ISpace: Interactive Volume Data Classification Techniques Using Independent Component Analysis
|
|
|
- Susanna Welch
- 5 years ago
- Views:
Transcription
1 ISpace: Interactive Volume Data Classification Techniques Using Independent Component Analysis z Ikuko Takanashi y Λ Eric B. Lum x Kwan-Liu Ma x Shigeru Muraki z yinformation Technology R&D Center, Mitsubishi Electric Corporation, Japan xcipic & Department of Computer Science, University of California, Davis, U.S.A. National Institute of Advanced Industrial Science and Technology (AIST), Tsukuba, Japan Abstract This paper introduces an interactive classification technique for volume data, called ISpace, which uses Independent Component Analysis (ICA) and a multidimensional histogram of the volume data in a transformed space. Essentially, classification in the volume domain becomes equivalent to interactive clipping in the ICA space, which as demonstrated using several examples is more intuitive and direct for the user to classify data. The result is an opacity transfer function defined for rendering multivariate scalar volume data. Keywords: Histogram, image processing, independent component analysis, medical imaging, multivariate data analysis, multimodality data, scientific visualization, segmentation, volume rendering 1 Introduction In scientific and medical investigations, often the goal is to identify unique features, trends, or structures in some sampled data domain. Segmentation is the core problem for many applications in medical imaging. Fully automatic segmentation methods are generally unavailable. Instead, interactive, supervised segmentation is often done with thresholding followed by binary mathematical morphology and connected component labeling [7]. For color images, interactive classification is often done in a color space, e.g., RGB [17], YIQ [19], or others. With parallel supercomputers, scientists are able to model physical phenomena at sufficient accuracy and complexity. Visualizing the resulting simulation data fully and directly can be a challenge. A general practice is to extract Λ Research performed while the author was in residence at CIPIC, University of California, Davis, CA ikuko@isl.melco.co.jp, {lume,ma}@cs.ucdavis.edu, s-muraki@aist.go.jp a subset of the data through either interactive exploration or a batch-mode feature extraction process. For example, for aerodynamic applications, scientists are interested in vortices, shocks, and boundary layers. Many of the existing techniques either have been designed for a particular application problem or are labor-intensive. Several techniques have been suggested for generating good transfer functions for classifying volume data [1, 5, 18]. These techniques work quite well for single-value scalar volume. More recently research has been devoted toward the development of transfer functions appropriate for multivariate data sets [13, 21] and time-varying data sets [9]. Kniss et al. describe the use of graphical widgets for multi-dimensional transfer function specification [12], where opacity is set based on several values such as the original scalar value, gradient magnitude and second derivative. Although this type of technique is effective for producing better classification, methods for the intuitive specification of higher dimensional transfer functions remains an open problem. In particular, the specification the three-dimensional transfer functions is complicated by the two-dimensional display typically found in visualization systems, with the specification of transfer functions beyond three dimensions becoming even more difficult. It is becoming increasingly necessary, however, to specify transfer functions with high dimensionality. Take for example a data set containing pressure and vorticity magnitude for each volumetric sample. If first and second derivative magnitudes are incorporated in classification, it becomes necessary to specify a sixth dimensional transfer function. In this paper, we introduces an intuitive interactive classification technique for multivariate volume data, in particular, multi-channel color data generated from different image modalities. We call the technique ISpace, which uses Independent Component Analysis(ICA) [14] to transform the original data space into a space better facilitating classification. The different variables in a data set, such as the RGB colors in a cryosection slice, are typically highly cor-
2 related with one another. Using ICA, however, it is possible to convert the data into a set of statistically (usually also physically) independent components[16]. Since ICA can separate the statistically independent components, we can apply ICA to a data set to classify the material in a volume based on statistically meaningful differences. Even if a volume has similar characteristics (e.g., brain tissues), ICA can be used to separate materials. Therefore we use ICA to transform these data sets into ICA space to facilitate interactive classification based on a volume s physical characteristics. The advantages of using ISpace are two fold. First, ICA can be used to transform a volume s histogram into a coordinate system that is better suited for segmentation via clipping along axis aligned plains, thus making transfer function specification more intuitive. Second, ICA is effective in reducing transfer function dimensionality, simplifying the multi-variable transfer function specification problem. Dimensionality is reduced since the transformed axis in ISpace are effective in separating statistically independent components, thus requiring fewer axis (dimensions) along which clipping must occur. We demonstrate the effectiveness of ISpace using the Visible Human data set, a cryosliced human brain data set, and a multi-modality MRI/CT mouse brain data set. The rest of the paper is organized as follows. Section 2 introduces ICA. Section 3 describes the interactive classification followed by case studies using data sets with different characteristics. Finally, we conclude our study and suggest directions for further research. Figure 1. Left: the 3D histogram for an RGB, cryosliced human brain data set. Right: volume rendering of the same data set using a constant opacity map. 2 Independent Component Analysis Multivariate data analysis has been an active research area. Generally, the data are preprocessed to derive relevant relationships between the variables or to reduce the dimensionality before we can better understand different aspects of the data. Many statistical methods have been developed and can be coupled with visualization techniques to help analyze multivariate data. For example, principal component analysis (PCA) is a statistical technique for data reduction. Independent component analysis (ICA) [14], on the other hand, allows us to reveal interesting information in a multivariate data set by giving access to its independent components. If the variables are independent, they are uncorrelated. ICA has many application in data analysis, source separation, and feature extraction. For example, Muraki et al. [16] use ICA to color multichannel MRI data. We can also think of ICA as representing arbitrary multivariate data in terms of a linear combination of non- Gaussian random variables that are as independent as possible. Non-Gaussian means that the data distribution is not symmetric. Let s assume that an L-dimensional multivariate vector X =[x 1 ; :::;x L ] T is observed in an M-dimensional space S =[s 1 ; :::;s M ] T spanned by M statistically independent parameters as a linear combination X = AS where M» L and A is an L M matrix. By assuming that X is a zero-mean vector, ICA is considered to be the problem of finding the L M matrix A and the component vector S by making each element of S as independent as possible. Assuming L = M, we can compute the inverse of A, say W, and obtain the independent component simply by S = WX. A (stochastic) gradient decent method may be used to modify W interactively so that each element of S becomes as independent as possible. There are several ways to make each element of S independent, including mutual information minimization [2, 22] and the nonlinear PCA [10]. The gradient descent method can be very expensive due to its slow convergence. Hyvärinen proposed a fixed-point algorithm, called FastICA, for performing the estimation of ICA [8]. Instead of modifying W, FastICA finds independent component one by one as s i = w T X, where the vector w is one of the rows of matrix W. Furthermore, instead of using every data point immediately, FastICA uses sample averages of the data in a single step. Consequently, the convergence speed of FastICA is much faster than that of the gradient descent method. Various experiments show that FastICA is about times faster than the conventional gradient descent methods. In this research, we thus use the FastICA package for MatLab [4] Although both ICA and PCA can be used for reducing data dimensionality, they have several differences worth noting. ICA and PCA can decorrelate components, and thus allows the user to extract specific features more easily. PCA decorrelates the data based on the magnitude of the distribution and the perpendicularity, while ICA finds the statistically independent components without regard to those properties (the magnitude of the distribution and the perpendicularity) so that it can be used to classify features even if its distribution is small.
3 Since PCA assumes a Gaussian distribution (in many cases, mixed signal) and maximizes this distribution, the features of the multi-dimensional data are only found in the first component. With ICA a non-gaussian distribution is assumed (in many cases, independent signal), and thus can be used to classify statistically independent components, according to the deflection of the data characteristics. Therefore ICA can enhance the small differences in voxel values depending on the data by taking advantage of the characteristics of the factors of the multi-dimensional data. B W2 W 1 G W 0 3 An Interactive Classification Technique In direct volume rendering, classification is equivalent to the problem of finding an appropriate opacity transfer function. Considerable research has looked into the generation of transfer functions for volume visualization. Recently, Fujishiro, Azuma, and Takeshima [5] use topological information from a hyper-reed graph to generate color and opacity transfer functions. Kindlmann and Durkin [11] use information from first- and second-order directional derivatives to generate a volume of derivative histograms for generating opacity maps emphasizing boundary surfaces. Bajaj et al. [1] discuss using other volume function information to find isovalues of interest; these include the volume enclosed by an isosurface, the isosurface area, and the isosurface gradient. Isovalues chosen by this method can be emphasized in a corresponding opacity map. For color transfer functions, Bergman et al. [3] lay out procedural rules for informative color choices. Both He et al. [18] and Marks et al. [15] describe systems for color and opacity parameter generation. In [18], genetic algorithms are used to breed transfer functions. The user can either select functions from generated images or allow the system to be fully automated; in the later case, images are evaluated using statistical qualities such as their entropy and variance. The Design Galleries system [15] addresses parameter manipulation in general by rendering a multidimensional space of those parameters. The user then navigates this space to discover a parameter setting. A transfer function is often specified and displayed as a 2D curve. When making a transfer function, it is helpful to also see the histogram of the volume data since the shape of the histogram shows the distribution of data values and gives hints about how to segment the data into parts. Given a single-value scalar volume V, let f (i) denote the relative frequency with which intensity level i occurs in V, for all i in the overall intensity level range [i min ;i max ] of V. The graph of f (i) as a function of i is called the histogram. If f is quantized, and has intensity levels i 1 ; :::;i k ; its histogram can be represented as a bar graph having k bars. For a multivariate scale volume data set M, such as a RGB volume data set, a three-dimensional histogram can be made Figure 2. A sketch of a 3D histogram with both RGB and ICA coordinate axes shown. by plotting f as a function of (R; G; B). Figure 1 shows the three-dimensional histogram of an RGB brain data set. The three main axes (R, G, and B) are colored accordingly. One way to segment such a data set is to derive threshold RGB values by operating directly on this three-dimensional histogram. The other way is to first convert the data from the RGB space to a different color space, e.g., YIQ, and then look for good threshold values [19]. We have developed an interactive technique based on ICA, permitting more intuitive classification of multivariate scalar volume data. 3.1 The classification procedure The proposed classification procedure starts by applying ICA to the volume data. Assume we are classifying an RGB volume data set. The operation can be characterized by: 2 4 s s 1 5 = 4 w 32 0;0 w 1;0 w 2;0 w 0;1 w 1;1 w 2;154 r 3 i g i 5 s 2 w 0;2 w 1;2 w 2;2 b i for every voxel. [s 0 ;s 1 ;s 2 ] T is the resulting three independent components. The three vectors [w 0;0 ;w 1;0 ;w 2;0] T, [w 0;1 ;w 1;1 ;w 2;1] T, and [w 0;2 ;w 1;2 ;w 2;2] T are essentially the three coordinate axes of the new space, i.e. the ICA space. A three-dimensional histogram of the RGB volume is constructed and displayed. Often, the distribution of voxels in the RGB space is an ellipsoid, as also shown in Figure 1. Figure 2 sketches such a histogram with both the RGB and ICA coordinate axes drawn. Typically, the long side of the ellipsoid is aligned with a particular coordinate axis of the ICA space. Each point is colored by using the RGB values, and transparency may be used to show the number of voxels has the corresponding RGB values. When transparency R
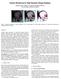
4 is used, this three-dimensional histogram should be volume rendered. Classification is then performed by the user moving clipping planes along the main axes. Interactive rendering permits the user to almost immediately see the resulting classified volume to quickly fine tune the positions of these planes. Note that the clipping becomes intuitive exactly because the three coordinate axes are oriented in space in a way such that each of them is independently aligned with the shape of the histogram. The transfer function used for rendering can be defined in several different ways. The simplest approach is to map all values of the voxels in the unclipped region to the same opacity, and make those voxels in the clipped region transparent (i.e. invisible). The histogram clipping technique has the advantage of being visually intuitive, but is limited to transfer function specification of three or fewer dimensions. As an alternative approach that avoids this limitation, we have also developed a second ISpace approach where a transfer function is specified as several 1D transfer functions, one for each ICA axis. The resulting opacity of each voxel is the product of the opacity specified along different axes. By showing the histogram along each axis, the user is also able to see value distributions to aid in classification. The dimension reducing properties ICA typically means that fewer curves need to be specified than there are dimensions, simplifying the specification process. Thus users can use a few relatively simple 1D transfer functions that effectively traverse the entire data space. 4 Test Results To evaluate the effectiveness of ISpace, we have experimented with three different data sets. Since ISpace must be integrated with interactive rendering to work well, we have utilized multi-texture graphics hardware to accelerate volume rendering. With an AMD 1.2 GHz Athlon with 768MB of main memory and a 64MB Nvidia GeForce 3 video card, we use 3D texture mapping [6] and can render up to volume interactively. It is particularly important for the user to be able interactively modify transfer functions and to immediately see the resulting changes in the rendered images. This is accomplished using the multi-texturing, where each ICA component is stored in a separate paletted texture. Changes to the classification function simply require the changing of texture palettes, rather that the computationally expensive manipulation of volume itself. Using the three data sets, we are able to show that operating in the ICA space with interactive rendering capability can greatly simplify the segmentation tasks. However, in this study, we had to downsize two of the data sets due to the limited video memory size. Figure 3. Two different views of the histogram of the human brain data set in ICA space. 4.1 A cryosliced human brain data set A brain data set provided by the UCLA Brain Research Institute consists of 753 cryosections of a human brain. Each slice has a resolution of pixels, and each pixel stores real-color RGB information with 16 bits per channel. The size of each slice is 7 MB. The cryoslice process used to create this data set consisted of taking photographs of the human brain after removing successive thin slices from the top to the bottom. Photos were taken of the unsliced portion of the brain rather than the removed slice itself. This results in a number of unique segmentation challenges not present in MRI or CT data since each image can contain data that is part of the deeper slices. In particular cavities or gaps in the brain reveal structures that are actually located in "deeper" layers. These parts of the image need to be eliminated. The brain must also be separated from the surrounding matter, i.e., from the ice and the background. Figure 3 shows two different views of the 3D histogram of the brain data in ICA space. The white-color region represents the ice, reddish region the brain, and the dark-color region the tray. In contrast to in the RGB space as shown in Figure 1, note that clipping now becomes straightforward to do in the ICA space since the three coordinate axes is independently aligned with the color distribution and the shape of the histogram. Figure 4 display two views of the clipped histogram to extract the only the brain from the whole volume. Figure 5 shows the corresponding volume rendering of the segmented data. The cut-away rendering shows that the cavity corresponding to the lateral ventricle, near the center of a brain, has been correctly removed. As a comparison, Figure 6 shows that clipping can be very difficult, if not impossible, in the RGB space. In order to obtain a good segmentation, the clipping must be done from oblique angles and separately for different regions. As shown in Figure 6, orthogonal clipping results in bad segmentation.
5 Figure 4. Two different views to show clipping of the histogram in ICA space to extract the brain. Figure 6. Segmentation of the brain volume in RGB space. Left: clipping the histogram; right: the corresponding volume rendered image. Figure 7. Left: histogram of extracted brain volume data as a result of repeating ICA; right: clipped to remove the gray matter (dark brown color). Figure 5. Rendering of the segmented result. Left: full brain; right: cut away to see inside. Further segmentation may be done to the extracted brain data by applying ICA multiple times. Figure 7 shows the histogram of the extracted brain volume data as a result of applying ICA the second time. This new histogram in the ICA space allows us to easily remove the darker brown color material representing the gray matter of the brain. The corresponding rendered result is displayed in Figure 8. We are able to segment the brain in ISpace by using our second technique that requires the specification of multiple 1D transfer functions which specifying an opacity distribution along each transformed ICA axis. The left side of Figure 9 shows the segmented brain that results from using the set of transfer functions shown on the right of the figure. For each transfer function, the histogram along that ICA axis is shown in beige while the opacity map is shown in blue. Notice that for the third axis the transfer function is constant across the distribution, demonstrating the dimension reducing properties found when working with ISpace; only two axis require transfer functions to segment the volume.
6 Figure 10. Segmentation of the Visible Female data is not effective using clipping in RGB space. Left: clipped histogram in RGB space; right: a volume rendered image. Figure 8. Volume rendered image of the segmented human brain using ICA twice. Figure 9. The segmented brain on the left was rendered using the three 1D transfer functions shown in blue on the right. Figure 11. Histogram of the Visible Female data in ICA space. Left: the complete histogram; right: clipped histogram.
7 4.2 The Visible Female data set The Visible Female data set has a resolution of Figure 10 shows its three-dimensional histogram in RGB space, and a volume rendered image of the data. The histogram was clipped to reveal the body. As shown in the right image of Figure 10, the body cannot be cleanly segmented in the RGB Space without also removing some portions inside the body. Figure 11(a) shows the 3D histogram in ICA space and Figure 11(b) shows a clipped histogram, in which the clipped part is colored in purple. Images of the corresponding segmented results are displayed in Figure 12, in which the right image reveals the internal of the body to show all parts are retained. 4.3 A mouse brain data set ISpace may be used for visualizing multivolume data; that is data from multiple imaging modalities. A mouse data set [20] consisting of two volumes, one from MRI (Magnetic Resonance Images) and one from PET (Positron Emission Tomography) of a mouse brain are used. Each volume consists of 47 slices at pixels per slice. Prior to visualization, the data was cropped to remove the significant amount of empty space, and resampled to 100 slices at The MRI data provides a structural information of the brain, and the PET volume contains functional data about the mouse brain. In the past, volumes from different modalities have been mainly visualized individually or side by side. It is beneficial to visualize them simultaneously by superimposing the volumes and rendering them into a single image. In this way, it is possible to relate the functional data to particular anatomical structures. However, we found it is very difficult to directly specify individual transfer functions for the PET and MRI volumes in order to derive desirable images. We thus treated the PET and MRI volumes as a bivariate volume and applied ICA to it. Transfer functions were made for the resulting first and second ICA components instead. The key advantage of this approach is that the user can explore the data set as a single entity rather than as two or three separate volumes (i.e. MRI, PET, CT, etc.) In some sense, a combined transfer function can be interactively defined with ease to reveal dominant features in the PET and MRI volumes. Figure 13 contrasts segmenting in the PET/MRI space (left) and the ICA space (right). As shown in two different views, some isolated features revealed in the ICA are almost impossible to capture in the PET/MRI space. The volume can also be rendered using two onedimensional transfer functions as shown in Figure 14. This methods provides the advantages of working in ISpace, while permitting the specification semi-transparent PET/MRI voxels. Although some of the intuition provided by the unified 2D histogram and clipping planes is lost, the user is able to work with the more familiar 1D histogram and transfer function user interface. 5 Conclusions We have introduced a new intuitive interactive technique for the often tedious classification problem. While we have demonstrated the effectiveness of ISpace using medical data sets, we believe it is possible to generalize ISpace for multivariate data visualization. Further work is also needed to improve multimodality volume visualization. Our segmentation of the mouse data set shows promising results. We will integrate this interface with a parallel volume renderer running on a PC cluster such that we are able to segment large data set at the original resolution. We will also investigate if visualization of the I-components (i.e. s 0 ;s 1 ;s 2 ) would offer other hints for classification. The shape of each of the 3D histograms we have seen so far are usually ellipsoids. If the shapes were spherical (i.e. Gaussian), then ICA would not be applicable. However, the histogram of physical data sets appear to almost always have these non-spherical shapes. We are currently implementing a VR interface for ISpace which we believe will make interactive clipping even more intuitive. Finally, we plan to conduct more studies using other types of data sets. Acknowledgments This work was supported by NSF (CAREER Awards: ACI , ACI , LSSDSV: ACI , NPACI). The authors are grateful to Dr. Juan Jose Vaquero and Dr. Michael Green at the National Institutes of Health, Dr. Arthur Toga of UCLA Brain Research Institute, and the Visible Human Project for providing the test data sets. References [1] C. L. Bajaj, V. Pascucci, and D. R. Schikore. The contour spectrum. In Proceedings of IEEE Visualization 1997, pages IEEE, [2] A. J. Bell and T. Sejnowski. An information-maximization approach to blind separation and blind deconvolution. Neural Computation, 7: , [3] L. D. Bergman, B. E. Rogowitz, and L. A. Treinish. A rulebased tool for assiting colormap selection. In Proceedings of IEEE Visualization 1995, pages IEEE, [4] The FastICA MatLab Package. projects/ica/fastica/. Laboratory of Computer and Information Science, Helsinki University of Technology.
8 Figure 12. Volume rendered images of the segmented Visible Female data using ICA. Left: external view; right: cut-way view. [5] I. Fujishiro, T.zuma, and Y. Takeshima. Automating transfer function design for comprehensible volume rendering based on 3d field topology analysis. In Proceedings of IEEE Visualization 1999, pages IEEE, [6] A. V. Gelder and K. Kim. Direct volume rendering with shading via three-dimension textures. In Proceedings of 1996 Symposium on Volume Visualizatrion, pages 23 30, October [7] K. H. Höhne and W. Hanson. Interactive 3d-segmentation of mri and ct volume using morphological operations. Journal of Computer Assisted Tomography, 16(2): , [8] A. Hyvärinen. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. on Neural Networks, 10(3): , [9] T. J. Jankun-Kelly and K.-L. Ma. Transfer function generation for time-varying volume data. In Proceedings of the International Workshop on Volume Graphics 2001, pages 33 43, [10] J. Karhunen, E. Oja, L. Wang, R. Vigario, and J. Joutsensalo. A class of neural networks for independent component analysis. IEEE Trans. on Neural Networks, 8(3): , [11] G. Kindlmann and J. Durkin. Semi-automatic generation of transfer functions for direct volume rendering. In Proceedings of the IEEE Symposium on Volume Visualization, pages IEEE, ACM SIGGRAPH, [12] J. Kniss, G. Kindlmann, and C. Hansen. Interactive volume rendering using multi-dimensional transfer functions and direct manipulation widgets. In Proceedings of IEEE Visualization 2001, pages IEEE, [13] J. Kniss, G. Kindlmann, and C. Hansen. Multi-dimensional transfer functions for interactive volume rendering. to appear in Transactions on Visualization and Computer Graphics, [14] T.-W. Lee. Independent Component Analysis. Kluwer Academic Publishers, Boston, October [15] J. Marks, B. Andalman, P. A. Beardsley, W. Freeman, S. Gibson, J. Hodgins, T. Kang, B. Mirtich, H. Pfister, W. Ruml, K. Ryall, J. Seims, and S. Shieber. Design galleries: A general approach to setting parameters for computer graphics and animation. In Proceedings of SIGGRAPH97, pages ACM SIGGRAPH, August [16] S. Muraki, T. Nakai, Y. Kita, and K. Tsuda. An attempt for coloring multichannel mr imaging data. IEEE Trans. Visualization and Computer Graphics, 7(3): , July- September [17] T. Schiemann, U. Tiede, and K. Höhne. Segmentation of the visible human for high quality volume based visualization. Medical Image Analysis, 1(4): , [18] L. H. T. He, A. Kaufman, and H. Pfister. Generation of transfer functions with stochastic search techniques. In Proceedings of IEEE Visualization 1996, pages IEEE, [19] I. Takanashi, E. B. Lum, K.-L. Ma, J. Meyer, B. Hamann, and A. J. Olson. Segmentation and 3d visualization of high resolution human brain. In Visualization and Data Analysis 2002, Electronic Imaging 2002, IS&T/SPIE, volume , January [20] J. J. Vaquero, M. Desco, J. Pascau, A. Santos, I. Lee, J. Seidel, and M. V. Green1. Pet, ct and mr image registration of the rat brain and skull. IEEE Transactions on Nuclear Science, 48(4): , August [21] B. Wilson, E. Lum, and K.-L. Ma. Interactive multi-volume visualization. In ICCS 2002 Workshop on Computer Graphics and Geometric Modeling, April [22] H. Yang and S. Amari. Adaptive online learning algorithms for blind separation - maximum entropy and minimum mutual information. Neural Computation, 9: , 1997.
9 Figure 13. Left: segmented mouse brain in PET&MRI space; right: segmented mouse brain in ICA space. Figure 14. The fusion of MRI and PET data shown on the left was rendered using the pair of ISpace aligned 1D transfer functions appearing on the right.
Mirrored LH Histograms for the Visualization of Material Boundaries
 Mirrored LH Histograms for the Visualization of Material Boundaries Petr Šereda 1, Anna Vilanova 1 and Frans A. Gerritsen 1,2 1 Department of Biomedical Engineering, Technische Universiteit Eindhoven,
Mirrored LH Histograms for the Visualization of Material Boundaries Petr Šereda 1, Anna Vilanova 1 and Frans A. Gerritsen 1,2 1 Department of Biomedical Engineering, Technische Universiteit Eindhoven,
A Study of Transfer Function Generation for Time-Varying Volume Data
 A Study of Transfer Function Generation for Time-Varying Volume Data T.J. Jankun-Kelly and Kwan-Liu Ma Visualization and Graphics Research Group, Center for Image Processing and Integrated Computing, Department
A Study of Transfer Function Generation for Time-Varying Volume Data T.J. Jankun-Kelly and Kwan-Liu Ma Visualization and Graphics Research Group, Center for Image Processing and Integrated Computing, Department
Scalar Data. CMPT 467/767 Visualization Torsten Möller. Weiskopf/Machiraju/Möller
 Scalar Data CMPT 467/767 Visualization Torsten Möller Weiskopf/Machiraju/Möller Overview Basic strategies Function plots and height fields Isolines Color coding Volume visualization (overview) Classification
Scalar Data CMPT 467/767 Visualization Torsten Möller Weiskopf/Machiraju/Möller Overview Basic strategies Function plots and height fields Isolines Color coding Volume visualization (overview) Classification
Scalar Data. Visualization Torsten Möller. Weiskopf/Machiraju/Möller
 Scalar Data Visualization Torsten Möller Weiskopf/Machiraju/Möller Overview Basic strategies Function plots and height fields Isolines Color coding Volume visualization (overview) Classification Segmentation
Scalar Data Visualization Torsten Möller Weiskopf/Machiraju/Möller Overview Basic strategies Function plots and height fields Isolines Color coding Volume visualization (overview) Classification Segmentation
cs6630 November TRANSFER FUNCTIONS Alex Bigelow University of Utah
 cs6630 November 14 2014 TRANSFER FUNCTIONS Alex Bigelow University of Utah 1 cs6630 November 13 2014 TRANSFER FUNCTIONS Alex Bigelow University of Utah slide acknowledgements: Miriah Meyer, University
cs6630 November 14 2014 TRANSFER FUNCTIONS Alex Bigelow University of Utah 1 cs6630 November 13 2014 TRANSFER FUNCTIONS Alex Bigelow University of Utah slide acknowledgements: Miriah Meyer, University
Interactive Multi-Volume Visualization
 Interactive Multi-Volume Visualization Brett Wilson, Eric B. Lum, and Kwan-Liu Ma Department of Computer Science University of California at Davis One Shields Avenue, Davis, CA 95616, U.S.A. fwilson,lume,mag@cs.ucdavis.edu,
Interactive Multi-Volume Visualization Brett Wilson, Eric B. Lum, and Kwan-Liu Ma Department of Computer Science University of California at Davis One Shields Avenue, Davis, CA 95616, U.S.A. fwilson,lume,mag@cs.ucdavis.edu,
2. Review of current methods
 Transfer Functions for Direct Volume Rendering Gordon Kindlmann gk@cs.utah.edu http://www.cs.utah.edu/~gk Scientific Computing and Imaging Institute School of Computing University of Utah Contributions:
Transfer Functions for Direct Volume Rendering Gordon Kindlmann gk@cs.utah.edu http://www.cs.utah.edu/~gk Scientific Computing and Imaging Institute School of Computing University of Utah Contributions:
Feature-Enhanced Visualization of Multidimensional, Multivariate Volume Data Using Non-photorealistic Rendering Techniques
 Feature-Enhanced Visualization of Multidimensional, Multivariate Volume Data Using Non-photorealistic Rendering Techniques Aleksander Stompel Eric B. Lum Kwan-Liu Ma University of California at Davis fastompel,lume,mag@cs.ucdavis.edu
Feature-Enhanced Visualization of Multidimensional, Multivariate Volume Data Using Non-photorealistic Rendering Techniques Aleksander Stompel Eric B. Lum Kwan-Liu Ma University of California at Davis fastompel,lume,mag@cs.ucdavis.edu
Abstract Feature Space Representation for Volumetric Transfer Function Exploration
 Abstract Feature Space Representation for Volumetric Transfer Function Exploration Ross Maciejewski 1, Yun Jang 2, David S. Ebert 3, and Kelly P. Gaither 4 1,3 Purdue University Visual Analytics Center
Abstract Feature Space Representation for Volumetric Transfer Function Exploration Ross Maciejewski 1, Yun Jang 2, David S. Ebert 3, and Kelly P. Gaither 4 1,3 Purdue University Visual Analytics Center
Interactive Boundary Detection for Automatic Definition of 2D Opacity Transfer Function
 Interactive Boundary Detection for Automatic Definition of 2D Opacity Transfer Function Martin Rauberger, Heinrich Martin Overhoff Medical Engineering Laboratory, University of Applied Sciences Gelsenkirchen,
Interactive Boundary Detection for Automatic Definition of 2D Opacity Transfer Function Martin Rauberger, Heinrich Martin Overhoff Medical Engineering Laboratory, University of Applied Sciences Gelsenkirchen,
Data Visualization (DSC 530/CIS )
 Data Visualization (DSC 530/CIS 60-0) Isosurfaces & Volume Rendering Dr. David Koop Fields & Grids Fields: - Values come from a continuous domain, infinitely many values - Sampled at certain positions
Data Visualization (DSC 530/CIS 60-0) Isosurfaces & Volume Rendering Dr. David Koop Fields & Grids Fields: - Values come from a continuous domain, infinitely many values - Sampled at certain positions
Interactive multi-scale exploration for volume classification
 Visual Comput (2006) 22: 622 630 DOI 10.1007/s00371-006-0049-8 ORIGINAL ARTICLE Eric B. Lum James Shearer Kwan-Liu Ma Interactive multi-scale exploration for volume classification Published online: 24
Visual Comput (2006) 22: 622 630 DOI 10.1007/s00371-006-0049-8 ORIGINAL ARTICLE Eric B. Lum James Shearer Kwan-Liu Ma Interactive multi-scale exploration for volume classification Published online: 24
Emissive Clip Planes for Volume Rendering Supplement.
 Emissive Clip Planes for Volume Rendering Supplement. More material than fit on the one page version for the SIGGRAPH 2003 Sketch by Jan Hardenbergh & Yin Wu of TeraRecon, Inc. Left Image: The clipped
Emissive Clip Planes for Volume Rendering Supplement. More material than fit on the one page version for the SIGGRAPH 2003 Sketch by Jan Hardenbergh & Yin Wu of TeraRecon, Inc. Left Image: The clipped
A Graph Based Interface for Representing Volume Visualization Results
 NASA/CR-1998-208468 ICASE Report No. 98-35 A Graph Based Interface for Representing Volume Visualization Results James M. Patten University of Virginia, Charlottesville, Virginia Kwan-Liu Ma ICASE, Hampton,
NASA/CR-1998-208468 ICASE Report No. 98-35 A Graph Based Interface for Representing Volume Visualization Results James M. Patten University of Virginia, Charlottesville, Virginia Kwan-Liu Ma ICASE, Hampton,
Volume visualization. Volume visualization. Volume visualization methods. Sources of volume visualization. Sources of volume visualization
 Volume visualization Volume visualization Volumes are special cases of scalar data: regular 3D grids of scalars, typically interpreted as density values. Each data value is assumed to describe a cubic
Volume visualization Volume visualization Volumes are special cases of scalar data: regular 3D grids of scalars, typically interpreted as density values. Each data value is assumed to describe a cubic
Advanced Visual Medicine: Techniques for Visual Exploration & Analysis
 Advanced Visual Medicine: Techniques for Visual Exploration & Analysis Interactive Visualization of Multimodal Volume Data for Neurosurgical Planning Felix Ritter, MeVis Research Bremen Multimodal Neurosurgical
Advanced Visual Medicine: Techniques for Visual Exploration & Analysis Interactive Visualization of Multimodal Volume Data for Neurosurgical Planning Felix Ritter, MeVis Research Bremen Multimodal Neurosurgical
CIS 467/602-01: Data Visualization
 CIS 467/60-01: Data Visualization Isosurfacing and Volume Rendering Dr. David Koop Fields and Grids Fields: values come from a continuous domain, infinitely many values - Sampled at certain positions to
CIS 467/60-01: Data Visualization Isosurfacing and Volume Rendering Dr. David Koop Fields and Grids Fields: values come from a continuous domain, infinitely many values - Sampled at certain positions to
RGVis: Region Growing Based Techniques for Volume Visualization
 RGVis: Region Growing Based Techniques for Volume Visualization Runzhen Huang Kwan-Liu Ma Department of Computer Science University of California at Davis {huangru,ma}@cs.ucdavis.edu Abstract Interactive
RGVis: Region Growing Based Techniques for Volume Visualization Runzhen Huang Kwan-Liu Ma Department of Computer Science University of California at Davis {huangru,ma}@cs.ucdavis.edu Abstract Interactive
Visualization Viewpoints: Interacting with Direct Volume Rendering
 Visualization Viewpoints: Interacting with Direct Volume Rendering Mike Bailey San Diego Supercomputer Center University of California San Diego mjb@sdsc.edu Visualization of volume data has been an active
Visualization Viewpoints: Interacting with Direct Volume Rendering Mike Bailey San Diego Supercomputer Center University of California San Diego mjb@sdsc.edu Visualization of volume data has been an active
A Study of Medical Image Analysis System
 Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
Prostate Detection Using Principal Component Analysis
 Prostate Detection Using Principal Component Analysis Aamir Virani (avirani@stanford.edu) CS 229 Machine Learning Stanford University 16 December 2005 Introduction During the past two decades, computed
Prostate Detection Using Principal Component Analysis Aamir Virani (avirani@stanford.edu) CS 229 Machine Learning Stanford University 16 December 2005 Introduction During the past two decades, computed
Palette-Style Volume Visualization
 Volume Graphics (2007) H. - C. Hege, R. Machiraju (Editors) Palette-Style Volume Visualization Yingcai Wu 1, Anbang Xu 1, Ming-Yuen Chan 1, Huamin Qu 1, and Ping Guo 2 1 The Hong Kong University of Science
Volume Graphics (2007) H. - C. Hege, R. Machiraju (Editors) Palette-Style Volume Visualization Yingcai Wu 1, Anbang Xu 1, Ming-Yuen Chan 1, Huamin Qu 1, and Ping Guo 2 1 The Hong Kong University of Science
An independent component analysis based tool for exploring functional connections in the brain
 An independent component analysis based tool for exploring functional connections in the brain S. M. Rolfe a, L. Finney b, R. F. Tungaraza b, J. Guan b, L.G. Shapiro b, J. F. Brinkely b, A. Poliakov c,
An independent component analysis based tool for exploring functional connections in the brain S. M. Rolfe a, L. Finney b, R. F. Tungaraza b, J. Guan b, L.G. Shapiro b, J. F. Brinkely b, A. Poliakov c,
Data Visualization (CIS/DSC 468)
 Data Visualization (CIS/DSC 46) Volume Rendering Dr. David Koop Visualizing Volume (3D) Data 2D visualization slice images (or multi-planar reformating MPR) Indirect 3D visualization isosurfaces (or surface-shaded
Data Visualization (CIS/DSC 46) Volume Rendering Dr. David Koop Visualizing Volume (3D) Data 2D visualization slice images (or multi-planar reformating MPR) Indirect 3D visualization isosurfaces (or surface-shaded
Norbert Schuff VA Medical Center and UCSF
 Norbert Schuff Medical Center and UCSF Norbert.schuff@ucsf.edu Medical Imaging Informatics N.Schuff Course # 170.03 Slide 1/67 Objective Learn the principle segmentation techniques Understand the role
Norbert Schuff Medical Center and UCSF Norbert.schuff@ucsf.edu Medical Imaging Informatics N.Schuff Course # 170.03 Slide 1/67 Objective Learn the principle segmentation techniques Understand the role
3D Surface Reconstruction of the Brain based on Level Set Method
 3D Surface Reconstruction of the Brain based on Level Set Method Shijun Tang, Bill P. Buckles, and Kamesh Namuduri Department of Computer Science & Engineering Department of Electrical Engineering University
3D Surface Reconstruction of the Brain based on Level Set Method Shijun Tang, Bill P. Buckles, and Kamesh Namuduri Department of Computer Science & Engineering Department of Electrical Engineering University
Calculating the Distance Map for Binary Sampled Data
 MITSUBISHI ELECTRIC RESEARCH LABORATORIES http://www.merl.com Calculating the Distance Map for Binary Sampled Data Sarah F. Frisken Gibson TR99-6 December 999 Abstract High quality rendering and physics-based
MITSUBISHI ELECTRIC RESEARCH LABORATORIES http://www.merl.com Calculating the Distance Map for Binary Sampled Data Sarah F. Frisken Gibson TR99-6 December 999 Abstract High quality rendering and physics-based
Volume Rendering for High Dynamic Range Displays
 Volume Graphics (25) E. Gröller, I. Fujishiro (Editors) Volume Rendering for High Dynamic Range Displays Abhijeet Ghosh, Matthew Trentacoste, Wolfgang Heidrich The University of British Columbia Abstract
Volume Graphics (25) E. Gröller, I. Fujishiro (Editors) Volume Rendering for High Dynamic Range Displays Abhijeet Ghosh, Matthew Trentacoste, Wolfgang Heidrich The University of British Columbia Abstract
Visualization. Images are used to aid in understanding of data. Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26]
![Visualization. Images are used to aid in understanding of data. Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26] Visualization. Images are used to aid in understanding of data. Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26]](/thumbs/74/70771954.jpg) Visualization Images are used to aid in understanding of data Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26] Tumor SCI, Utah Scientific Visualization Visualize large
Visualization Images are used to aid in understanding of data Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [chapter 26] Tumor SCI, Utah Scientific Visualization Visualize large
Independent Component Analysis of fmri Data
 Independent Component Analysis of fmri Data Denise Miller April 2005 Introduction Techniques employed to analyze functional magnetic resonance imaging (fmri) data typically use some form of univariate
Independent Component Analysis of fmri Data Denise Miller April 2005 Introduction Techniques employed to analyze functional magnetic resonance imaging (fmri) data typically use some form of univariate
Computational Medical Imaging Analysis Chapter 4: Image Visualization
 Computational Medical Imaging Analysis Chapter 4: Image Visualization Jun Zhang Laboratory for Computational Medical Imaging & Data Analysis Department of Computer Science University of Kentucky Lexington,
Computational Medical Imaging Analysis Chapter 4: Image Visualization Jun Zhang Laboratory for Computational Medical Imaging & Data Analysis Department of Computer Science University of Kentucky Lexington,
Visualization Computer Graphics I Lecture 20
 15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 15, 2003 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 15, 2003 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University
![Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University](/thumbs/90/102611276.jpg) 15-462 Computer Graphics I Lecture 21 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
15-462 Computer Graphics I Lecture 21 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] April 23, 2002 Frank Pfenning Carnegie Mellon University http://www.cs.cmu.edu/~fp/courses/graphics/
Clipping. CSC 7443: Scientific Information Visualization
 Clipping Clipping to See Inside Obscuring critical information contained in a volume data Contour displays show only exterior visible surfaces Isosurfaces can hide other isosurfaces Other displays can
Clipping Clipping to See Inside Obscuring critical information contained in a volume data Contour displays show only exterior visible surfaces Isosurfaces can hide other isosurfaces Other displays can
Modern Medical Image Analysis 8DC00 Exam
 Parts of answers are inside square brackets [... ]. These parts are optional. Answers can be written in Dutch or in English, as you prefer. You can use drawings and diagrams to support your textual answers.
Parts of answers are inside square brackets [... ]. These parts are optional. Answers can be written in Dutch or in English, as you prefer. You can use drawings and diagrams to support your textual answers.
Image Registration. Prof. Dr. Lucas Ferrari de Oliveira UFPR Informatics Department
 Image Registration Prof. Dr. Lucas Ferrari de Oliveira UFPR Informatics Department Introduction Visualize objects inside the human body Advances in CS methods to diagnosis, treatment planning and medical
Image Registration Prof. Dr. Lucas Ferrari de Oliveira UFPR Informatics Department Introduction Visualize objects inside the human body Advances in CS methods to diagnosis, treatment planning and medical
Visualizing Industrial CT Volume Data for Nondestructive Testing Applications
 Visualizing Industrial CT Volume Data for Nondestructive Testing Applications Runzhen Huang Kwan-Liu Ma University of California at Davis {huangru,ma}@cs.ucdavis.edu Patrick McCormick William Ward Los
Visualizing Industrial CT Volume Data for Nondestructive Testing Applications Runzhen Huang Kwan-Liu Ma University of California at Davis {huangru,ma}@cs.ucdavis.edu Patrick McCormick William Ward Los
Fast Interactive Region of Interest Selection for Volume Visualization
 Fast Interactive Region of Interest Selection for Volume Visualization Dominik Sibbing and Leif Kobbelt Lehrstuhl für Informatik 8, RWTH Aachen, 20 Aachen Email: {sibbing,kobbelt}@informatik.rwth-aachen.de
Fast Interactive Region of Interest Selection for Volume Visualization Dominik Sibbing and Leif Kobbelt Lehrstuhl für Informatik 8, RWTH Aachen, 20 Aachen Email: {sibbing,kobbelt}@informatik.rwth-aachen.de
Chapter 11 Representation & Description
 Chain Codes Chain codes are used to represent a boundary by a connected sequence of straight-line segments of specified length and direction. The direction of each segment is coded by using a numbering
Chain Codes Chain codes are used to represent a boundary by a connected sequence of straight-line segments of specified length and direction. The direction of each segment is coded by using a numbering
Real-time Monitoring of Multi-mode Industrial Processes using Feature-extraction Tools
 Real-time Monitoring of Multi-mode Industrial Processes using Feature-extraction Tools Y. S. Manjili *, M. Niknamfar, M. Jamshidi Department of Electrical and Computer Engineering The University of Texas
Real-time Monitoring of Multi-mode Industrial Processes using Feature-extraction Tools Y. S. Manjili *, M. Niknamfar, M. Jamshidi Department of Electrical and Computer Engineering The University of Texas
Chapter 8 Visualization and Optimization
 Chapter 8 Visualization and Optimization Recommended reference books: [1] Edited by R. S. Gallagher: Computer Visualization, Graphics Techniques for Scientific and Engineering Analysis by CRC, 1994 [2]
Chapter 8 Visualization and Optimization Recommended reference books: [1] Edited by R. S. Gallagher: Computer Visualization, Graphics Techniques for Scientific and Engineering Analysis by CRC, 1994 [2]
Interactive Visualization of Very Large Medical Datasets using Point-based Rendering
 Interactive Visualization of Very Large Medical Datasets using Point-based Rendering Christof Nuber, Ralph W. Bruckschen, Bernd Hamann and Kenneth I. Joy Center for Image Processing and Integrated Computing,
Interactive Visualization of Very Large Medical Datasets using Point-based Rendering Christof Nuber, Ralph W. Bruckschen, Bernd Hamann and Kenneth I. Joy Center for Image Processing and Integrated Computing,
Image Segmentation. Ross Whitaker SCI Institute, School of Computing University of Utah
 Image Segmentation Ross Whitaker SCI Institute, School of Computing University of Utah What is Segmentation? Partitioning images/volumes into meaningful pieces Partitioning problem Labels Isolating a specific
Image Segmentation Ross Whitaker SCI Institute, School of Computing University of Utah What is Segmentation? Partitioning images/volumes into meaningful pieces Partitioning problem Labels Isolating a specific
Volume Visualization. Part 1 (out of 3) Volume Data. Where do the data come from? 3D Data Space How are volume data organized?
 Volume Data Volume Visualization Part 1 (out of 3) Where do the data come from? Medical Application Computed Tomographie (CT) Magnetic Resonance Imaging (MR) Materials testing Industrial-CT Simulation
Volume Data Volume Visualization Part 1 (out of 3) Where do the data come from? Medical Application Computed Tomographie (CT) Magnetic Resonance Imaging (MR) Materials testing Industrial-CT Simulation
Data Visualization (DSC 530/CIS )
 Data Visualization (DSC 530/CIS 60-01) Scalar Visualization Dr. David Koop Online JavaScript Resources http://learnjsdata.com/ Good coverage of data wrangling using JavaScript Fields in Visualization Scalar
Data Visualization (DSC 530/CIS 60-01) Scalar Visualization Dr. David Koop Online JavaScript Resources http://learnjsdata.com/ Good coverage of data wrangling using JavaScript Fields in Visualization Scalar
Volume Rendering for High Dynamic Range Displays
 Volume Rendering for High Dynamic Range Displays Abhijeet Ghosh, Matthew Trentacoste, Wolfgang Heidrich The University of British Columbia Figure : Screen photographs of volume rendering of the CT head
Volume Rendering for High Dynamic Range Displays Abhijeet Ghosh, Matthew Trentacoste, Wolfgang Heidrich The University of British Columbia Figure : Screen photographs of volume rendering of the CT head
Image Segmentation. Ross Whitaker SCI Institute, School of Computing University of Utah
 Image Segmentation Ross Whitaker SCI Institute, School of Computing University of Utah What is Segmentation? Partitioning images/volumes into meaningful pieces Partitioning problem Labels Isolating a specific
Image Segmentation Ross Whitaker SCI Institute, School of Computing University of Utah What is Segmentation? Partitioning images/volumes into meaningful pieces Partitioning problem Labels Isolating a specific
Interactivity is the Key to Expressive Visualization
 VlSFILES Interactivity is the Key to Expressive Visualization Eric B. Lum and Kwan-Liu Ma University of California, Davis Scientific visualization is widely used for gaining insight into phenomena through
VlSFILES Interactivity is the Key to Expressive Visualization Eric B. Lum and Kwan-Liu Ma University of California, Davis Scientific visualization is widely used for gaining insight into phenomena through
An Efficient Approach for Emphasizing Regions of Interest in Ray-Casting based Volume Rendering
 An Efficient Approach for Emphasizing Regions of Interest in Ray-Casting based Volume Rendering T. Ropinski, F. Steinicke, K. Hinrichs Institut für Informatik, Westfälische Wilhelms-Universität Münster
An Efficient Approach for Emphasizing Regions of Interest in Ray-Casting based Volume Rendering T. Ropinski, F. Steinicke, K. Hinrichs Institut für Informatik, Westfälische Wilhelms-Universität Münster
Computer-Aided Diagnosis in Abdominal and Cardiac Radiology Using Neural Networks
 Computer-Aided Diagnosis in Abdominal and Cardiac Radiology Using Neural Networks Du-Yih Tsai, Masaru Sekiya and Yongbum Lee Department of Radiological Technology, School of Health Sciences, Faculty of
Computer-Aided Diagnosis in Abdominal and Cardiac Radiology Using Neural Networks Du-Yih Tsai, Masaru Sekiya and Yongbum Lee Department of Radiological Technology, School of Health Sciences, Faculty of
Medical Image Processing using MATLAB
 Medical Image Processing using MATLAB Emilia Dana SELEŢCHI University of Bucharest, Romania ABSTRACT 2. 3. 2. IMAGE PROCESSING TOOLBOX MATLAB and the Image Processing Toolbox provide a wide range of advanced
Medical Image Processing using MATLAB Emilia Dana SELEŢCHI University of Bucharest, Romania ABSTRACT 2. 3. 2. IMAGE PROCESSING TOOLBOX MATLAB and the Image Processing Toolbox provide a wide range of advanced
Volume Visualization
 Volume Visualization Part 1 (out of 3) Overview: Volume Visualization Introduction to volume visualization On volume data Surface vs. volume rendering Overview: Techniques Simple methods Slicing, cuberille
Volume Visualization Part 1 (out of 3) Overview: Volume Visualization Introduction to volume visualization On volume data Surface vs. volume rendering Overview: Techniques Simple methods Slicing, cuberille
Segmentation and Modeling of the Spinal Cord for Reality-based Surgical Simulator
 Segmentation and Modeling of the Spinal Cord for Reality-based Surgical Simulator Li X.C.,, Chui C. K.,, and Ong S. H.,* Dept. of Electrical and Computer Engineering Dept. of Mechanical Engineering, National
Segmentation and Modeling of the Spinal Cord for Reality-based Surgical Simulator Li X.C.,, Chui C. K.,, and Ong S. H.,* Dept. of Electrical and Computer Engineering Dept. of Mechanical Engineering, National
INTERACTIVE ENVIRONMENT FOR INTUITIVE UNDERSTANDING OF 4D DATA. M. Murata and S. Hashimoto Humanoid Robotics Institute, Waseda University, Japan
 1 INTRODUCTION INTERACTIVE ENVIRONMENT FOR INTUITIVE UNDERSTANDING OF 4D DATA M. Murata and S. Hashimoto Humanoid Robotics Institute, Waseda University, Japan Abstract: We present a new virtual reality
1 INTRODUCTION INTERACTIVE ENVIRONMENT FOR INTUITIVE UNDERSTANDING OF 4D DATA M. Murata and S. Hashimoto Humanoid Robotics Institute, Waseda University, Japan Abstract: We present a new virtual reality
8. Tensor Field Visualization
 8. Tensor Field Visualization Tensor: extension of concept of scalar and vector Tensor data for a tensor of level k is given by t i1,i2,,ik (x 1,,x n ) Second-order tensor often represented by matrix Examples:
8. Tensor Field Visualization Tensor: extension of concept of scalar and vector Tensor data for a tensor of level k is given by t i1,i2,,ik (x 1,,x n ) Second-order tensor often represented by matrix Examples:
Scalar Visualization
 Scalar Visualization 5-1 Motivation Visualizing scalar data is frequently encountered in science, engineering, and medicine, but also in daily life. Recalling from earlier, scalar datasets, or scalar fields,
Scalar Visualization 5-1 Motivation Visualizing scalar data is frequently encountered in science, engineering, and medicine, but also in daily life. Recalling from earlier, scalar datasets, or scalar fields,
11/1/13. Visualization. Scientific Visualization. Types of Data. Height Field. Contour Curves. Meshes
 CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 2.11] Jernej Barbic University of Southern California Scientific Visualization
CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 2.11] Jernej Barbic University of Southern California Scientific Visualization
Visualization. CSCI 420 Computer Graphics Lecture 26
 CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 11] Jernej Barbic University of Southern California 1 Scientific Visualization
CSCI 420 Computer Graphics Lecture 26 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 11] Jernej Barbic University of Southern California 1 Scientific Visualization
Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging
 1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
1 CS 9 Final Project Classification of Subject Motion for Improved Reconstruction of Dynamic Magnetic Resonance Imaging Feiyu Chen Department of Electrical Engineering ABSTRACT Subject motion is a significant
BOSS. Quick Start Guide For research use only. Blackrock Microsystems, LLC. Blackrock Offline Spike Sorter. User s Manual. 630 Komas Drive Suite 200
 BOSS Quick Start Guide For research use only Blackrock Microsystems, LLC 630 Komas Drive Suite 200 Salt Lake City UT 84108 T: +1 801 582 5533 www.blackrockmicro.com support@blackrockmicro.com 1 2 1.0 Table
BOSS Quick Start Guide For research use only Blackrock Microsystems, LLC 630 Komas Drive Suite 200 Salt Lake City UT 84108 T: +1 801 582 5533 www.blackrockmicro.com support@blackrockmicro.com 1 2 1.0 Table
Semiautomatic Cardiac CT Scan Classification with a Semantic User Interface
 Semiautomatic Cardiac CT Scan Classification with a Semantic User Interface Andreas Elsner 1, Dženan Zukić 2, Gitta Domik 1, Zikrija Avdagić 2, Wilhelm Schäfer 1, Eva Fricke 3, Dušanka Bošković 2 1 Department
Semiautomatic Cardiac CT Scan Classification with a Semantic User Interface Andreas Elsner 1, Dženan Zukić 2, Gitta Domik 1, Zikrija Avdagić 2, Wilhelm Schäfer 1, Eva Fricke 3, Dušanka Bošković 2 1 Department
Discovering High-level Parameters for Visualization Design
 EUROGRAPHICS - IEEE VGTC Symposium on Visualization (2005), pp. 1 8 K. W. Brodlie, D. J. Duke, K. I. Joy (Editors) Discovering High-level Parameters for Visualization Design Submission 26 Abstract In most
EUROGRAPHICS - IEEE VGTC Symposium on Visualization (2005), pp. 1 8 K. W. Brodlie, D. J. Duke, K. I. Joy (Editors) Discovering High-level Parameters for Visualization Design Submission 26 Abstract In most
A Design Toolbox to Generate Complex Phantoms for the Evaluation of Medical Image Processing Algorithms
 A Design Toolbox to Generate Complex Phantoms for the Evaluation of Medical Image Processing Algorithms Omar Hamo, Georg Nelles, Gudrun Wagenknecht Central Institute for Electronics, Research Center Juelich,
A Design Toolbox to Generate Complex Phantoms for the Evaluation of Medical Image Processing Algorithms Omar Hamo, Georg Nelles, Gudrun Wagenknecht Central Institute for Electronics, Research Center Juelich,
CIS 4930/ SCIENTIFICVISUALIZATION
 CIS 4930/6930-902 SCIENTIFICVISUALIZATION ISOSURFACING Paul Rosen Assistant Professor University of South Florida slides credits Tricoche and Meyer ADMINISTRATIVE Read (or watch video): Kieffer et al,
CIS 4930/6930-902 SCIENTIFICVISUALIZATION ISOSURFACING Paul Rosen Assistant Professor University of South Florida slides credits Tricoche and Meyer ADMINISTRATIVE Read (or watch video): Kieffer et al,
Classification of Hyperspectral Breast Images for Cancer Detection. Sander Parawira December 4, 2009
 1 Introduction Classification of Hyperspectral Breast Images for Cancer Detection Sander Parawira December 4, 2009 parawira@stanford.edu In 2009 approximately one out of eight women has breast cancer.
1 Introduction Classification of Hyperspectral Breast Images for Cancer Detection Sander Parawira December 4, 2009 parawira@stanford.edu In 2009 approximately one out of eight women has breast cancer.
The design of medical image transfer function using multi-feature fusion and improved k-means clustering
 Available online www.ocpr.com Journal of Chemical and Pharmaceutical Research, 04, 6(7):008-04 Research Article ISSN : 0975-7384 CODEN(USA) : JCPRC5 The design of medical image transfer function using
Available online www.ocpr.com Journal of Chemical and Pharmaceutical Research, 04, 6(7):008-04 Research Article ISSN : 0975-7384 CODEN(USA) : JCPRC5 The design of medical image transfer function using
MR IMAGE SEGMENTATION
 MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
Perception-Based Transfer Function Design
 Perception-Based Transfer Function Design Huamin Qu Department of Computer Science and Engineering The Hong Kong University of Science and Technology Outline Introduction Transfer Function Design Based
Perception-Based Transfer Function Design Huamin Qu Department of Computer Science and Engineering The Hong Kong University of Science and Technology Outline Introduction Transfer Function Design Based
Image Segmentation and Registration
 Image Segmentation and Registration Dr. Christine Tanner (tanner@vision.ee.ethz.ch) Computer Vision Laboratory, ETH Zürich Dr. Verena Kaynig, Machine Learning Laboratory, ETH Zürich Outline Segmentation
Image Segmentation and Registration Dr. Christine Tanner (tanner@vision.ee.ethz.ch) Computer Vision Laboratory, ETH Zürich Dr. Verena Kaynig, Machine Learning Laboratory, ETH Zürich Outline Segmentation
K-Means Clustering Using Localized Histogram Analysis
 K-Means Clustering Using Localized Histogram Analysis Michael Bryson University of South Carolina, Department of Computer Science Columbia, SC brysonm@cse.sc.edu Abstract. The first step required for many
K-Means Clustering Using Localized Histogram Analysis Michael Bryson University of South Carolina, Department of Computer Science Columbia, SC brysonm@cse.sc.edu Abstract. The first step required for many
Advances in Neural Information Processing Systems, 1999, In press. Unsupervised Classication with Non-Gaussian Mixture Models using ICA Te-Won Lee, Mi
 Advances in Neural Information Processing Systems, 1999, In press. Unsupervised Classication with Non-Gaussian Mixture Models using ICA Te-Won Lee, Michael S. Lewicki and Terrence Sejnowski Howard Hughes
Advances in Neural Information Processing Systems, 1999, In press. Unsupervised Classication with Non-Gaussian Mixture Models using ICA Te-Won Lee, Michael S. Lewicki and Terrence Sejnowski Howard Hughes
3-D MRI Brain Scan Classification Using A Point Series Based Representation
 3-D MRI Brain Scan Classification Using A Point Series Based Representation Akadej Udomchaiporn 1, Frans Coenen 1, Marta García-Fiñana 2, and Vanessa Sluming 3 1 Department of Computer Science, University
3-D MRI Brain Scan Classification Using A Point Series Based Representation Akadej Udomchaiporn 1, Frans Coenen 1, Marta García-Fiñana 2, and Vanessa Sluming 3 1 Department of Computer Science, University
Saturn User Manual. Rubén Cárdenes. 29th January 2010 Image Processing Laboratory, University of Valladolid. Abstract
 Saturn User Manual Rubén Cárdenes 29th January 2010 Image Processing Laboratory, University of Valladolid Abstract Saturn is a software package for DTI processing and visualization, provided with a graphic
Saturn User Manual Rubén Cárdenes 29th January 2010 Image Processing Laboratory, University of Valladolid Abstract Saturn is a software package for DTI processing and visualization, provided with a graphic
An ICA-Based Multivariate Discretization Algorithm
 An ICA-Based Multivariate Discretization Algorithm Ye Kang 1,2, Shanshan Wang 1,2, Xiaoyan Liu 1, Hokyin Lai 1, Huaiqing Wang 1, and Baiqi Miao 2 1 Department of Information Systems, City University of
An ICA-Based Multivariate Discretization Algorithm Ye Kang 1,2, Shanshan Wang 1,2, Xiaoyan Liu 1, Hokyin Lai 1, Huaiqing Wang 1, and Baiqi Miao 2 1 Department of Information Systems, City University of
Transform Introduction page 96 Spatial Transforms page 97
 Transform Introduction page 96 Spatial Transforms page 97 Pad page 97 Subregion page 101 Resize page 104 Shift page 109 1. Correcting Wraparound Using the Shift Tool page 109 Flip page 116 2. Flipping
Transform Introduction page 96 Spatial Transforms page 97 Pad page 97 Subregion page 101 Resize page 104 Shift page 109 1. Correcting Wraparound Using the Shift Tool page 109 Flip page 116 2. Flipping
ICA vs. PCA Active Appearance Models: Application to Cardiac MR Segmentation
 ICA vs. PCA Active Appearance Models: Application to Cardiac MR Segmentation M. Üzümcü 1, A.F. Frangi 2, M. Sonka 3, J.H.C. Reiber 1, B.P.F. Lelieveldt 1 1 Div. of Image Processing, Dept. of Radiology
ICA vs. PCA Active Appearance Models: Application to Cardiac MR Segmentation M. Üzümcü 1, A.F. Frangi 2, M. Sonka 3, J.H.C. Reiber 1, B.P.F. Lelieveldt 1 1 Div. of Image Processing, Dept. of Radiology
Robust PDF Table Locator
 Robust PDF Table Locator December 17, 2016 1 Introduction Data scientists rely on an abundance of tabular data stored in easy-to-machine-read formats like.csv files. Unfortunately, most government records
Robust PDF Table Locator December 17, 2016 1 Introduction Data scientists rely on an abundance of tabular data stored in easy-to-machine-read formats like.csv files. Unfortunately, most government records
Visualization Computer Graphics I Lecture 20
 15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] November 20, 2003 Doug James Carnegie Mellon University http://www.cs.cmu.edu/~djames/15-462/fall03
15-462 Computer Graphics I Lecture 20 Visualization Height Fields and Contours Scalar Fields Volume Rendering Vector Fields [Angel Ch. 12] November 20, 2003 Doug James Carnegie Mellon University http://www.cs.cmu.edu/~djames/15-462/fall03
Region-based Segmentation
 Region-based Segmentation Image Segmentation Group similar components (such as, pixels in an image, image frames in a video) to obtain a compact representation. Applications: Finding tumors, veins, etc.
Region-based Segmentation Image Segmentation Group similar components (such as, pixels in an image, image frames in a video) to obtain a compact representation. Applications: Finding tumors, veins, etc.
An Intelligent Clustering Algorithm for High Dimensional and Highly Overlapped Photo-Thermal Infrared Imaging Data
 An Intelligent Clustering Algorithm for High Dimensional and Highly Overlapped Photo-Thermal Infrared Imaging Data Nian Zhang and Lara Thompson Department of Electrical and Computer Engineering, University
An Intelligent Clustering Algorithm for High Dimensional and Highly Overlapped Photo-Thermal Infrared Imaging Data Nian Zhang and Lara Thompson Department of Electrical and Computer Engineering, University
Iso-surface cell search. Iso-surface Cells. Efficient Searching. Efficient search methods. Efficient iso-surface cell search. Problem statement:
 Iso-Contouring Advanced Issues Iso-surface cell search 1. Efficiently determining which cells to examine. 2. Using iso-contouring as a slicing mechanism 3. Iso-contouring in higher dimensions 4. Texturing
Iso-Contouring Advanced Issues Iso-surface cell search 1. Efficiently determining which cells to examine. 2. Using iso-contouring as a slicing mechanism 3. Iso-contouring in higher dimensions 4. Texturing
Biometrics Technology: Image Processing & Pattern Recognition (by Dr. Dickson Tong)
 Biometrics Technology: Image Processing & Pattern Recognition (by Dr. Dickson Tong) References: [1] http://homepages.inf.ed.ac.uk/rbf/hipr2/index.htm [2] http://www.cs.wisc.edu/~dyer/cs540/notes/vision.html
Biometrics Technology: Image Processing & Pattern Recognition (by Dr. Dickson Tong) References: [1] http://homepages.inf.ed.ac.uk/rbf/hipr2/index.htm [2] http://www.cs.wisc.edu/~dyer/cs540/notes/vision.html
Skull Segmentation of MR images based on texture features for attenuation correction in PET/MR
 Skull Segmentation of MR images based on texture features for attenuation correction in PET/MR CHAIBI HASSEN, NOURINE RACHID ITIO Laboratory, Oran University Algeriachaibih@yahoo.fr, nourine@yahoo.com
Skull Segmentation of MR images based on texture features for attenuation correction in PET/MR CHAIBI HASSEN, NOURINE RACHID ITIO Laboratory, Oran University Algeriachaibih@yahoo.fr, nourine@yahoo.com
Analysis of Functional MRI Timeseries Data Using Signal Processing Techniques
 Analysis of Functional MRI Timeseries Data Using Signal Processing Techniques Sea Chen Department of Biomedical Engineering Advisors: Dr. Charles A. Bouman and Dr. Mark J. Lowe S. Chen Final Exam October
Analysis of Functional MRI Timeseries Data Using Signal Processing Techniques Sea Chen Department of Biomedical Engineering Advisors: Dr. Charles A. Bouman and Dr. Mark J. Lowe S. Chen Final Exam October
Topology Correction for Brain Atlas Segmentation using a Multiscale Algorithm
 Topology Correction for Brain Atlas Segmentation using a Multiscale Algorithm Lin Chen and Gudrun Wagenknecht Central Institute for Electronics, Research Center Jülich, Jülich, Germany Email: l.chen@fz-juelich.de
Topology Correction for Brain Atlas Segmentation using a Multiscale Algorithm Lin Chen and Gudrun Wagenknecht Central Institute for Electronics, Research Center Jülich, Jülich, Germany Email: l.chen@fz-juelich.de
A User-friendly Tool for Semi-automated Segmentation and Surface Extraction from Color Volume Data Using Geometric Feature-space Operations
 A User-friendly Tool for Semi-automated Segmentation and Surface Extraction from Color Volume Data Using Geometric Feature-space Operations Tetyana Ivanovska and Lars Linsen School of Engineering and Science
A User-friendly Tool for Semi-automated Segmentation and Surface Extraction from Color Volume Data Using Geometric Feature-space Operations Tetyana Ivanovska and Lars Linsen School of Engineering and Science
Shear-Warp Volume Rendering. Volume Rendering Overview
 Shear-Warp Volume Rendering R. Daniel Bergeron Department of Computer Science University of New Hampshire Durham, NH 03824 From: Lacroute and Levoy, Fast Volume Rendering Using a Shear-Warp- Factorization
Shear-Warp Volume Rendering R. Daniel Bergeron Department of Computer Science University of New Hampshire Durham, NH 03824 From: Lacroute and Levoy, Fast Volume Rendering Using a Shear-Warp- Factorization
Chapter 3 Set Redundancy in Magnetic Resonance Brain Images
 16 Chapter 3 Set Redundancy in Magnetic Resonance Brain Images 3.1 MRI (magnetic resonance imaging) MRI is a technique of measuring physical structure within the human anatomy. Our proposed research focuses
16 Chapter 3 Set Redundancy in Magnetic Resonance Brain Images 3.1 MRI (magnetic resonance imaging) MRI is a technique of measuring physical structure within the human anatomy. Our proposed research focuses
Pedestrian Detection with Improved LBP and Hog Algorithm
 Open Access Library Journal 2018, Volume 5, e4573 ISSN Online: 2333-9721 ISSN Print: 2333-9705 Pedestrian Detection with Improved LBP and Hog Algorithm Wei Zhou, Suyun Luo Automotive Engineering College,
Open Access Library Journal 2018, Volume 5, e4573 ISSN Online: 2333-9721 ISSN Print: 2333-9705 Pedestrian Detection with Improved LBP and Hog Algorithm Wei Zhou, Suyun Luo Automotive Engineering College,
CoE4TN4 Image Processing
 CoE4TN4 Image Processing Chapter 11 Image Representation & Description Image Representation & Description After an image is segmented into regions, the regions are represented and described in a form suitable
CoE4TN4 Image Processing Chapter 11 Image Representation & Description Image Representation & Description After an image is segmented into regions, the regions are represented and described in a form suitable
INDEPENDENT COMPONENT ANALYSIS WITH FEATURE SELECTIVE FILTERING
 INDEPENDENT COMPONENT ANALYSIS WITH FEATURE SELECTIVE FILTERING Yi-Ou Li 1, Tülay Adalı 1, and Vince D. Calhoun 2,3 1 Department of Computer Science and Electrical Engineering University of Maryland Baltimore
INDEPENDENT COMPONENT ANALYSIS WITH FEATURE SELECTIVE FILTERING Yi-Ou Li 1, Tülay Adalı 1, and Vince D. Calhoun 2,3 1 Department of Computer Science and Electrical Engineering University of Maryland Baltimore
Texture Analysis. Selim Aksoy Department of Computer Engineering Bilkent University
 Texture Analysis Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr Texture An important approach to image description is to quantify its texture content. Texture
Texture Analysis Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr Texture An important approach to image description is to quantify its texture content. Texture
Surface Projection Method for Visualizing Volumetric Data
 Surface Projection Method for Visualizing Volumetric Data by Peter Lincoln A senior thesis submitted in partial fulfillment of the requirements for the degree of Bachelor of Science With Departmental Honors
Surface Projection Method for Visualizing Volumetric Data by Peter Lincoln A senior thesis submitted in partial fulfillment of the requirements for the degree of Bachelor of Science With Departmental Honors
Locating ego-centers in depth for hippocampal place cells
 204 5th Joint Symposium on Neural Computation Proceedings UCSD (1998) Locating ego-centers in depth for hippocampal place cells Kechen Zhang,' Terrence J. Sejeowski112 & Bruce L. ~cnau~hton~ 'Howard Hughes
204 5th Joint Symposium on Neural Computation Proceedings UCSD (1998) Locating ego-centers in depth for hippocampal place cells Kechen Zhang,' Terrence J. Sejeowski112 & Bruce L. ~cnau~hton~ 'Howard Hughes
Isosurface Rendering. CSC 7443: Scientific Information Visualization
 Isosurface Rendering What is Isosurfacing? An isosurface is the 3D surface representing the locations of a constant scalar value within a volume A surface with the same scalar field value Isosurfaces form
Isosurface Rendering What is Isosurfacing? An isosurface is the 3D surface representing the locations of a constant scalar value within a volume A surface with the same scalar field value Isosurfaces form
MEDICAL IMAGE ANALYSIS
 SECOND EDITION MEDICAL IMAGE ANALYSIS ATAM P. DHAWAN g, A B IEEE Engineering in Medicine and Biology Society, Sponsor IEEE Press Series in Biomedical Engineering Metin Akay, Series Editor +IEEE IEEE PRESS
SECOND EDITION MEDICAL IMAGE ANALYSIS ATAM P. DHAWAN g, A B IEEE Engineering in Medicine and Biology Society, Sponsor IEEE Press Series in Biomedical Engineering Metin Akay, Series Editor +IEEE IEEE PRESS
This exercise uses one anatomical data set (ANAT1) and two functional data sets (FUNC1 and FUNC2).
 Exploring Brain Anatomy This week s exercises will let you explore the anatomical organization of the brain to learn some of its basic properties, as well as the location of different structures. The human
Exploring Brain Anatomy This week s exercises will let you explore the anatomical organization of the brain to learn some of its basic properties, as well as the location of different structures. The human
Multicriteria Image Thresholding Based on Multiobjective Particle Swarm Optimization
 Applied Mathematical Sciences, Vol. 8, 2014, no. 3, 131-137 HIKARI Ltd, www.m-hikari.com http://dx.doi.org/10.12988/ams.2014.3138 Multicriteria Image Thresholding Based on Multiobjective Particle Swarm
Applied Mathematical Sciences, Vol. 8, 2014, no. 3, 131-137 HIKARI Ltd, www.m-hikari.com http://dx.doi.org/10.12988/ams.2014.3138 Multicriteria Image Thresholding Based on Multiobjective Particle Swarm
Ensemble registration: Combining groupwise registration and segmentation
 PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
PURWANI, COOTES, TWINING: ENSEMBLE REGISTRATION 1 Ensemble registration: Combining groupwise registration and segmentation Sri Purwani 1,2 sri.purwani@postgrad.manchester.ac.uk Tim Cootes 1 t.cootes@manchester.ac.uk
Available Online through
 Available Online through www.ijptonline.com ISSN: 0975-766X CODEN: IJPTFI Research Article ANALYSIS OF CT LIVER IMAGES FOR TUMOUR DIAGNOSIS BASED ON CLUSTERING TECHNIQUE AND TEXTURE FEATURES M.Krithika
Available Online through www.ijptonline.com ISSN: 0975-766X CODEN: IJPTFI Research Article ANALYSIS OF CT LIVER IMAGES FOR TUMOUR DIAGNOSIS BASED ON CLUSTERING TECHNIQUE AND TEXTURE FEATURES M.Krithika
