Press Release. Introduction. Prerequisites for LORETA
|
|
|
- Shavonne Suzanna Holt
- 5 years ago
- Views:
Transcription
1 Press Release Support & Tips A guided tour through LORETA Source localization in BrainVision Analyzer 2 by Dr.-Ing. Kidist Mideksa, Scientific Consultant at Brain Products Scientific Support What do I need in order to include source analysis as part of my analysis pipeline? This article provides you with the solution mainly focusing on LORETA. We will also walk you through the requirements prior to applying LORETA so that you will be able to make the best usage of LORETA in BrainVision Analyzer 2. Introduction Prerequisites for LORETA Accessing LORETA Creating Regions of Interest (ROIs) Output Measures Measure Extraction and Export Summary References Introduction Why source analysis? EEG recordings at the surface of the scalp represent the summed electrical activity coming from the different parts of the brain. This electrical activity is generated by large numbers of neurons with electrical charges moving within the central nervous system and causing small electrical currents. The amount of current flow is characterized by the volume conduction, i.e., the electromagnetic properties of the tissues inside the brain. There have been many algorithms developed so far for processing these signals at the scalp-level. The operations include, but are not limited to, time-domain analysis, frequency-domain analysis, and time-frequency analysis. Signals measured on the scalp, however, do not necessarily represent the true neuronal activity in the brain. That is, due to the volume conduction effect, electric currents spread over a relatively large distance from the sources to the electrodes, making EEG measurements possible in the first place. However, the underlying neural dynamics cannot be revealed because of considerable attenuation, mixing and smearing of the neuroelectric fields arising from different sources while spreading through the tissues of the brain until they reach at the scalp. Thus, there is a need for analysis techniques that enable backward tracing of the active brain regions. Using source analysis you will be able to estimate the location and dynamics of the underlying neuronal generators responsible for the electrical potentials measured on the scalp. Source analysis consists of solving the so-called forward problem and inverse problem. Given the physiological sources, one can calculate the generated electrical potentials using classical electrodynamics equations. This is known as solving the forward problem. When proceeding in the opposite direction one can infer where the underlying physiological sources are located by estimating the current distribution from the measured electrical potential. This is known as solving the inverse problem. Different forward and inverse methods offer an opportunity to select the most appropriate algorithm for a given experiment or research paradigm. In this article we will mainly focus on LORETA (Low Resolution Brain Electromagnetic Tomography), which is one of the most established and widely used current density reconstruction algorithms (Pascual-Marqui et al., 2002). It enables you to estimate distributed activity throughout the brain volume by decomposing the overlapping EEG voltage patterns into their underlying sources and localizing them within the brain. LORETA is not only answering the question of where the sources are located, but it also provides you with the time courses of each source. Prerequisites for LORETA As in any other inverse method, source estimation with the help of LORETA also requires several sophisticated steps, grouped into the forward and inverse problem. No worries, all these steps are implicitly taken care of in the implementation of LORETA in Analyzer 2. Thus, what is expected from you are mainly two things: Valid head coordinates. If your data does not contain valid head coordinates (standard or realistic), you can adopt the default coordinates as defined internally in Analyzer 2, as long as your channel names match the international electrode system and its derivatives (i.e., and 10-5 systems). Moreover, it is also recommended to have a uniform distribution of the electrodes throughout the scalp with the number of electrodes being at least 64 or higher (Michel CM et al., 2004). The coordinate system used by Analyzer 2 is illustrated in the User Manual (Appendix C Electrode coordinate system).
2 Voltage measurements (EEG data). The most important pre-requisite which is highly specific to Analyzer s LORETA is that only timedomain voltage values are valid input for the algorithm. Moreover, as in any kind of scalp-level analyses, for e.g., ERP analysis, source analysis also benefits from a high signal-to-noise ratio, which can be achieved by having well pre-processed data. This is the only prior information that is required from your side. However, if you are curious to know more about the implicit mechanisms of the forward and inverse problem, we will provide you with the basic ingredients without going too deeply into the mathematics behind it. To start with, consider that you have two domains: one being the electrode space where you have the voltage measurements and the second one being the source space, where the neuronal current sources are confined inside the brain. As mentioned above, the forward problem computes the electrical potentials based on the information about channel coordinates from the electrode space and the source parameters (position, orientation and magnitude of the neuronal current sources) from the source space. This computation entails modeling of 1) the neuronal current sources and its spatial distribution within the brain as well as 2) the volume conductor medium. Source model: a pre-defined source space is important to model sources. In Analyzer 2, the source space is restricted to the regions of the cortical gray matter and hippocampus in the Talairach atlas, discretized into a total of 2394 voxels at 7 mm spatial resolution. These sources are then modeled using equivalent current dipoles. Volume conductor model (anatomical and head models): it describes the geometrical and conductive properties of the head. In Analyzer 2, we offer a standard brain which is based on the MNI-305 brain template. The MNI images are co-registered to the Talairach brain atlas to map the detailed brain structures into the MNI space. Moreover, our LORETA implementation is based on a 3-shell spherical head model. This head model is fitted to the MNI brain template, which has already been co-registered to the Talairach brain atlas. In addition, the electrodes shall be projected to the co-registered spherical head model. Once all these ingredients are brought to the same coordinate system, the lead-field matrix, [N electrodes x 3M sources ] (3 represents the X, Y and Z components of the sources), is computed. This matrix serves as a mapping mechanism from the neuronal current sources within the brain to the electrical potentials on the head surface. All of the above steps are necessary pre-requisites for solving the inverse problem. The inverse problem aims to minimize the difference between the true voltage measurements and the calculated electrical potentials, as provided by the forward solution. Strikingly, there is an infinite number of source spatial configurations giving rise to the same voltage distribution. Therefore, the inverse problem in general does not have a unique solution, unless certain assumptions and mathematical constraints are imposed. In the case of LORETA, a unique inverse solution is achieved by imposing a constraint on the spatial configurations of the neuronal current sources, i.e., LORETA aims to find the smoothest spatial distributon within the source space. (Pascual-Marqui et al., 1994 and Pascual-Marqui et al., 1999). Finally, the unique inverse matrix, [3M sources x N electrodes ], fulfills all the conditions mentioned above, which relates the true voltage measurements to the estimated neuronal current sources. Accessing LORETA There are mainly two approaches to access LORETA in Analyzer 2. To decide which approach suits you best, let s start asking the following questions: Are you aiming at performing an exploratory analysis to identify the main brain areas underlying a given ERP/EEG pattern? Are you aiming to infer the Regions of Interest (ROIs)? In this case, you are looking for the LORETA transient view. The transient LORETA is available via the right-click Transient Views menu. After inferring your desired ROIs in the source space with the transient LORETA, are you now aiming at extracting the underlying neural dynamics as ROI-specific time series? Do you want to create ROIs based on other (e.g., fmri) studies and/or load ROIs from the predefined source space (the MNI voxels)? Then you are looking for the LORETA transform which is available via the Transformations ribbon (Transformations > Special Signal Processing > LORETA). Creating Regions of Interest (ROI) In this section, we will show you how to create ROIs and the possible computations that can be performed on the created ROIs. As mentioned above, the LORETA transform enables you to create and/or load ROIs. ROIs can be created by clicking on the Insert ROI button in the LORETA dialog, which will generate nodes automatically labeled as NewROI01, NewROI02, depending on the number of times you insert a new ROI. You then have the possibility to edit the ROI name.
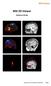
3 A single ROI node can be constructed in various different ways: Only one voxel associated to a specific brain region: A single voxel can be created with the option Add Nearest Voxel. Let s say during the exploratory analysis using the transient LORETA, you found out that the strongest source activation was located at the MNI coordinate (X = -3, Y = -74, Z = 29) and now you want to explore the dynamics of this single voxel. To do that, you first insert a ROI node and then using your mouse, fine tune the position of the yellow sliders to the desired position in the MNI brain image. Then click on the option Add Nearest Voxel button as shown in Fig. 1. It will give you the best matching location of what you have defined using the yellow sliders. The other possibility is that you already have a-priori information about the desired voxel from other studies and would like to create it. This is possible by making use of the pre-defined source space database. This database can be accessed from the LORETA dialog by clicking on the Export MNI Voxels File button. If you then open the saved *.csv file, you will see detailed information about all the pre-defined 2394 voxels. So, let s say that the a-priori information Fig. 1: Creating a single ROI associated to a single voxel from a predefined source space database. you have is the MNI coordinate (X = -52, Y = 3, Z = -41). You can search for this coordinate in the file and once you find the desired voxel, change its corresponding ROI-number (last column) to 1 as shown in Fig. 2. Please also make sure to change the total number of ROIs (2nd line of the *.csv file) from 0 to the desired number of ROIs (for e.g., to 1 in this case). You can then save it and go back to the LORETA dialog to load the modified *.csv file using the button Load ROIs from File. Generally, you can create K number of ROIs associated to a single voxel by simply incrementing the ROI-numbering from 0 to 1,2,3, K and finally changing the total number of ROIs (2nd line of the *.csv file) to K. Fig. 3 illustrates how to create 4 ROIs, each associated with a single voxel. Fig. 2: Creating a single ROI associated to a single voxel from a pre-defined source space database. Fig. 3: Creating 4 ROIs, each associated with a single voxel from the pre-defined source space database. The voxel associated with the ROI labeled as NewROI04 can be visualized with the red area in the MNI brain image.
4 Cluster of voxels associated with a specific brain region: In a similar way, you can also create several voxels associated with a single ROI. This is achieved by simply labeling all desired voxels with the same ROI-numbering scheme. Then change the total number of ROIs from 0 to 1, if a single ROI is desired as shown in Fig. 4. For instance, you can select L voxels and group them to the first ROI, another M voxels to the second ROI and so on. This can be achieved by labeling all the L voxels with the same ROI-numbering scheme (e.g., labeled as 1 to group them in the first ROI) and all the M voxels (e.g., labeled as 2 to group them in the second ROI) and so on. Finally, change the total number of ROIs to the desired number of ROIs you would like to create. Fig. 4: Creating a single ROI associated to 5 voxels from the pre-defined source space database. Cluster of voxels associated to different regions of the brain: Different structures can also be associated to a single ROI either based on lobes, gyri or Brodmann areas by clicking on the Add Lobe, Add Gyrus, Add Brodmann Area button, respectively. CAUTION! Unless there is a rationale behind creating such a ROI, it is not recommended to combine different regions of the brain in a single ROI due to the computational reasons explained below in the Output Measures section. Once you are done with defining your ROIs, you can make use of the Save ROIs to File option, which will enable you to save the created ROI tree to an *.xml file. This will allow you to conveniently share your ROI tree across different workspaces in Analyzer 2. See more, do more: Make use of our 88-ROIs file! So far, you may have noticed that creating and labeling ROIs, as well as associating brain areas to existing ROIs entails a considerable amount of manual work. How would you proceed if you are interested in an automatic creation of ROIs associated to a particular brain parcellation? You can imagine how challenging it is to manually define a ROI for each parcel either by editing the pre-defined source space database, or using the options in the group Edit Brain Regions of the LORETA transform. For your convenience, we have taken over this tedious job and created an *.xml file containing 88 ROIs/parcels associated to Brodmann areas, Amygdala and Hippocampus, separately for the left and right hemisphere. You can download the file from and with the help of the option Load ROIs from File you will be able to import all the 88 ROIs in one go. In the same way automatic ROI definition for other brain parcellations can be achieved. In case you need further assistance on how to do it, please feel free to contact us at support@brainproducts.com.
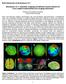
5 Output Measures After the successful creation of ROIs, the next step will be to estimate the current source density within the specified ROI. The LORETA dialog provides you with several output options. You have the possibility to compute for each time point the mean of the (X, Y, Z) component of the current density vector separately, the Magnitude of the current source density, or its Squared Magnitude within the specified ROI. Finishing the LORETA dialog will create a new history node named LORETA in the History Tree. It contains new LORETA channel(s), representing the temporal dynamics of the neuronal current source(s) corresponding to the created ROI(s). >> Tip: The default output option is the Current Density (Magnitude). In case you want different output options to be applied for the different ROIs, please make sure to select the desired output option for each ROI, separately. >> Tip: Don t worry if you see the resulting source time-series looking like a flat line. This is due to the fact that current source density values have indeed very small but non-zero values. In addition to that, the scale used in Analyzer 2 to display signals basically fits the voltage value of a typical EEG, and is not appropriate to display the source strength values. Thus, please make sure to increase the scaling of the amplitude either by using ctrl+up arrow keys of your keyboard or using the Scale Up Amplitude button from the toolbar. >> Tip: If you selected the check box Keep Old Channels in the LORETA dialog, this will add the LORETA channel(s) to the original set of EEG channels. This enables you to compare the EEG signal(s) to the corresponding LORETA source signal(s). To visualize both signals, you can specifically select and rescale only the LORETA channel(s) by clicking on the Scaling Range button from the toolbar and choose the option Scale Selected. The current density values obtained from the transient LORETA may differ from the results of the LORETA transform. Estimation of a source with the transient LORETA is done for the selected time-interval. So, what you will obtain for each displayed voxel is the magnitude of the vectorial mean of all current density vectors associated with all time points included in the selected time-interval. As illustrated in Fig. 5, what you see in the status bar of the LORETA transient view is the maximum current density (magnitude) ( µa/mm 2 ), i.e., the mean of all current density vectors within the selected time-interval (61 ms 131 ms) associated to a specific voxel located at (X = 4, Y = -81, Z = 8). >> Tip: To make sure that the maximum current density is displayed, right-click anywhere in the dockable window of the transient LORETA and make sure that the option Lock Navigation to Maximum is checked. By default, this option is selected. With this setting, the yellow sliders will always point to the position of the voxel with the maximal activation, and at the same time its current density value is displayed in the status bar. >> Tip: The option Show Horizontal Slices gives you the opportunity to explore the complete 3D current source density distribution in a 2D by 2D manner. >> Tip: You have the possibility of adjusting the scale of the current source density values via the Settings dialog box (accessible by clicking on the option Settings). The option Automatic Scaling helps you detect where the focal point of the maximum current source density value lies. Based on this information, you can then use the Manual Scaling option to clip the current source density values above a certain threshold (i.e., concentrated around the maximum current source density value). This will enable you to display the largest current source density value only. Fig. 5: Dockable window of the transient LORETA showing source activation map for the selected time-interval.
6 On the other hand, the LORETA transform provides you with the output measures for each time point. Analogous to the computation of the vectorial mean across time points based on the selection of the time-interval, the LORETA transform also computes the vectorial mean across multiple voxels, when multiple voxels are included within a single ROI. So, what you will obtain for a single LORETA channel is a source signal containing, for each time point, the average current density across all the voxels within a ROI, as shown in Fig. 6. This is the reason why we said to take care when adding different brain areas to a single ROI. Especially when creating ROIs comprising large brain areas, the current density vectors across all voxels may point in different opposing directions, and thus their contribution to the vectorial mean cancels out, which might not be desired. So, if extracting source signals from different regions of the brain is desired, we do recommend to create separate ROIs for each region and aggregate only the desired voxels to a ROI associated to a particular region, as illustrated in Fig. 7. Fig. 6: Effect of including different structures of the brain within a single ROI along with its corresponding source signal depicted in one LORETA channel (NewROI01). Fig. 7: Separate ROIs for the different regions of the brain along with its corresponding source signal depicted in separate LORETA channels (Occipital and Frontal). Measure Extraction and Export At this final stage you might be interested in exporting the results obtained from the LORETA transform for further statistical analyses. We will briefly provide you with some valuable tips about the measures that can be extracted from the source time series and the ways to extract them. Exporting current source density values: Current source density values for each time point can be easily exported by applying the Generic Data Export module (Export > Node Export > Generic Data) on the LORETA node. The exported file will be stored under the Export folder of the current workspace. >> Tip: Since current source density values are typically very small, in order to make sure that you don t loose any value by the rounding effect while exporting, please make sure to increase the precision (e.g., 10 or more). The precision can be adjusted using the option Use Custom Precision in the Generic Data Export dialog. Area measures: Are you looking for a way to export area information or the mean activity around the maximum source signal? The Area Information Export module (Export > Multiple Export > Area Information) enables you to compute and export area measures for the desired time interval. Finding the peak current source density value: Are you interested in pinpointing the maximum and/or minimum current source density value of the source signal and its corresponding latency for each ROI? If so, you can make use of the Peak Detection transformation (Transformations > Segment Analysis Functions > Result Evaluation > Peak Detection). It enables you to detect and mark the maximum and/or minimum current source density value within the desired time interval. After detection, you might want to export the corresponding current source density value and latency for each ROI. The Peak Information Export module (Export > Multiple Export > Peak Information) enables you to export the detected maximum and/or minimum current source density value(s) and time point(s), separately for each ROI.
7 Summary It is our pleasure to invite you to make use of the two modes of LORETA, i.e., the LORETA transient view and LORETA transform in Analyzer 2. As you can see they complement each other. With the help of the transient LORETA you can inspect your EEG data and make a first impression about the location of the activated area. Once you have an impression about the location, you can infer your ROIs and use the LORETA transform to create the ROIs and estimate the current source density, as well as extract the source time series. We hope that you will broaden your research questions beyond scalp domain analyses by diving into the source domain analysis. Further detailed information about the two modes of LORETA as well as the different Export modules can be found in the BrainVision Analyzer 2 User Manual. For a step by step protocol on how to use LORETA, we would like to invite you to have a look at the pdf slides of our webinar on Introduction to Source Analysis Distributed Source Imaging Using LORETA ( If you still can t find what you are looking for, drop us a line at support@brainproducts.com. We will be happy to help! References [1] Pascual-Marqui RD, Michel CM, Lehmann D (1994). Low resolution electromagnetic tomography: a new method for localizing electrical activity in the brain. International Journal of Psychophysiology, 18: [2] Pascual-Marqui RD, Lehmann D, Koenig T, Kochi K, Merlo MCG, et al. (1999). Low resolution brain electromagnetic tomography (LORETA) functional imaging in acute, neuroleptic-naive, first-episode, productive schizophrenia. Psychiatry Res., 90: [3] Pascual-Marqui RD, Esslen M, Kochi K, Lehmann D (2002). Functional imaging with low resolution brain electromagnetic tomography (LORETA): a review. Methods & Findings in Experimental & Clinical Pharmacology, 24C: [4] Michel CM, Murray MM, Lantz G, Gonzalez S, Spinelli L, et al. (2004). EEG source imaging. Clinical Neurophysiology, 115(10),
May Brain Products Teaching Material. Introduction to Source Analysis Distributed Source Imaging using LORETA. BrainVision Analyzer 2 Webinar
 Introduction to Source Analysis Distributed Source Imaging using LORETA BrainVision Analyzer 2 Webinar Scientific Support Team Brain Products GmbH Webinar Team Presenter BP Moderator BP Tech Support Kidist
Introduction to Source Analysis Distributed Source Imaging using LORETA BrainVision Analyzer 2 Webinar Scientific Support Team Brain Products GmbH Webinar Team Presenter BP Moderator BP Tech Support Kidist
BESA Research. CE certified software package for comprehensive, fast, and user-friendly analysis of EEG and MEG
 BESA Research CE certified software package for comprehensive, fast, and user-friendly analysis of EEG and MEG BESA Research choose the best analysis tool for your EEG and MEG data BESA Research is the
BESA Research CE certified software package for comprehensive, fast, and user-friendly analysis of EEG and MEG BESA Research choose the best analysis tool for your EEG and MEG data BESA Research is the
- Graphical editing of user montages for convenient data review - Import of user-defined file formats using generic reader
 Data review and processing Source montages and 3D whole-head mapping Onset of epileptic seizure with 3D whole-head maps and hemispheric comparison of density spectral arrays (DSA) Graphical display of
Data review and processing Source montages and 3D whole-head mapping Onset of epileptic seizure with 3D whole-head maps and hemispheric comparison of density spectral arrays (DSA) Graphical display of
Group (Level 2) fmri Data Analysis - Lab 4
 Group (Level 2) fmri Data Analysis - Lab 4 Index Goals of this Lab Before Getting Started The Chosen Ten Checking Data Quality Create a Mean Anatomical of the Group Group Analysis: One-Sample T-Test Examine
Group (Level 2) fmri Data Analysis - Lab 4 Index Goals of this Lab Before Getting Started The Chosen Ten Checking Data Quality Create a Mean Anatomical of the Group Group Analysis: One-Sample T-Test Examine
Surface-based Analysis: Inter-subject Registration and Smoothing
 Surface-based Analysis: Inter-subject Registration and Smoothing Outline Exploratory Spatial Analysis Coordinate Systems 3D (Volumetric) 2D (Surface-based) Inter-subject registration Volume-based Surface-based
Surface-based Analysis: Inter-subject Registration and Smoothing Outline Exploratory Spatial Analysis Coordinate Systems 3D (Volumetric) 2D (Surface-based) Inter-subject registration Volume-based Surface-based
The organization of the human cerebral cortex estimated by intrinsic functional connectivity
 1 The organization of the human cerebral cortex estimated by intrinsic functional connectivity Journal: Journal of Neurophysiology Author: B. T. Thomas Yeo, et al Link: https://www.ncbi.nlm.nih.gov/pubmed/21653723
1 The organization of the human cerebral cortex estimated by intrinsic functional connectivity Journal: Journal of Neurophysiology Author: B. T. Thomas Yeo, et al Link: https://www.ncbi.nlm.nih.gov/pubmed/21653723
Theory: modeling, localization and imaging
 Electromagnetic Brain Mapping with MEG/EEG Theory: modeling, localization and imaging Sylvain Baillet Imaging Group Cognitive Neuroscience & Brain Imaging Lab. Hôpital de la Salpêtrière CNRS UPR640 - LENA
Electromagnetic Brain Mapping with MEG/EEG Theory: modeling, localization and imaging Sylvain Baillet Imaging Group Cognitive Neuroscience & Brain Imaging Lab. Hôpital de la Salpêtrière CNRS UPR640 - LENA
Section 9. Human Anatomy and Physiology
 Section 9. Human Anatomy and Physiology 9.1 MR Neuroimaging 9.2 Electroencephalography Overview As stated throughout, electrophysiology is the key tool in current systems neuroscience. However, single-
Section 9. Human Anatomy and Physiology 9.1 MR Neuroimaging 9.2 Electroencephalography Overview As stated throughout, electrophysiology is the key tool in current systems neuroscience. However, single-
Update ASA 4.8. Expand your research potential with ASA 4.8. Highly advanced 3D display of single channel coherence
 Update ASA 4.8 Expand your research potential with ASA 4.8. The ASA 4.8 software has everything needed for a complete analysis of EEG / ERP and MEG data. From features like (pre)processing of data, co-registration
Update ASA 4.8 Expand your research potential with ASA 4.8. The ASA 4.8 software has everything needed for a complete analysis of EEG / ERP and MEG data. From features like (pre)processing of data, co-registration
FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING. Francesca Pizzorni Ferrarese
 FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING Francesca Pizzorni Ferrarese Pipeline overview WM and GM Segmentation Registration Data reconstruction Tractography
FROM IMAGE RECONSTRUCTION TO CONNECTIVITY ANALYSIS: A JOURNEY THROUGH THE BRAIN'S WIRING Francesca Pizzorni Ferrarese Pipeline overview WM and GM Segmentation Registration Data reconstruction Tractography
This exercise uses one anatomical data set (ANAT1) and two functional data sets (FUNC1 and FUNC2).
 Exploring Brain Anatomy This week s exercises will let you explore the anatomical organization of the brain to learn some of its basic properties, as well as the location of different structures. The human
Exploring Brain Anatomy This week s exercises will let you explore the anatomical organization of the brain to learn some of its basic properties, as well as the location of different structures. The human
Effect of age and dementia on topology of brain functional networks. Paul McCarthy, Luba Benuskova, Liz Franz University of Otago, New Zealand
 Effect of age and dementia on topology of brain functional networks Paul McCarthy, Luba Benuskova, Liz Franz University of Otago, New Zealand 1 Structural changes in aging brain Age-related changes in
Effect of age and dementia on topology of brain functional networks Paul McCarthy, Luba Benuskova, Liz Franz University of Otago, New Zealand 1 Structural changes in aging brain Age-related changes in
ASA Getting Started. ANT BV, Enschede, Netherlands Advanced Neuro Technology
 ASA Getting Started ANT BV, Enschede, Netherlands Advanced Neuro Technology www.ant-neuro.com asa@ant-neuro.com TABLE OF CONTENTS DISCLAIMER... 3 NOTICE... 3 INTRODUCTION... 5 ASA-LAB... 6 ASA-LAB RECORDING
ASA Getting Started ANT BV, Enschede, Netherlands Advanced Neuro Technology www.ant-neuro.com asa@ant-neuro.com TABLE OF CONTENTS DISCLAIMER... 3 NOTICE... 3 INTRODUCTION... 5 ASA-LAB... 6 ASA-LAB RECORDING
BrainSpace V2.1: Geometric mapping and diffusion-based software for cross-subject multimodality brain imaging informatics
 Brief Introduction to BrainSpace V2.1 BrainSpace V2.1: Geometric mapping and diffusion-based software for cross-subject multimodality brain imaging informatics Graphics and Imaging Laboratory Wayne State
Brief Introduction to BrainSpace V2.1 BrainSpace V2.1: Geometric mapping and diffusion-based software for cross-subject multimodality brain imaging informatics Graphics and Imaging Laboratory Wayne State
Analysis of Functional MRI Timeseries Data Using Signal Processing Techniques
 Analysis of Functional MRI Timeseries Data Using Signal Processing Techniques Sea Chen Department of Biomedical Engineering Advisors: Dr. Charles A. Bouman and Dr. Mark J. Lowe S. Chen Final Exam October
Analysis of Functional MRI Timeseries Data Using Signal Processing Techniques Sea Chen Department of Biomedical Engineering Advisors: Dr. Charles A. Bouman and Dr. Mark J. Lowe S. Chen Final Exam October
Single Subject Demo Data Instructions 1) click "New" and answer "No" to the "spatially preprocess" question.
 (1) conn - Functional connectivity toolbox v1.0 Single Subject Demo Data Instructions 1) click "New" and answer "No" to the "spatially preprocess" question. 2) in "Basic" enter "1" subject, "6" seconds
(1) conn - Functional connectivity toolbox v1.0 Single Subject Demo Data Instructions 1) click "New" and answer "No" to the "spatially preprocess" question. 2) in "Basic" enter "1" subject, "6" seconds
Statistical Analysis of Neuroimaging Data. Phebe Kemmer BIOS 516 Sept 24, 2015
 Statistical Analysis of Neuroimaging Data Phebe Kemmer BIOS 516 Sept 24, 2015 Review from last time Structural Imaging modalities MRI, CAT, DTI (diffusion tensor imaging) Functional Imaging modalities
Statistical Analysis of Neuroimaging Data Phebe Kemmer BIOS 516 Sept 24, 2015 Review from last time Structural Imaging modalities MRI, CAT, DTI (diffusion tensor imaging) Functional Imaging modalities
BioEST ver. 1.5beta User Manual Chang-Hwan Im (Ph.D.)
 BioEST ver. 1.5beta User Manual Chang-Hwan Im (Ph.D.) http://www.bioest.com E-mail: ichism@elecmech.snu.ac.kr imxxx010@umn.edu 1. What is BioEST? BioEST is a special edition of SNUEEG(http://www.bioinverse.com),
BioEST ver. 1.5beta User Manual Chang-Hwan Im (Ph.D.) http://www.bioest.com E-mail: ichism@elecmech.snu.ac.kr imxxx010@umn.edu 1. What is BioEST? BioEST is a special edition of SNUEEG(http://www.bioinverse.com),
Automated MR Image Analysis Pipelines
 Automated MR Image Analysis Pipelines Andy Simmons Centre for Neuroimaging Sciences, Kings College London Institute of Psychiatry. NIHR Biomedical Research Centre for Mental Health at IoP & SLAM. Neuroimaging
Automated MR Image Analysis Pipelines Andy Simmons Centre for Neuroimaging Sciences, Kings College London Institute of Psychiatry. NIHR Biomedical Research Centre for Mental Health at IoP & SLAM. Neuroimaging
Analysis of fmri data within Brainvisa Example with the Saccades database
 Analysis of fmri data within Brainvisa Example with the Saccades database 18/11/2009 Note : All the sentences in italic correspond to informations relative to the specific dataset under study TP participants
Analysis of fmri data within Brainvisa Example with the Saccades database 18/11/2009 Note : All the sentences in italic correspond to informations relative to the specific dataset under study TP participants
An artificial neural network approach to the classification of inferred intracranial signals
 An artificial neural network approach to the classification of inferred intracranial signals CHRISTOS E. VASIOS 1 and GEORGE K. MATSOPOULOS 2 1 Athinoula A. Martinos Center for Biomedical Imaging, Department
An artificial neural network approach to the classification of inferred intracranial signals CHRISTOS E. VASIOS 1 and GEORGE K. MATSOPOULOS 2 1 Athinoula A. Martinos Center for Biomedical Imaging, Department
SDC. Engineering Analysis with COSMOSWorks. Paul M. Kurowski Ph.D., P.Eng. SolidWorks 2003 / COSMOSWorks 2003
 Engineering Analysis with COSMOSWorks SolidWorks 2003 / COSMOSWorks 2003 Paul M. Kurowski Ph.D., P.Eng. SDC PUBLICATIONS Design Generator, Inc. Schroff Development Corporation www.schroff.com www.schroff-europe.com
Engineering Analysis with COSMOSWorks SolidWorks 2003 / COSMOSWorks 2003 Paul M. Kurowski Ph.D., P.Eng. SDC PUBLICATIONS Design Generator, Inc. Schroff Development Corporation www.schroff.com www.schroff-europe.com
MSI 2D Viewer Software Guide
 MSI 2D Viewer Software Guide Page:1 DISCLAIMER We have used reasonable effort to include accurate and up-to-date information in this manual; it does not, however, make any warranties, conditions or representations
MSI 2D Viewer Software Guide Page:1 DISCLAIMER We have used reasonable effort to include accurate and up-to-date information in this manual; it does not, however, make any warranties, conditions or representations
Manual image registration in BrainVoyager QX Table of Contents
 Manual image registration in BrainVoyager QX Table of Contents Manual image registration in BrainVoyager QX......1 Performing manual alignment for functional to anatomical images......2 Step 1: preparation......2
Manual image registration in BrainVoyager QX Table of Contents Manual image registration in BrainVoyager QX......1 Performing manual alignment for functional to anatomical images......2 Step 1: preparation......2
Brain Explorer for Connectomic Analysis (BECA) Software Manual
 Brain Explorer for Connectomic Analysis (BECA) Software Manual Table of Contents 1. Launch the application... 2 2. Tractography Visualization... 3 3. Gray Matter Visualization... 5 4. Fiber classification
Brain Explorer for Connectomic Analysis (BECA) Software Manual Table of Contents 1. Launch the application... 2 2. Tractography Visualization... 3 3. Gray Matter Visualization... 5 4. Fiber classification
CS/NEUR125 Brains, Minds, and Machines. Due: Wednesday, April 5
 CS/NEUR125 Brains, Minds, and Machines Lab 8: Using fmri to Discover Language Areas in the Brain Due: Wednesday, April 5 In this lab, you will analyze fmri data from an experiment that was designed to
CS/NEUR125 Brains, Minds, and Machines Lab 8: Using fmri to Discover Language Areas in the Brain Due: Wednesday, April 5 In this lab, you will analyze fmri data from an experiment that was designed to
INDEPENDENT COMPONENT ANALYSIS APPLIED TO fmri DATA: A GENERATIVE MODEL FOR VALIDATING RESULTS
 INDEPENDENT COMPONENT ANALYSIS APPLIED TO fmri DATA: A GENERATIVE MODEL FOR VALIDATING RESULTS V. Calhoun 1,2, T. Adali, 2 and G. Pearlson 1 1 Johns Hopkins University Division of Psychiatric Neuro-Imaging,
INDEPENDENT COMPONENT ANALYSIS APPLIED TO fmri DATA: A GENERATIVE MODEL FOR VALIDATING RESULTS V. Calhoun 1,2, T. Adali, 2 and G. Pearlson 1 1 Johns Hopkins University Division of Psychiatric Neuro-Imaging,
This Time. fmri Data analysis
 This Time Reslice example Spatial Normalization Noise in fmri Methods for estimating and correcting for physiologic noise SPM Example Spatial Normalization: Remind ourselves what a typical functional image
This Time Reslice example Spatial Normalization Noise in fmri Methods for estimating and correcting for physiologic noise SPM Example Spatial Normalization: Remind ourselves what a typical functional image
Contents. Introduction... iii. CHAPTER 1 - Getting Started... 5
 Transducer Characterization User s Guide Version 5 5 1 build 12 or later UTEX Scientific Instruments Inc support@utex com Winspect Transducer Characterization User s Guide Contents Table of Contents and
Transducer Characterization User s Guide Version 5 5 1 build 12 or later UTEX Scientific Instruments Inc support@utex com Winspect Transducer Characterization User s Guide Contents Table of Contents and
Multi-voxel pattern analysis: Decoding Mental States from fmri Activity Patterns
 Multi-voxel pattern analysis: Decoding Mental States from fmri Activity Patterns Artwork by Leon Zernitsky Jesse Rissman NITP Summer Program 2012 Part 1 of 2 Goals of Multi-voxel Pattern Analysis Decoding
Multi-voxel pattern analysis: Decoding Mental States from fmri Activity Patterns Artwork by Leon Zernitsky Jesse Rissman NITP Summer Program 2012 Part 1 of 2 Goals of Multi-voxel Pattern Analysis Decoding
COMPUTER SIMULATION OF COMPLEX SYSTEMS USING AUTOMATA NETWORKS K. Ming Leung
 POLYTECHNIC UNIVERSITY Department of Computer and Information Science COMPUTER SIMULATION OF COMPLEX SYSTEMS USING AUTOMATA NETWORKS K. Ming Leung Abstract: Computer simulation of the dynamics of complex
POLYTECHNIC UNIVERSITY Department of Computer and Information Science COMPUTER SIMULATION OF COMPLEX SYSTEMS USING AUTOMATA NETWORKS K. Ming Leung Abstract: Computer simulation of the dynamics of complex
The Allen Human Brain Atlas offers three types of searches to allow a user to: (1) obtain gene expression data for specific genes (or probes) of
 Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
Microarray Data MICROARRAY DATA Gene Search Boolean Syntax Differential Search Mouse Differential Search Search Results Gene Classification Correlative Search Download Search Results Data Visualization
Fmri Spatial Processing
 Educational Course: Fmri Spatial Processing Ray Razlighi Jun. 8, 2014 Spatial Processing Spatial Re-alignment Geometric distortion correction Spatial Normalization Smoothing Why, When, How, Which Why is
Educational Course: Fmri Spatial Processing Ray Razlighi Jun. 8, 2014 Spatial Processing Spatial Re-alignment Geometric distortion correction Spatial Normalization Smoothing Why, When, How, Which Why is
BrainMask. Quick Start
 BrainMask Quick Start Segmentation of the brain from three-dimensional MR images is a crucial pre-processing step in morphological and volumetric brain studies. BrainMask software implements a fully automatic
BrainMask Quick Start Segmentation of the brain from three-dimensional MR images is a crucial pre-processing step in morphological and volumetric brain studies. BrainMask software implements a fully automatic
MultiVariate Bayesian (MVB) decoding of brain images
 MultiVariate Bayesian (MVB) decoding of brain images Alexa Morcom Edinburgh SPM course 2015 With thanks to J. Daunizeau, K. Brodersen for slides stimulus behaviour encoding of sensorial or cognitive state?
MultiVariate Bayesian (MVB) decoding of brain images Alexa Morcom Edinburgh SPM course 2015 With thanks to J. Daunizeau, K. Brodersen for slides stimulus behaviour encoding of sensorial or cognitive state?
ERPEEG Tutorial. Version 1.0. This tutorial was written by: Sravya Atluri, Matthew Frehlich and Dr. Faranak Farzan.
 ERPEEG Tutorial Version 1.0 This tutorial was written by: Sravya Atluri, Matthew Frehlich and Dr. Faranak Farzan. Contact: faranak.farzan@sfu.ca Temerty Centre for Therapeutic Brain Stimulation Centre
ERPEEG Tutorial Version 1.0 This tutorial was written by: Sravya Atluri, Matthew Frehlich and Dr. Faranak Farzan. Contact: faranak.farzan@sfu.ca Temerty Centre for Therapeutic Brain Stimulation Centre
Conference Biomedical Engineering
 Automatic Medical Image Analysis for Measuring Bone Thickness and Density M. Kovalovs *, A. Glazs Image Processing and Computer Graphics Department, Riga Technical University, Latvia * E-mail: mihails.kovalovs@rtu.lv
Automatic Medical Image Analysis for Measuring Bone Thickness and Density M. Kovalovs *, A. Glazs Image Processing and Computer Graphics Department, Riga Technical University, Latvia * E-mail: mihails.kovalovs@rtu.lv
FMA901F: Machine Learning Lecture 6: Graphical Models. Cristian Sminchisescu
 FMA901F: Machine Learning Lecture 6: Graphical Models Cristian Sminchisescu Graphical Models Provide a simple way to visualize the structure of a probabilistic model and can be used to design and motivate
FMA901F: Machine Learning Lecture 6: Graphical Models Cristian Sminchisescu Graphical Models Provide a simple way to visualize the structure of a probabilistic model and can be used to design and motivate
MR IMAGE SEGMENTATION
 MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
MR IMAGE SEGMENTATION Prepared by : Monil Shah What is Segmentation? Partitioning a region or regions of interest in images such that each region corresponds to one or more anatomic structures Classification
New Approaches for EEG Source Localization and Dipole Moment Estimation. Shun Chi Wu, Yuchen Yao, A. Lee Swindlehurst University of California Irvine
 New Approaches for EEG Source Localization and Dipole Moment Estimation Shun Chi Wu, Yuchen Yao, A. Lee Swindlehurst University of California Irvine Outline Motivation why EEG? Mathematical Model equivalent
New Approaches for EEG Source Localization and Dipole Moment Estimation Shun Chi Wu, Yuchen Yao, A. Lee Swindlehurst University of California Irvine Outline Motivation why EEG? Mathematical Model equivalent
BrainMask. Quick Start
 BrainMask Quick Start Segmentation of the brain from three-dimensional MR images is a crucial preprocessing step in morphological and volumetric brain studies. BrainMask software implements a fully automatic
BrainMask Quick Start Segmentation of the brain from three-dimensional MR images is a crucial preprocessing step in morphological and volumetric brain studies. BrainMask software implements a fully automatic
DesignDirector Version 1.0(E)
 Statistical Design Support System DesignDirector Version 1.0(E) User s Guide NHK Spring Co.,Ltd. Copyright NHK Spring Co.,Ltd. 1999 All Rights Reserved. Copyright DesignDirector is registered trademarks
Statistical Design Support System DesignDirector Version 1.0(E) User s Guide NHK Spring Co.,Ltd. Copyright NHK Spring Co.,Ltd. 1999 All Rights Reserved. Copyright DesignDirector is registered trademarks
Quick Start Guide - Contents. Opening Word Locating Big Lottery Fund Templates The Word 2013 Screen... 3
 Quick Start Guide - Contents Opening Word... 1 Locating Big Lottery Fund Templates... 2 The Word 2013 Screen... 3 Things You Might Be Looking For... 4 What s New On The Ribbon... 5 The Quick Access Toolbar...
Quick Start Guide - Contents Opening Word... 1 Locating Big Lottery Fund Templates... 2 The Word 2013 Screen... 3 Things You Might Be Looking For... 4 What s New On The Ribbon... 5 The Quick Access Toolbar...
The LabScribe Tutorial
 The LabScribe Tutorial LabScribe allows data to be accumulated, displayed and analyzed on a computer screen in a format similar to a laboratory strip chart recorder. Equipment Setup 1 Place the iworx unit
The LabScribe Tutorial LabScribe allows data to be accumulated, displayed and analyzed on a computer screen in a format similar to a laboratory strip chart recorder. Equipment Setup 1 Place the iworx unit
Quality Checking an fmri Group Result (art_groupcheck)
 Quality Checking an fmri Group Result (art_groupcheck) Paul Mazaika, Feb. 24, 2009 A statistical parameter map of fmri group analyses relies on the assumptions of the General Linear Model (GLM). The assumptions
Quality Checking an fmri Group Result (art_groupcheck) Paul Mazaika, Feb. 24, 2009 A statistical parameter map of fmri group analyses relies on the assumptions of the General Linear Model (GLM). The assumptions
Rare Event Detection Algorithm. User s Guide
 Rare Event Detection Algorithm User s Guide Copyright 2008 Aperio Technologies, Inc. Part Number/Revision: MAN 0123, Revision A Date: September 2, 2008 This document applies to software versions Release
Rare Event Detection Algorithm User s Guide Copyright 2008 Aperio Technologies, Inc. Part Number/Revision: MAN 0123, Revision A Date: September 2, 2008 This document applies to software versions Release
Virtual Frap User Guide
 Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
Virtual Frap User Guide http://wiki.vcell.uchc.edu/twiki/bin/view/vcell/vfrap Center for Cell Analysis and Modeling University of Connecticut Health Center 2010-1 - 1 Introduction Flourescence Photobleaching
Resting state network estimation in individual subjects
 Resting state network estimation in individual subjects Data 3T NIL(21,17,10), Havard-MGH(692) Young adult fmri BOLD Method Machine learning algorithm MLP DR LDA Network image Correlation Spatial Temporal
Resting state network estimation in individual subjects Data 3T NIL(21,17,10), Havard-MGH(692) Young adult fmri BOLD Method Machine learning algorithm MLP DR LDA Network image Correlation Spatial Temporal
Rat 2D EPSI Dual Band Variable Flip Angle 13 C Dynamic Spectroscopy
 Rat 2D EPSI Dual Band Variable Flip Angle 13 C Dynamic Spectroscopy In this example you will load a dynamic MRS animal data set acquired on a GE 3T scanner. This data was acquired with an EPSI sequence
Rat 2D EPSI Dual Band Variable Flip Angle 13 C Dynamic Spectroscopy In this example you will load a dynamic MRS animal data set acquired on a GE 3T scanner. This data was acquired with an EPSI sequence
Figure 1. Comparison of the frequency of centrality values for central London and our region of Soho studied. The comparison shows that Soho falls
 A B C Figure 1. Comparison of the frequency of centrality values for central London and our region of Soho studied. The comparison shows that Soho falls well within the distribution for London s streets.
A B C Figure 1. Comparison of the frequency of centrality values for central London and our region of Soho studied. The comparison shows that Soho falls well within the distribution for London s streets.
Journal of Articles in Support of The Null Hypothesis
 Data Preprocessing Martin M. Monti, PhD UCLA Psychology NITP 2016 Typical (task-based) fmri analysis sequence Image Pre-processing Single Subject Analysis Group Analysis Journal of Articles in Support
Data Preprocessing Martin M. Monti, PhD UCLA Psychology NITP 2016 Typical (task-based) fmri analysis sequence Image Pre-processing Single Subject Analysis Group Analysis Journal of Articles in Support
Intensity Transformations and Spatial Filtering
 77 Chapter 3 Intensity Transformations and Spatial Filtering Spatial domain refers to the image plane itself, and image processing methods in this category are based on direct manipulation of pixels in
77 Chapter 3 Intensity Transformations and Spatial Filtering Spatial domain refers to the image plane itself, and image processing methods in this category are based on direct manipulation of pixels in
FSL Workshop Session 3 David Smith & John Clithero
 FSL Workshop 12.09.08 Session 3 David Smith & John Clithero What is MELODIC? Probabilistic ICA Improves upon standard ICA Allows for inference Avoids over-fitting Three stage process ( ppca ) 1.) Dimension
FSL Workshop 12.09.08 Session 3 David Smith & John Clithero What is MELODIC? Probabilistic ICA Improves upon standard ICA Allows for inference Avoids over-fitting Three stage process ( ppca ) 1.) Dimension
Neural Network based textural labeling of images in multimedia applications
 Neural Network based textural labeling of images in multimedia applications S.A. Karkanis +, G.D. Magoulas +, and D.A. Karras ++ + University of Athens, Dept. of Informatics, Typa Build., Panepistimiopolis,
Neural Network based textural labeling of images in multimedia applications S.A. Karkanis +, G.D. Magoulas +, and D.A. Karras ++ + University of Athens, Dept. of Informatics, Typa Build., Panepistimiopolis,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy
 The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
The Anatomical Equivalence Class Formulation and its Application to Shape-based Computational Neuroanatomy Sokratis K. Makrogiannis, PhD From post-doctoral research at SBIA lab, Department of Radiology,
Spatial Distribution of Temporal Lobe Subregions in First Episode Schizophrenia: Statistical Probability Atlas Approach
 Spatial Distribution of Temporal Lobe Subregions in First Episode Schizophrenia: Statistical Probability Atlas Approach Hae-Jeong Park, Martha E. Shenton, James Levitt, Dean Salisbury, Marek Kubicki, Ferenc
Spatial Distribution of Temporal Lobe Subregions in First Episode Schizophrenia: Statistical Probability Atlas Approach Hae-Jeong Park, Martha E. Shenton, James Levitt, Dean Salisbury, Marek Kubicki, Ferenc
GE Healthcare. Agile Ultrasound. The Next Revolution in Ultrasound Imaging
 Agile Ultrasound The Next Revolution in Ultrasound Imaging Abstract Diagnostic use of ultrasound has greatly expanded over the past couple of decades because it offers many advantages as an imaging modality.
Agile Ultrasound The Next Revolution in Ultrasound Imaging Abstract Diagnostic use of ultrasound has greatly expanded over the past couple of decades because it offers many advantages as an imaging modality.
Tutorial 01 Quick Start Tutorial
 Tutorial 01 Quick Start Tutorial Homogeneous single material slope No water pressure (dry) Circular slip surface search (Grid Search) Intro to multi scenario modeling Introduction Model This quick start
Tutorial 01 Quick Start Tutorial Homogeneous single material slope No water pressure (dry) Circular slip surface search (Grid Search) Intro to multi scenario modeling Introduction Model This quick start
Tutorial - Introduction to LabScribe and the IX-ELVIS
 Tutorial - Introduction to LabScribe and the IX-ELVIS Background In addition to its prototyping capabilities, the iworx Bioinstrumentation Breadboard for the National Instruments Educational Laboratory
Tutorial - Introduction to LabScribe and the IX-ELVIS Background In addition to its prototyping capabilities, the iworx Bioinstrumentation Breadboard for the National Instruments Educational Laboratory
Image Acquisition Systems
 Image Acquisition Systems Goals and Terminology Conventional Radiography Axial Tomography Computer Axial Tomography (CAT) Magnetic Resonance Imaging (MRI) PET, SPECT Ultrasound Microscopy Imaging ITCS
Image Acquisition Systems Goals and Terminology Conventional Radiography Axial Tomography Computer Axial Tomography (CAT) Magnetic Resonance Imaging (MRI) PET, SPECT Ultrasound Microscopy Imaging ITCS
Independent Component Analysis of fmri Data
 Independent Component Analysis of fmri Data Denise Miller April 2005 Introduction Techniques employed to analyze functional magnetic resonance imaging (fmri) data typically use some form of univariate
Independent Component Analysis of fmri Data Denise Miller April 2005 Introduction Techniques employed to analyze functional magnetic resonance imaging (fmri) data typically use some form of univariate
Definiens. Tissue Studio 4.4. Tutorial 4: Manual ROI Selection and Marker Area Detection
 Definiens Tissue Studio 4.4 Tutorial 4: Manual ROI Selection and Marker Area Detection Tutorial 4: Manual ROI Selection and Marker Area Detection Imprint and Version Copyright 2017 Definiens AG. All rights
Definiens Tissue Studio 4.4 Tutorial 4: Manual ROI Selection and Marker Area Detection Tutorial 4: Manual ROI Selection and Marker Area Detection Imprint and Version Copyright 2017 Definiens AG. All rights
Normalization for clinical data
 Normalization for clinical data Christopher Rorden, Leonardo Bonilha, Julius Fridriksson, Benjamin Bender, Hans-Otto Karnath (2012) Agespecific CT and MRI templates for spatial normalization. NeuroImage
Normalization for clinical data Christopher Rorden, Leonardo Bonilha, Julius Fridriksson, Benjamin Bender, Hans-Otto Karnath (2012) Agespecific CT and MRI templates for spatial normalization. NeuroImage
A Simple Generative Model for Single-Trial EEG Classification
 A Simple Generative Model for Single-Trial EEG Classification Jens Kohlmorgen and Benjamin Blankertz Fraunhofer FIRST.IDA Kekuléstr. 7, 12489 Berlin, Germany {jek, blanker}@first.fraunhofer.de http://ida.first.fraunhofer.de
A Simple Generative Model for Single-Trial EEG Classification Jens Kohlmorgen and Benjamin Blankertz Fraunhofer FIRST.IDA Kekuléstr. 7, 12489 Berlin, Germany {jek, blanker}@first.fraunhofer.de http://ida.first.fraunhofer.de
Hesheng Liu, Member, IEEE, Xiaorong Gao, Paul H. Schimpf, Member, IEEE, Fusheng Yang, and Shangkai Gao, Senior Member, IEEE
 1794 IEEE TRANSACTIONS ON BIOMEDICAL ENGINEERING, NO 10, VOL. 51, OCTOBER 2004 A Recursive Algorithm for the Three-Dimensional Imaging of Brain Electric Activity: Shrinking LORETA-FOCUSS Hesheng Liu, Member,
1794 IEEE TRANSACTIONS ON BIOMEDICAL ENGINEERING, NO 10, VOL. 51, OCTOBER 2004 A Recursive Algorithm for the Three-Dimensional Imaging of Brain Electric Activity: Shrinking LORETA-FOCUSS Hesheng Liu, Member,
Spatial Filtering Methods in MEG. Part 3: Template Normalization and Group Analysis"
 Spatial Filtering Methods in MEG Part 3: Template Normalization and Group Analysis" Douglas Cheyne, PhD" Program in Neurosciences and Mental Health" Hospital for Sick Children Research Institute " &" Department
Spatial Filtering Methods in MEG Part 3: Template Normalization and Group Analysis" Douglas Cheyne, PhD" Program in Neurosciences and Mental Health" Hospital for Sick Children Research Institute " &" Department
Image Transformation Techniques Dr. Rajeev Srivastava Dept. of Computer Engineering, ITBHU, Varanasi
 Image Transformation Techniques Dr. Rajeev Srivastava Dept. of Computer Engineering, ITBHU, Varanasi 1. Introduction The choice of a particular transform in a given application depends on the amount of
Image Transformation Techniques Dr. Rajeev Srivastava Dept. of Computer Engineering, ITBHU, Varanasi 1. Introduction The choice of a particular transform in a given application depends on the amount of
A Study of Medical Image Analysis System
 Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
Indian Journal of Science and Technology, Vol 8(25), DOI: 10.17485/ijst/2015/v8i25/80492, October 2015 ISSN (Print) : 0974-6846 ISSN (Online) : 0974-5645 A Study of Medical Image Analysis System Kim Tae-Eun
number Understand the equivalence between recurring decimals and fractions
 number Understand the equivalence between recurring decimals and fractions Using and Applying Algebra Calculating Shape, Space and Measure Handling Data Use fractions or percentages to solve problems involving
number Understand the equivalence between recurring decimals and fractions Using and Applying Algebra Calculating Shape, Space and Measure Handling Data Use fractions or percentages to solve problems involving
Locating ego-centers in depth for hippocampal place cells
 204 5th Joint Symposium on Neural Computation Proceedings UCSD (1998) Locating ego-centers in depth for hippocampal place cells Kechen Zhang,' Terrence J. Sejeowski112 & Bruce L. ~cnau~hton~ 'Howard Hughes
204 5th Joint Symposium on Neural Computation Proceedings UCSD (1998) Locating ego-centers in depth for hippocampal place cells Kechen Zhang,' Terrence J. Sejeowski112 & Bruce L. ~cnau~hton~ 'Howard Hughes
CHAPTER 6 DETECTION OF MASS USING NOVEL SEGMENTATION, GLCM AND NEURAL NETWORKS
 130 CHAPTER 6 DETECTION OF MASS USING NOVEL SEGMENTATION, GLCM AND NEURAL NETWORKS A mass is defined as a space-occupying lesion seen in more than one projection and it is described by its shapes and margin
130 CHAPTER 6 DETECTION OF MASS USING NOVEL SEGMENTATION, GLCM AND NEURAL NETWORKS A mass is defined as a space-occupying lesion seen in more than one projection and it is described by its shapes and margin
OPTIMIZING A VIDEO PREPROCESSOR FOR OCR. MR IBM Systems Dev Rochester, elopment Division Minnesota
 OPTIMIZING A VIDEO PREPROCESSOR FOR OCR MR IBM Systems Dev Rochester, elopment Division Minnesota Summary This paper describes how optimal video preprocessor performance can be achieved using a software
OPTIMIZING A VIDEO PREPROCESSOR FOR OCR MR IBM Systems Dev Rochester, elopment Division Minnesota Summary This paper describes how optimal video preprocessor performance can be achieved using a software
syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets
 syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets Julien Gervais; Lisa Chuah Siemens Healthcare, Magnetic
syngo.mr Neuro 3D: Your All-In-One Post Processing, Visualization and Reporting Engine for BOLD Functional and Diffusion Tensor MR Imaging Datasets Julien Gervais; Lisa Chuah Siemens Healthcare, Magnetic
Lecture 8 Object Descriptors
 Lecture 8 Object Descriptors Azadeh Fakhrzadeh Centre for Image Analysis Swedish University of Agricultural Sciences Uppsala University 2 Reading instructions Chapter 11.1 11.4 in G-W Azadeh Fakhrzadeh
Lecture 8 Object Descriptors Azadeh Fakhrzadeh Centre for Image Analysis Swedish University of Agricultural Sciences Uppsala University 2 Reading instructions Chapter 11.1 11.4 in G-W Azadeh Fakhrzadeh
Implemented by Valsamis Douskos Laboratoty of Photogrammetry, Dept. of Surveying, National Tehnical University of Athens
 An open-source toolbox in Matlab for fully automatic calibration of close-range digital cameras based on images of chess-boards FAUCCAL (Fully Automatic Camera Calibration) Implemented by Valsamis Douskos
An open-source toolbox in Matlab for fully automatic calibration of close-range digital cameras based on images of chess-boards FAUCCAL (Fully Automatic Camera Calibration) Implemented by Valsamis Douskos
The L&S LSS Podcaster s Tutorial for Audacity
 The L&S LSS Podcaster s Tutorial for Audacity The L&S LSS Podcaster s Tutorial for Audacity... 1 Audacity Quick Reference... 2 About this tutorial... 3 Some Thoughts Before You Get Started... 3 Do Academic
The L&S LSS Podcaster s Tutorial for Audacity The L&S LSS Podcaster s Tutorial for Audacity... 1 Audacity Quick Reference... 2 About this tutorial... 3 Some Thoughts Before You Get Started... 3 Do Academic
Advanced Visual Medicine: Techniques for Visual Exploration & Analysis
 Advanced Visual Medicine: Techniques for Visual Exploration & Analysis Interactive Visualization of Multimodal Volume Data for Neurosurgical Planning Felix Ritter, MeVis Research Bremen Multimodal Neurosurgical
Advanced Visual Medicine: Techniques for Visual Exploration & Analysis Interactive Visualization of Multimodal Volume Data for Neurosurgical Planning Felix Ritter, MeVis Research Bremen Multimodal Neurosurgical
Sparse Source EEG Imaging with the Variational Garrote
 Downloaded from orbit.dtu.dk on: Jun 8, 28 Sparse Source EEG Imaging with the Variational Garrote Hansen, Sofie Therese; Stahlhut, Carsten; Hansen, Lars Kai Published in: 23 International Workshop on Pattern
Downloaded from orbit.dtu.dk on: Jun 8, 28 Sparse Source EEG Imaging with the Variational Garrote Hansen, Sofie Therese; Stahlhut, Carsten; Hansen, Lars Kai Published in: 23 International Workshop on Pattern
Segmentation and Modeling of the Spinal Cord for Reality-based Surgical Simulator
 Segmentation and Modeling of the Spinal Cord for Reality-based Surgical Simulator Li X.C.,, Chui C. K.,, and Ong S. H.,* Dept. of Electrical and Computer Engineering Dept. of Mechanical Engineering, National
Segmentation and Modeling of the Spinal Cord for Reality-based Surgical Simulator Li X.C.,, Chui C. K.,, and Ong S. H.,* Dept. of Electrical and Computer Engineering Dept. of Mechanical Engineering, National
Mathematically Modeling Fetal Electrocardiograms
 Pursuit - The Journal of Undergraduate Research at the University of Tennessee Volume 5 Issue 1 Article 10 June 2014 Mathematically Modeling Fetal Electrocardiograms Samuel Estes University of Tennessee,
Pursuit - The Journal of Undergraduate Research at the University of Tennessee Volume 5 Issue 1 Article 10 June 2014 Mathematically Modeling Fetal Electrocardiograms Samuel Estes University of Tennessee,
An open source tool for automatic spatiotemporal assessment of calcium transients and local signal-close-to-noise activity in calcium imaging data
 Supplementary material: User manual An open source tool for automatic spatiotemporal assessment of calcium transients and local signal-close-to-noise activity in calcium imaging data Juan Prada, Manju
Supplementary material: User manual An open source tool for automatic spatiotemporal assessment of calcium transients and local signal-close-to-noise activity in calcium imaging data Juan Prada, Manju
SISCOM (Subtraction Ictal SPECT CO-registered to MRI)
 SISCOM (Subtraction Ictal SPECT CO-registered to MRI) Introduction A method for advanced imaging of epilepsy patients has been developed with Analyze at the Mayo Foundation which uses a combination of
SISCOM (Subtraction Ictal SPECT CO-registered to MRI) Introduction A method for advanced imaging of epilepsy patients has been developed with Analyze at the Mayo Foundation which uses a combination of
GE Dynamic 13 C Acquisition
 GE Dynamic 13 C Acquisition In this example you will load a human prostate dynamic MRS data set (single slice, 24 time points) acquired on a GE 3T scanner. You will generate metabolite maps for pyruvate
GE Dynamic 13 C Acquisition In this example you will load a human prostate dynamic MRS data set (single slice, 24 time points) acquired on a GE 3T scanner. You will generate metabolite maps for pyruvate
Chapter 7 Inserting Spreadsheets, Charts, and Other Objects
 Impress Guide Chapter 7 Inserting Spreadsheets, Charts, and Other Objects OpenOffice.org Copyright This document is Copyright 2007 by its contributors as listed in the section titled Authors. You can distribute
Impress Guide Chapter 7 Inserting Spreadsheets, Charts, and Other Objects OpenOffice.org Copyright This document is Copyright 2007 by its contributors as listed in the section titled Authors. You can distribute
TRAVELTIME TOMOGRAPHY (CONT)
 30 March 005 MODE UNIQUENESS The forward model TRAVETIME TOMOGRAPHY (CONT di = A ik m k d = Am (1 Data vecto r Sensitivit y matrix Model vector states the linearized relationship between data and model
30 March 005 MODE UNIQUENESS The forward model TRAVETIME TOMOGRAPHY (CONT di = A ik m k d = Am (1 Data vecto r Sensitivit y matrix Model vector states the linearized relationship between data and model
CONN fmri functional connectivity toolbox
 CONN fmri functional connectivity toolbox Gabrieli Lab. McGovern Institute for Brain Research Massachusetts Institute of Technology http://www.nitrc.org/projects/conn Susan Whitfield-Gabrieli Alfonso Nieto-Castanon
CONN fmri functional connectivity toolbox Gabrieli Lab. McGovern Institute for Brain Research Massachusetts Institute of Technology http://www.nitrc.org/projects/conn Susan Whitfield-Gabrieli Alfonso Nieto-Castanon
Datasets Size: Effect on Clustering Results
 1 Datasets Size: Effect on Clustering Results Adeleke Ajiboye 1, Ruzaini Abdullah Arshah 2, Hongwu Qin 3 Faculty of Computer Systems and Software Engineering Universiti Malaysia Pahang 1 {ajibraheem@live.com}
1 Datasets Size: Effect on Clustering Results Adeleke Ajiboye 1, Ruzaini Abdullah Arshah 2, Hongwu Qin 3 Faculty of Computer Systems and Software Engineering Universiti Malaysia Pahang 1 {ajibraheem@live.com}
Last Time. This Time. Thru-plane dephasing: worse at long TE. Local susceptibility gradients: thru-plane dephasing
 Motion Correction Last Time Mutual Information Optimiation Decoupling Translation & Rotation Interpolation SPM Example (Least Squares & MI) A Simple Derivation This Time Reslice example SPM Example : Remind
Motion Correction Last Time Mutual Information Optimiation Decoupling Translation & Rotation Interpolation SPM Example (Least Squares & MI) A Simple Derivation This Time Reslice example SPM Example : Remind
INCREASING CLASSIFICATION QUALITY BY USING FUZZY LOGIC
 JOURNAL OF APPLIED ENGINEERING SCIENCES VOL. 1(14), issue 4_2011 ISSN 2247-3769 ISSN-L 2247-3769 (Print) / e-issn:2284-7197 INCREASING CLASSIFICATION QUALITY BY USING FUZZY LOGIC DROJ Gabriela, University
JOURNAL OF APPLIED ENGINEERING SCIENCES VOL. 1(14), issue 4_2011 ISSN 2247-3769 ISSN-L 2247-3769 (Print) / e-issn:2284-7197 INCREASING CLASSIFICATION QUALITY BY USING FUZZY LOGIC DROJ Gabriela, University
RADIOMICS: potential role in the clinics and challenges
 27 giugno 2018 Dipartimento di Fisica Università degli Studi di Milano RADIOMICS: potential role in the clinics and challenges Dr. Francesca Botta Medical Physicist Istituto Europeo di Oncologia (Milano)
27 giugno 2018 Dipartimento di Fisica Università degli Studi di Milano RADIOMICS: potential role in the clinics and challenges Dr. Francesca Botta Medical Physicist Istituto Europeo di Oncologia (Milano)
DynaConn Users Guide. Dynamic Function Connectivity Graphical User Interface. Last saved by John Esquivel Last update: 3/27/14 4:26 PM
 DynaConn Users Guide Dynamic Function Connectivity Graphical User Interface Last saved by John Esquivel Last update: 3/27/14 4:26 PM Table Of Contents Chapter 1 Introduction... 1 1.1 Introduction to this
DynaConn Users Guide Dynamic Function Connectivity Graphical User Interface Last saved by John Esquivel Last update: 3/27/14 4:26 PM Table Of Contents Chapter 1 Introduction... 1 1.1 Introduction to this
CHAPTER 1 INTRODUCTION
 1 CHAPTER 1 INTRODUCTION 1.1 Advance Encryption Standard (AES) Rijndael algorithm is symmetric block cipher that can process data blocks of 128 bits, using cipher keys with lengths of 128, 192, and 256
1 CHAPTER 1 INTRODUCTION 1.1 Advance Encryption Standard (AES) Rijndael algorithm is symmetric block cipher that can process data blocks of 128 bits, using cipher keys with lengths of 128, 192, and 256
NeuroMaps Mapper. The Mapper is a stand-alone application for mapping data to brain atlases for presentation and publication.
 NeuroMaps Mapper The Mapper is a stand-alone application for mapping data to brain atlases for presentation and publication. Image data are mapped to a stereotaxic MRI atlas of the macaque brain or the
NeuroMaps Mapper The Mapper is a stand-alone application for mapping data to brain atlases for presentation and publication. Image data are mapped to a stereotaxic MRI atlas of the macaque brain or the
Neurophysical Model by Barten and Its Development
 Chapter 14 Neurophysical Model by Barten and Its Development According to the Barten model, the perceived foveal image is corrupted by internal noise caused by statistical fluctuations, both in the number
Chapter 14 Neurophysical Model by Barten and Its Development According to the Barten model, the perceived foveal image is corrupted by internal noise caused by statistical fluctuations, both in the number
Mid-movement Force (N) Mid-movement Force (N) Mid-movement Force (N)
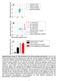 Mid-movement Force (N) Mid-movement Force (N) Mid-movement Force (N) 6 4 A Individual Trials Subject Average Velocity Average Ideal Performance 2 0 0 0.2 0.4 6 4 2 B 0 0 0.2 0.4 Movement Velocity (m/s)
Mid-movement Force (N) Mid-movement Force (N) Mid-movement Force (N) 6 4 A Individual Trials Subject Average Velocity Average Ideal Performance 2 0 0 0.2 0.4 6 4 2 B 0 0 0.2 0.4 Movement Velocity (m/s)
Dendrogram export options
 BioNumerics Tutorial: Dendrogram export options 1 Introduction In this tutorial, the export options of a dendrogram, displayed in the Dendrogram panel of the Comparison window is covered. This tutorial
BioNumerics Tutorial: Dendrogram export options 1 Introduction In this tutorial, the export options of a dendrogram, displayed in the Dendrogram panel of the Comparison window is covered. This tutorial
USER GUIDE MADCAP CAPTURE 7. Getting Started
 USER GUIDE MADCAP CAPTURE 7 Getting Started Copyright 2018 MadCap Software. All rights reserved. Information in this document is subject to change without notice. The software described in this document
USER GUIDE MADCAP CAPTURE 7 Getting Started Copyright 2018 MadCap Software. All rights reserved. Information in this document is subject to change without notice. The software described in this document
Temperature Distribution Measurement Based on ML-EM Method Using Enclosed Acoustic CT System
 Sensors & Transducers 2013 by IFSA http://www.sensorsportal.com Temperature Distribution Measurement Based on ML-EM Method Using Enclosed Acoustic CT System Shinji Ohyama, Masato Mukouyama Graduate School
Sensors & Transducers 2013 by IFSA http://www.sensorsportal.com Temperature Distribution Measurement Based on ML-EM Method Using Enclosed Acoustic CT System Shinji Ohyama, Masato Mukouyama Graduate School
MARS: Multiple Atlases Robust Segmentation
 Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Software Release (1.0.1) Last updated: April 30, 2014. MARS: Multiple Atlases Robust Segmentation Guorong Wu, Minjeong Kim, Gerard Sanroma, and Dinggang Shen {grwu, mjkim, gerard_sanroma, dgshen}@med.unc.edu
Unit 21 - Creating a Navigation Bar in Macromedia Fireworks
 Unit 21 - Creating a Navigation Bar in Macromedia Fireworks Items needed to complete the Navigation Bar: Unit 21 - House Style Unit 21 - Graphics Sketch Diagrams Document ------------------------------------------------------------------------------------------------
Unit 21 - Creating a Navigation Bar in Macromedia Fireworks Items needed to complete the Navigation Bar: Unit 21 - House Style Unit 21 - Graphics Sketch Diagrams Document ------------------------------------------------------------------------------------------------
