from a Single Atlas
 Semi-supervised Segmentation Using Multiple Segmentation Hypotheses from a Single Atlas Tobias Gass, Gabor Szekely, and Orcun Goksel Computer Vision Lab, Dep. of Electrical Engineering, ETH Zurich, Switzerland.
Semi-supervised Segmentation Using Multiple Segmentation Hypotheses from a Single Atlas Tobias Gass, Gabor Szekely, and Orcun Goksel Computer Vision Lab, Dep. of Electrical Engineering, ETH Zurich, Switzerland.
Segmentation of Brain MR Images via Sparse Patch Representation
 Segmentation of Brain MR Images via Sparse Patch Representation Tong Tong 1, Robin Wolz 1, Joseph V. Hajnal 2, and Daniel Rueckert 1 1 Department of Computing, Imperial College London, London, UK 2 MRC
Segmentation of Brain MR Images via Sparse Patch Representation Tong Tong 1, Robin Wolz 1, Joseph V. Hajnal 2, and Daniel Rueckert 1 1 Department of Computing, Imperial College London, London, UK 2 MRC
Automatic Rapid Segmentation of Human Lung from 2D Chest X-Ray Images
 Automatic Rapid Segmentation of Human Lung from 2D Chest X-Ray Images Abstract. In this paper, we propose a complete framework that segments lungs from 2D Chest X-Ray (CXR) images automatically and rapidly.
Automatic Rapid Segmentation of Human Lung from 2D Chest X-Ray Images Abstract. In this paper, we propose a complete framework that segments lungs from 2D Chest X-Ray (CXR) images automatically and rapidly.
REAL-TIME ADAPTIVITY IN HEAD-AND-NECK AND LUNG CANCER RADIOTHERAPY IN A GPU ENVIRONMENT
 REAL-TIME ADAPTIVITY IN HEAD-AND-NECK AND LUNG CANCER RADIOTHERAPY IN A GPU ENVIRONMENT Anand P Santhanam Assistant Professor, Department of Radiation Oncology OUTLINE Adaptive radiotherapy for head and
REAL-TIME ADAPTIVITY IN HEAD-AND-NECK AND LUNG CANCER RADIOTHERAPY IN A GPU ENVIRONMENT Anand P Santhanam Assistant Professor, Department of Radiation Oncology OUTLINE Adaptive radiotherapy for head and
Towards image-guided pancreas and biliary endoscopy: automatic multi-organ segmentation on abdominal CT with dense dilated networks
 Towards image-guided pancreas and biliary endoscopy: automatic multi-organ segmentation on abdominal CT with dense dilated networks Eli Gibson 1, Francesco Giganti 2,3, Yipeng Hu 1, Ester Bonmati 1, Steve
Towards image-guided pancreas and biliary endoscopy: automatic multi-organ segmentation on abdominal CT with dense dilated networks Eli Gibson 1, Francesco Giganti 2,3, Yipeng Hu 1, Ester Bonmati 1, Steve
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy
 The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
8/3/2017. Contour Assessment for Quality Assurance and Data Mining. Objective. Outline. Tom Purdie, PhD, MCCPM
 Contour Assessment for Quality Assurance and Data Mining Tom Purdie, PhD, MCCPM Objective Understand the state-of-the-art in contour assessment for quality assurance including data mining-based techniques
Contour Assessment for Quality Assurance and Data Mining Tom Purdie, PhD, MCCPM Objective Understand the state-of-the-art in contour assessment for quality assurance including data mining-based techniques
Is deformable image registration a solved problem?
 Is deformable image registration a solved problem? Marcel van Herk On behalf of the imaging group of the RT department of NKI/AVL Amsterdam, the Netherlands DIR 1 Image registration Find translation.deformation
Is deformable image registration a solved problem? Marcel van Herk On behalf of the imaging group of the RT department of NKI/AVL Amsterdam, the Netherlands DIR 1 Image registration Find translation.deformation
Hierarchical Segmentation and Identification of Thoracic Vertebra Using Learning-Based Edge Detection and Coarse-to-Fine Deformable Model
 Hierarchical Segmentation and Identification of Thoracic Vertebra Using Learning-Based Edge Detection and Coarse-to-Fine Deformable Model Jun Ma, Le Lu, Yiqiang Zhan, Xiang Zhou, Marcos Salganicoff, and
Hierarchical Segmentation and Identification of Thoracic Vertebra Using Learning-Based Edge Detection and Coarse-to-Fine Deformable Model Jun Ma, Le Lu, Yiqiang Zhan, Xiang Zhou, Marcos Salganicoff, and
Neighbourhood Approximation Forests
 Neighbourhood Approximation Forests Ender Konukoglu, Ben Glocker, Darko Zikic and Antonio Criminisi Microsoft Research Cambridge Abstract. Methods that leverage neighbourhood structures in highdimensional
Neighbourhood Approximation Forests Ender Konukoglu, Ben Glocker, Darko Zikic and Antonio Criminisi Microsoft Research Cambridge Abstract. Methods that leverage neighbourhood structures in highdimensional
Vertebrae Segmentation in 3D CT Images based on a Variational Framework
 Vertebrae Segmentation in 3D CT Images based on a Variational Framework Kerstin Hammernik, Thomas Ebner, Darko Stern, Martin Urschler, and Thomas Pock Abstract Automatic segmentation of 3D vertebrae is
Vertebrae Segmentation in 3D CT Images based on a Variational Framework Kerstin Hammernik, Thomas Ebner, Darko Stern, Martin Urschler, and Thomas Pock Abstract Automatic segmentation of 3D vertebrae is
A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation
 A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation Xiahai Zhuang (PhD) Centre for Medical Image Computing University College London Fields-MITACS Conference on Mathematics
A Registration-Based Atlas Propagation Framework for Automatic Whole Heart Segmentation Xiahai Zhuang (PhD) Centre for Medical Image Computing University College London Fields-MITACS Conference on Mathematics
Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs
 Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs Yonghong Shi 1 and Dinggang Shen 2,*1 1 Digital Medical Research Center, Fudan University, Shanghai, 232, China
Hierarchical Shape Statistical Model for Segmentation of Lung Fields in Chest Radiographs Yonghong Shi 1 and Dinggang Shen 2,*1 1 Digital Medical Research Center, Fudan University, Shanghai, 232, China
Whole Body MRI Intensity Standardization
 Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Whole Body MRI Intensity Standardization Florian Jäger 1, László Nyúl 1, Bernd Frericks 2, Frank Wacker 2 and Joachim Hornegger 1 1 Institute of Pattern Recognition, University of Erlangen, {jaeger,nyul,hornegger}@informatik.uni-erlangen.de
Deep Fusion Net for Multi-Atlas Segmentation: Application to Cardiac MR Images
 Deep Fusion Net for Multi-Atlas Segmentation: Application to Cardiac MR Images Heran Yang 1, Jian Sun 1, Huibin Li 1, Lisheng Wang 2, and Zongben Xu 1 1 School of Mathematics and Statistics, Xi an Jiaotong
Deep Fusion Net for Multi-Atlas Segmentation: Application to Cardiac MR Images Heran Yang 1, Jian Sun 1, Huibin Li 1, Lisheng Wang 2, and Zongben Xu 1 1 School of Mathematics and Statistics, Xi an Jiaotong
Joint Tumor Segmentation and Dense Deformable Registration of Brain MR Images
 Joint Tumor Segmentation and Dense Deformable Registration of Brain MR Images Sarah Parisot 1,2,3, Hugues Duffau 4, Stéphane Chemouny 3, Nikos Paragios 1,2 1. Center for Visual Computing, Ecole Centrale
Joint Tumor Segmentation and Dense Deformable Registration of Brain MR Images Sarah Parisot 1,2,3, Hugues Duffau 4, Stéphane Chemouny 3, Nikos Paragios 1,2 1. Center for Visual Computing, Ecole Centrale
VALIDATION OF DIR. Raj Varadhan, PhD, DABMP Minneapolis Radiation Oncology
 VALIDATION OF DIR Raj Varadhan, PhD, DABMP Minneapolis Radiation Oncology Overview Basics: Registration Framework, Theory Discuss Validation techniques Using Synthetic CT data & Phantoms What metrics to
VALIDATION OF DIR Raj Varadhan, PhD, DABMP Minneapolis Radiation Oncology Overview Basics: Registration Framework, Theory Discuss Validation techniques Using Synthetic CT data & Phantoms What metrics to
Atlas Based Segmentation of the prostate in MR images
 Atlas Based Segmentation of the prostate in MR images Albert Gubern-Merida and Robert Marti Universitat de Girona, Computer Vision and Robotics Group, Girona, Spain {agubern,marly}@eia.udg.edu Abstract.
Atlas Based Segmentation of the prostate in MR images Albert Gubern-Merida and Robert Marti Universitat de Girona, Computer Vision and Robotics Group, Girona, Spain {agubern,marly}@eia.udg.edu Abstract.
Hierarchical Patch Generation for Multi-level Statistical Shape Analysis by Principal Factor Analysis Decomposition
 Hierarchical Patch Generation for Multi-level Statistical Shape Analysis by Principal Factor Analysis Decomposition Mauricio Reyes 1, Miguel A. González Ballester 2, Nina Kozic 1, Jesse K. Sandberg 3,
Hierarchical Patch Generation for Multi-level Statistical Shape Analysis by Principal Factor Analysis Decomposition Mauricio Reyes 1, Miguel A. González Ballester 2, Nina Kozic 1, Jesse K. Sandberg 3,
Manifold Learning: Applications in Neuroimaging
 Your own logo here Manifold Learning: Applications in Neuroimaging Robin Wolz 23/09/2011 Overview Manifold learning for Atlas Propagation Multi-atlas segmentation Challenges LEAP Manifold learning for
Your own logo here Manifold Learning: Applications in Neuroimaging Robin Wolz 23/09/2011 Overview Manifold learning for Atlas Propagation Multi-atlas segmentation Challenges LEAP Manifold learning for
Algorithms for medical image registration and segmentation
 Algorithms for medical image registration and segmentation Multi-atlas methods ernst.schwartz@meduniwien.ac.at www.cir.meduniwien.ac.at Overview Medical imaging hands-on Data formats: DICOM, NifTI Software:
Algorithms for medical image registration and segmentation Multi-atlas methods ernst.schwartz@meduniwien.ac.at www.cir.meduniwien.ac.at Overview Medical imaging hands-on Data formats: DICOM, NifTI Software:
NIH Public Access Author Manuscript Proc Soc Photo Opt Instrum Eng. Author manuscript; available in PMC 2014 October 07.
 NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
NIH Public Access Author Manuscript Published in final edited form as: Proc Soc Photo Opt Instrum Eng. 2014 March 21; 9034: 903442. doi:10.1117/12.2042915. MRI Brain Tumor Segmentation and Necrosis Detection
Atlas-Based Under-Segmentation
 Atlas-Based Under-Segmentation Christian Wachinger 1,2 and Polina Golland 1 1 Computer Science and Artificial Intelligence Lab, MIT 2 Massachusetts General Hospital, Harvard Medical School Abstract. We
Atlas-Based Under-Segmentation Christian Wachinger 1,2 and Polina Golland 1 1 Computer Science and Artificial Intelligence Lab, MIT 2 Massachusetts General Hospital, Harvard Medical School Abstract. We
Detecting Bone Lesions in Multiple Myeloma Patients using Transfer Learning
 Detecting Bone Lesions in Multiple Myeloma Patients using Transfer Learning Matthias Perkonigg 1, Johannes Hofmanninger 1, Björn Menze 2, Marc-André Weber 3, and Georg Langs 1 1 Computational Imaging Research
Detecting Bone Lesions in Multiple Myeloma Patients using Transfer Learning Matthias Perkonigg 1, Johannes Hofmanninger 1, Björn Menze 2, Marc-André Weber 3, and Georg Langs 1 1 Computational Imaging Research
MEDICAL IMAGE COMPUTING (CAP 5937) LECTURE 10: Medical Image Segmentation as an Energy Minimization Problem
 SPRING 06 MEDICAL IMAGE COMPUTING (CAP 97) LECTURE 0: Medical Image Segmentation as an Energy Minimization Problem Dr. Ulas Bagci HEC, Center for Research in Computer Vision (CRCV), University of Central
SPRING 06 MEDICAL IMAGE COMPUTING (CAP 97) LECTURE 0: Medical Image Segmentation as an Energy Minimization Problem Dr. Ulas Bagci HEC, Center for Research in Computer Vision (CRCV), University of Central
Shape-Based Kidney Detection and Segmentation in Three-Dimensional Abdominal Ultrasound Images
 University of Toronto Shape-Based Kidney Detection and Segmentation in Three-Dimensional Abdominal Ultrasound Images Authors: M. Marsousi, K. N. Plataniotis, S. Stergiopoulos Presenter: M. Marsousi, M.
University of Toronto Shape-Based Kidney Detection and Segmentation in Three-Dimensional Abdominal Ultrasound Images Authors: M. Marsousi, K. N. Plataniotis, S. Stergiopoulos Presenter: M. Marsousi, M.
Sparsity Based Spectral Embedding: Application to Multi-Atlas Echocardiography Segmentation!
 Sparsity Based Spectral Embedding: Application to Multi-Atlas Echocardiography Segmentation Ozan Oktay, Wenzhe Shi, Jose Caballero, Kevin Keraudren, and Daniel Rueckert Department of Compu.ng Imperial
Sparsity Based Spectral Embedding: Application to Multi-Atlas Echocardiography Segmentation Ozan Oktay, Wenzhe Shi, Jose Caballero, Kevin Keraudren, and Daniel Rueckert Department of Compu.ng Imperial
Comparison of Vessel Segmentations using STAPLE
 Comparison of Vessel Segmentations using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel Hill, Department
Comparison of Vessel Segmentations using STAPLE Julien Jomier, Vincent LeDigarcher, and Stephen R. Aylward Computer-Aided Diagnosis and Display Lab The University of North Carolina at Chapel Hill, Department
MARS: Multiple Atlases Robust Segmentation
 Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Generation of Triangle Meshes from Time-of-Flight Data for Surface Registration
 Generation of Triangle Meshes from Time-of-Flight Data for Surface Registration Thomas Kilgus, Thiago R. dos Santos, Alexander Seitel, Kwong Yung, Alfred M. Franz, Anja Groch, Ivo Wolf, Hans-Peter Meinzer,
Generation of Triangle Meshes from Time-of-Flight Data for Surface Registration Thomas Kilgus, Thiago R. dos Santos, Alexander Seitel, Kwong Yung, Alfred M. Franz, Anja Groch, Ivo Wolf, Hans-Peter Meinzer,
Accurate Intervertebral Disc Localisation and Segmentation in MRI using Vantage Point Hough Forests and Multi-Atlas Fusion
 Accurate Intervertebral Disc Localisation and Segmentation in MRI using Vantage Point Hough Forests and Multi-Atlas Fusion Mattias P. Heinrich 1 and Ozan Oktay 2 1 Institute of Medical Informatics, University
Accurate Intervertebral Disc Localisation and Segmentation in MRI using Vantage Point Hough Forests and Multi-Atlas Fusion Mattias P. Heinrich 1 and Ozan Oktay 2 1 Institute of Medical Informatics, University
Initialising Groupwise Non-rigid Registration Using Multiple Parts+Geometry Models
 Initialising Groupwise Non-rigid Registration Using Multiple Parts+Geometry Models Pei Zhang 1,Pew-ThianYap 1, Dinggang Shen 1, and Timothy F. Cootes 2 1 Department of Radiology and Biomedical Research
Initialising Groupwise Non-rigid Registration Using Multiple Parts+Geometry Models Pei Zhang 1,Pew-ThianYap 1, Dinggang Shen 1, and Timothy F. Cootes 2 1 Department of Radiology and Biomedical Research
Supplementary Material: Adaptive and Move Making Auxiliary Cuts for Binary Pairwise Energies
 Supplementary Material: Adaptive and Move Making Auxiliary Cuts for Binary Pairwise Energies Lena Gorelick Yuri Boykov Olga Veksler Computer Science Department University of Western Ontario Abstract Below
Supplementary Material: Adaptive and Move Making Auxiliary Cuts for Binary Pairwise Energies Lena Gorelick Yuri Boykov Olga Veksler Computer Science Department University of Western Ontario Abstract Below
Multi-Atlas Brain MRI Segmentation with Multiway Cut
 Multi-Atlas Brain MRI Segmentation with Multiway Cut Duygu Sarikaya, Liang Zhao, and Jason J. Corso SUNY Buffalo, Computer Science and Engineering Department, 338 Davis Hall- Buffalo, New York, USA 14260-2500
Multi-Atlas Brain MRI Segmentation with Multiway Cut Duygu Sarikaya, Liang Zhao, and Jason J. Corso SUNY Buffalo, Computer Science and Engineering Department, 338 Davis Hall- Buffalo, New York, USA 14260-2500
Segmentation of the Pectoral Muscle in Breast MRI Using Atlas-Based Approaches
 Segmentation of the Pectoral Muscle in Breast MRI Using Atlas-Based Approaches Albert Gubern-Mérida 1, Michiel Kallenberg 2, Robert Martí 1, and Nico Karssemeijer 2 1 University of Girona, Spain {agubern,marly}@eia.udg.edu
Segmentation of the Pectoral Muscle in Breast MRI Using Atlas-Based Approaches Albert Gubern-Mérida 1, Michiel Kallenberg 2, Robert Martí 1, and Nico Karssemeijer 2 1 University of Girona, Spain {agubern,marly}@eia.udg.edu
Discrete Optimization of Ray Potentials for Semantic 3D Reconstruction
 Discrete Optimization of Ray Potentials for Semantic 3D Reconstruction Marc Pollefeys Joined work with Nikolay Savinov, Christian Haene, Lubor Ladicky 2 Comparison to Volumetric Fusion Higher-order ray
Discrete Optimization of Ray Potentials for Semantic 3D Reconstruction Marc Pollefeys Joined work with Nikolay Savinov, Christian Haene, Lubor Ladicky 2 Comparison to Volumetric Fusion Higher-order ray
Supervised Learning for Image Segmentation
 Supervised Learning for Image Segmentation Raphael Meier 06.10.2016 Raphael Meier MIA 2016 06.10.2016 1 / 52 References A. Ng, Machine Learning lecture, Stanford University. A. Criminisi, J. Shotton, E.
Supervised Learning for Image Segmentation Raphael Meier 06.10.2016 Raphael Meier MIA 2016 06.10.2016 1 / 52 References A. Ng, Machine Learning lecture, Stanford University. A. Criminisi, J. Shotton, E.
Registration Techniques
 EMBO Practical Course on Light Sheet Microscopy Junior-Prof. Dr. Olaf Ronneberger Computer Science Department and BIOSS Centre for Biological Signalling Studies University of Freiburg Germany O. Ronneberger,
EMBO Practical Course on Light Sheet Microscopy Junior-Prof. Dr. Olaf Ronneberger Computer Science Department and BIOSS Centre for Biological Signalling Studies University of Freiburg Germany O. Ronneberger,
Atlas-Based Auto-segmentation of Head and Neck CT Images
 Atlas-Based Auto-segmentation of Head and Neck CT Images Xiao Han 1,MischaS.Hoogeman 2, Peter C. Levendag 2, Lyndon S. Hibbard 1, David N. Teguh 2, Peter Voet 2, Andrew C. Cowen 1, and Theresa K. Wolf
Atlas-Based Auto-segmentation of Head and Neck CT Images Xiao Han 1,MischaS.Hoogeman 2, Peter C. Levendag 2, Lyndon S. Hibbard 1, David N. Teguh 2, Peter Voet 2, Andrew C. Cowen 1, and Theresa K. Wolf
Deformable Segmentation via Sparse Shape Representation
 Deformable Segmentation via Sparse Shape Representation Shaoting Zhang 2, Yiqiang Zhan 1, Maneesh Dewan 1, Junzhou Huang 2, Dimitris N. Metaxas 2, and Xiang Sean Zhou 1 1 Siemens Medical Solutions, Malvern,
Deformable Segmentation via Sparse Shape Representation Shaoting Zhang 2, Yiqiang Zhan 1, Maneesh Dewan 1, Junzhou Huang 2, Dimitris N. Metaxas 2, and Xiang Sean Zhou 1 1 Siemens Medical Solutions, Malvern,
